Abstract
Undifferentiated pleomorphic sarcoma (UPS) is an aggressive mesenchymal malignancy requiring novel therapeutic approaches to improve clinical outcome. Patient-derived cancer cell lines are an essential tool for investigating molecular mechanisms underlying cancer initiation and development; however, there is a lack of patient-derived cell lines of UPS available for research. The objective of this study was to develop a patient-derived cell model of UPS. A cell line designated NCC-UPS2-C1 was established from the primary tumor tissue of an 84-yr-old female patient with UPS. The short tandem repeat pattern of NCC-UPS2-C1 cells was identical to that of the original tumor and distinct from that of any other cell lines deposited in public cell banks. NCC-UPS2-C1 cells were maintained as a monolayer culture for over 80 passages during 30 mo and exhibited spindle-like morphology, continuous growth, and ability for spheroid formation and invasion. Proteomic profiling using mass spectrometry and functional treemap analysis revealed that the original tumor and the derived NCC-UPS2-C1 cells had similar but distinct protein expression patterns. Our results indicate that a novel UPS cell line was successfully established and could be used to study UPS development and effects of anti-cancer drugs. However, the revealed difference between proteomes of the original tumor and NCC-UPS2-C1 cells should be further investigated to determine the appropriate applications of this cell line in UPS research.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Undifferentiated pleomorphic sarcoma (UPS) is a common aggressive soft tissue cancer, representing a pleomorphic variant of undifferentiated sarcomas which have a highly variable morphology without identifiable differentiation (Fletcher et al. 2013). The therapeutic strategy for UPS is surgery with wide margins, and adjuvant treatment is usually considered. The clinical outcome of UPS patients remains dismal, as evidenced by the local recurrence rate of 19–31%, metastatic rate of 31–35%, and 5-yr survival of 65–70% (Goldblum 2014). Therefore, novel more effective therapeutic approaches are required for the treatment of UPS.
Patient-derived cancer models are crucial for the elucidation of molecular mechanisms underlying carcinogenesis and for the development of novel therapeutic strategies. Previous studies indicated the considerable potentials of use of cell lines in the development of predictive biomarkers and the identification of therapeutic targets (Barretina et al. 2012; Iorio et al. 2016). In this respect, tumor cell lines provide means to screen numerous anti-cancer drugs and investigate their effects. UPS has been classified as malignant fibrous histiocytoma (MFH), and several previous studies have reported the establishment of UPS and MFH cell lines (Krause et al. 1997; Mairal et al. 2000; Nakatani et al. 2001; Hakozaki et al. 2006; Nishio et al. 2010; Becker et al. 2016). However, considering the clinical and biological heterogeneity of UPS, the number of the obtained cell lines is insufficient, and more of them need to be established directly from UPS patients.
Here, we report a novel patient-derived cell line obtained from tumor tissue of an UPS patient. The cell line designated as NCC-UPS2-C1 showed constant growth in culture, and the ability for spheroid formation and invasion. Our results indicate that NCC-UPS2-C1 cells should be a useful tool for in vitro studies of UPS.
Methods
Patient background
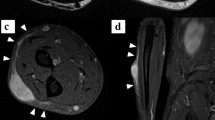
The donor patient was an 84-yr-old woman who was referred to the National Cancer Center Hospital, Tokyo, Japan, for a painful soft tissue tumor on the left thigh, which was rapidly increasing in size without distant metastases (Fig. 1). A needle biopsy resulted in a primary diagnosis of pleomorphic spindle cell sarcoma. Wide excision of the tumor was performed and the tissue was used as a cell source. The definitive diagnosis made after analysis of the surgical specimen was UPS. A carcinoma was detected 5 mo later in the gallbladder. At the final observation 10 mo postoperatively, the patient had no metastasis or local recurrence of UPS. This study was approved by the ethics committee of the National Cancer Center, and written informed consent was obtained from the patient.
Clinical imaging data of an UPS patient who was the donor of tumor tissue. Axial (A) and coronal (B) views obtained by gadolinium-enhanced fat-suppressed magnetic resonance imaging at the initial presentation to National Cancer Center Hospital. The tumor was located in the left quadriceps muscle, was 100 mm in size, and presented a heterogeneous solid mass with contrast effect in most of the area.
Cell line establishment
Tumor tissue was immersed in DMEM (Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal bovine serum (FBS) (Gibco, Grand Island, NY), 100 U penicillin G, and 100 μg/mL streptomycin (Gibco) and cut into small pieces, which were mechanically dissociated by passing through an 18-gauge needle. Cell suspension was filtered through a 40-μm nylon mesh (BD Falcon, Franklin Lakes, NJ) and seeded into a 10-cm culture plate at 37°C in a humidified atmosphere of 5% CO2.
Short tandem repeat analysis for authentication
To authenticate the established cell line, we analyzed short tandem repeats (STRs) of the original tumor tissue and cultured cells. Genomic DNA was extracted from cells or tumor tissue using the AllPrep DNA/RNA mini kit (Qiagen, Venlo, Netherland), its concentration measured using NanoDrop 8000 (Thermo Fisher Scientific, Waltham, MA), and samples were stored at − 80°C until use. STR multiplex assays were performed using 500 pg genomic DNA and GenePrint 10 (Promega, Madison, WI) according to the manufacturer’s instructions, and the amplified DNA fragments were detected in a 3500xL Genetic Analyzer (Applied Biosystems, Waltham, MA). The data were analyzed using GeneMapper 5 (Applied Biosystems), and the STR profiles were compared with those recorded in public cell banks such as ATCC, DSMZ, or JRCB for reference matching.
Mycoplasma contamination screening
Mycoplasma contamination was assessed by examining the presence of mycoplasma DNA in cell culture medium according to the standard procedure of cell line establishment recommended by The International Cell Line Authentication Committee (The International Cell Line Authentication Committee 2018). DNA was extracted from culture medium when cells reached 70–90% confluence, heated at 95°C for 10 min, and amplified using an e-Myco Mycoplasma PCR Detection Kit (Intron Biotechnology, Gyeonggi-do, Korea). The amplified DNA was separated by electrophoresis in a 1.5% agarose gel, stained with Midori Green Advanced (Nippon Genetics, Tokyo, Japan), and detected using an Amersham Imager 600 (GE Healthcare Biosciences, Uppsala, Sweden).
Characterization of cell growth
Cells were seeded into 96-well culture plates at the concentration of 2, 4, or 8 × 103 cells/well in triplicate and grown at 37°C. The CCK-8 reagent (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was added to each well at 24, 48, 72, and 96 h after seeding, and cells were incubated for another 2 h. Absorbance was measured at a wavelength of 450 nm using a microplate reader (BioRad, Hercules, CA) and plotted against culture time to construct growth curves.
Spheroid formation assay
To explore the ability of the cultured UPS cells to form spheroids, 1 × 105 cells were seeded in DMEM containing 10% FBS in 6-cm plates (Ultra Low Culture Dish, Thermo Fisher Scientific) and incubated in a humidified atmosphere of 5% CO2 at 37°C for 3 wk. The presence of spheroids was confirmed under a microscope (Keyence, Osaka, Japan). All assays were performed in duplicate.
Cell invasion assay
The invasion ability of cultured cells was assessed using BD Biocoat Matrigel Invasion Chambers (BD Biosciences, Bedford, MA) according to the manufacturer’s instructions. In brief, 5 × 104 and 1 × 105 tumor cells in serum-free DMEM were plated in the upper chamber, whereas the lower chamber contained DMEM supplemented with 10% FBS. After 48 h of incubation, tumor cells passing through the filter separating the two chambers were stained and counted in nine separate areas at × 200 magnification.
Proteomic analysis
Protein expression profiles of tumor tissue and cultured cells were obtained by mass spectrometry (MS) according to our previous report (Oyama et al. 2017). In brief, 50 μg of proteins extracted from cells and tissue in urea lysis buffer was digested with trypsin using filter-aided sample preparation. The obtained peptides were separated in a nanoflow HPLC system (AMR, Tokyo, Japan) equipped with an analytical column (C18, 3 μm, 100 Å, 150 × 0.075 mm; AMR) at the flow rate of 250 nL/min. The following gradient elution conditions were used: 0.1% aqueous formic acid solution as eluent A and 0.1% formic acid in 90% aqueous acetonitrile solution as eluent B; 0–140 min: 5–45% B.
The peptides were ionized using a nanospray source (Thermo Fisher Scientific) and analyzed in a Finnigan LTQ Orbitrap XL mass spectrometer. Full MS scans were obtained using an orbitrap mass analyzer, and five most intense precursor ions were selected for MS/MS scans. MS data were analyzed by searching the SWISS-PROT database (Homo sapiens, 20,205 sequences in the Swiss prot_2015_09.fasta file) with MASCOT 2.5.1 (MatrixScience, London, UK).
Functional annotation
The proteomic data was analyzed using Kyoto Encyclopedia of Genes and Genomes (KEGG) to assign proteins into respective molecular pathways (Altermann and Klaenhammer 2005), and the annotations were loaded into the Database for Annotation, Visualization and Integration Discovery (DAVID) software (http://david.abcc.ncifcrf.gov/). Significance of enrichment for each category was evaluated according to the p value, which was considered significant at less than 0.05. The results of KEGG analysis were plotted using the R package “treemap” (Baehrecke et al. 2004).
Results
Establishment and authentication of the cell line from primary UPS tissue
We established a cell line from the primary tumor tissue of an UPS patient and designated it NCC-UPS2-C1. Adherent cells were maintained in culture for 80 passages over more than 30 mo. We did not clone the cells. To authenticate the established cell line, we compared the status of ten microsatellites (STRs) in the original tumor tissue and the cultured cells. We found that the examined STRs were similar but not identical between the two samples (Table 1). Possible loss of heterozygosity was observed in five out of nine loci. It may be attributable to the long-term culture. The STR pattern of NCC-UPS2-C1 cells did not match any of those for cell lines deposited in public cell banks such as ATCC, DSMZ, or JRCB. These results indicated that NCC-UPS2-C1 was a novel UPS cell line. Testing of the cell line for mycoplasma contamination was negative as no mycoplasma-specific DNA was found in cell-conditioned medium (data not shown).
Phenotypic characterization of cultured UPS cells
NCC-UPS2-C1 cells had spindle-shape morphology (Fig. 2A) and demonstrated the ability to spheroid formation on low attachment substrates (Fig. 2B). Analysis of the in vitro growth rate indicated that the doubling time of NCC-UPS2-C1 cells seeded at the density of 8 × 103 cells per well was 45 h. Furthermore, the Matrigel assay revealed that the cell line had invasive ability in vitro (Fig. 2D), which increased proportionally to the number of cells seeded in the upper chamber (Fig. 2E). Overall, these results showed that the established NCC-UPS2-C1 cell line demonstrated features of tumor cells.
Phenotypic characterization of NCC-UPS2-C1 cells. (A) Representative phase contrast micrograph; scale bar = 100 μm. (B) Spheroid formation examined on low-attachment substrates; scale bar = 200 μm. (C) Growth curves constructed using the indicated numbers of plated cells. (D, E) Cell invasion examined by the Matrigel invasion assay. Representative images of invaded cells (D) and quantitative analysis of cell invasion depending on the number of seeded cells (E).
Proteomic profiling
Protein expression spectra of the primary tumor and NCC-UPS2-C1 cells were analyzed by MS (Supplementary Table 1–3), and the identified proteins were classified according to their potential functions based on the KEGG data. The top 10 most enriched pathways are presented in a treemap format (Fig. 3 and Supplementary Table 3), where box areas indicated the number of allocated proteins and colors indicated the degree of statistical enrichment.
Proteomic profiling of the primary tumor tissue and the derived cell line. Proteins identified by MS were functionally classified according to the KEGG database. The results are presented as a treemap where box size reflects the number of proteins in the pathway and color indicates degree of enrichment. (A) The primary tumor tissue and (B) NCC-UPS2-C1 cell line had similar but distinct proteome structures. (C) Functional pathways common for the original tumor and the cell line.
Proteomic analysis revealed that in the primary tumor, proteins participating in the biosynthesis of antibiotics were predominantly detected and those involved in the complement and coagulation cascades were most significantly enriched (Fig. 3A). However, the proteome profile of NCC-UPS2-C1 cells was somewhat different from that of the primary tumor. Although, similar to the original tumor tissue, proteins of the biosynthesis of antibiotics pathway were mainly detected, the ribosome pathway was the most enriched (Fig. 3B). The enrichment scores for ribosome, carbon metabolism, spliceosome, biosynthesis of amino acids, and proteasome pathways were increased, whereas those for protein processing in endoplasmic reticulum and complement and coagulation cascades were decreased in the cell line compared to the original tumor (Fig. 3).
A treemap of pathways common for the two samples demonstrated that protein processing in the endoplasmic reticulum was both the most enriched and comprised the highest number of detected proteins (Fig. 3C), indicating that this pathway was representative for the original tumor and the derived cell line. Quantitative analysis of all pathways unique and common for the tumor and NCC-UPS2-C1 cells is presented in Supplementary Table 3.
Discussion
Patient-derived cell lines are an essential tool in both basic and translational research on cancer. They are indispensable for the investigation of phenotypic changes induced by ectopic expression of oncogenes and for screening of tumor-suppressing agents, and provide valuable information about carcinogenic mechanisms and potential treatment effects of novel anti-cancer drugs. The utility of cell line collections for clinical applications has been comprehensively analyzed in previous publications (Barretina et al. 2012; Domcke et al. 2013; Iorio et al. 2016). Although the versatile usage of cell lines is universally acknowledged, precise mechanisms of cell line establishment are not completely understood, and significant time and efforts are required to obtain a cell line. It particularly concerns the cell lines of rare tumors such as UPS, which are not readily available from public cell banks (Pan et al. 2016). Here, we report a successful establishment of a novel cell line, NCC-UPS2-C1, derived from the primary tumor of an UPS patients.
During cell line development, we did not select a specific tumor cell population; therefore, multiple cell clones may exist in NCC-UPS2-C1 culture. Clonally selected cells are needed for experiments requiring large cell numbers for a long period, such as screening of compound libraries; however, such cell lines have inherent drawbacks as they are established based on cell adaptation and fast expansion in culture conditions, but their characteristics may considerably differ from those of the original tumor. In contrast, the total, unselected population of tumor cells reflects the diversity of tumor tissue composition better than cloned cells. As NCC-UPS2-C1 may include multiple cell clones, we need to consider the possibility that the overall character of this cell line could change after long-term culture.
NCC-UPS2-C1 cells formed spheroids when seeded on a low-attachment substrate. Spheroids are 3-D cell structures that retain the biological characteristics of original tumors better than cell monolayers. Therefore, the ability to organize into spheroids is a very valuable feature of a cancer cell line, which thus presents a useful model for studying cancer drug sensitivity known to be distinct in 3-D and 2-D cultures (Arai et al. 2013). NCC-UPS2-C1 cells exhibited aggressive proliferation as evidenced by the population-doubling time of 45 h, and demonstrated invasion properties. These findings suggest that NCC-UPS2-C1 cells should be instrumental for the experiments where the growth rate, spheroid formation, and invasion are the parameters to evaluate the effects of various stimuli on UPS cells.
We also performed comparative proteomic analysis of the original UPS tissue and the derived NCC-UPS2-C1 cell line, which indicated that their proteome profiles were not identical. Our result is consistent with the findings of a previous large-scale genomic study showing that the genetic landscape of tumors was not fully captured by patient-derived cancer models such as cell lines and xenografts (Ben-David et al. 2017). These observations suggest that although patient-derived cell lines are a helpful research system, they do not represent a comprehensive model of clinical tumors. In this respect, it would be useful to compare genomic, proteomic, and other -omic profiles of patient-derived cancer cell lines with those of the original tumors in order to determine the degree of applicability of cell cancer models to clinical research.
Conclusion
Here, we report the establishment of a UPS cell line derived from resected tumor tissue of a patient with UPS. The NCC-UPS2-C1 cell line had the STR pattern identical to that of the primary tumor but different from those of the existing cell lines, and demonstrated continuous proliferation and the ability for invasion and formation of 3-D spheroids, which are the feature characteristics for tumor cells. Proteomic analysis showed that protein processing in the endoplasmic reticulum was the most enriched pathway common for the original tumor and the derived cell line, although several functional pathways showed distinct representation. The established NCC-UPS2-C1 cell line should be useful for studying the mechanisms of UPS development and screening of novel therapeutic compounds.
References
Altermann E, Klaenhammer TR (2005) PathwayVoyager: pathway mapping using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. BMC Genomics 6(1):1–7. https://doi.org/10.1186/1471-2164-6-60
Arai K, Sakamoto R, Kubota D, Kondo T (2013) Proteomic approach toward molecular backgrounds of drug resistance of osteosarcoma cells in spheroid culture system. Proteomics 13(15):2351–2360. https://doi.org/10.1002/pmic.201300053
Baehrecke EH, Dang N, Babaria K, Shneiderman B (2004) Visualization and analysis of microarray and gene ontology data with treemaps. BMC Bioinf 5(1):84. https://doi.org/10.1186/1471-2105-5-84
Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, GK Y, Yu J, Aspesi P Jr, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA (2012) The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483(7391):603–607. https://doi.org/10.1038/nature11003
Becker M, Graf C, Tonak M, Radsak MP, Bopp T, Bals R, Bohle RM, Theobald M, Rommens PM, Proschek D, Wehler TC (2016) Xenograft models for undifferentiated pleomorphic sarcoma not otherwise specified are essential for preclinical testing of therapeutic agents. Oncol Lett 12(2):1257–1264. https://doi.org/10.3892/ol.2016.4784
Ben-David U, Ha G, Tseng YY, Greenwald NF, Oh C, Shih J, McFarland JM, Wong B, Boehm JS, Beroukhim R, Golub TR (2017) Patient-derived xenografts undergo mouse-specific tumor evolution. Nat Genet 49(11):1567–1575. https://doi.org/10.1038/ng.3967
Domcke S, Sinha R, Levine DA, Sander C, Schultz N (2013) Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun 4:2126
Fletcher CDM, Bridge JA, Hogendoorn P, Mertens F (2013) WHO classification of tumours of soft tissue and bone, 4th edn. WHO Press, Geneva
Goldblum JR (2014) An approach to pleomorphic sarcomas: can we subclassify, and does it matter? Mod Pathol 27(Suppl 1):S39–S46. https://doi.org/10.1038/modpathol.2013.174
Hakozaki M, Hojo H, Sato M, Tajino T, Yamada H, Kikuchi S, Abe M (2006) Establishment and characterization of a new cell line, FPS-1, derived from human undifferentiated pleomorphic sarcoma, overexpressing epidermal growth factor receptor and cyclooxygenase-2. Anticancer Res 26(5A):3393–3401
Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, Aben N, Goncalves E, Barthorpe S, Lightfoot H, Cokelaer T, Greninger P, van Dyk E, Chang H, de Silva H, Heyn H, Deng X, Egan RK, Liu Q, Mironenko T, Mitropoulos X, Richardson L, Wang J, Zhang T, Moran S, Sayols S, Soleimani M, Tamborero D, Lopez-Bigas N, Ross-Macdonald P, Esteller M, Gray NS, Haber DA, Stratton MR, Benes CH, Wessels LF, Saez-Rodriguez J, McDermott U, Garnett MJ (2016) A landscape of pharmacogenomic interactions in cancer. Cell 166(3):740–754. https://doi.org/10.1016/j.cell.2016.06.017
Krause AK, Hinrichs SH, Orndal C, DeBoer J, Neff JR, Bridge JA (1997) Characterization of a human myxoid malignant fibrous histiocytoma cell line, OH931. Cancer Genet Cytogenet 94(2):138–143. https://doi.org/10.1016/S0165-4608(96)00223-3
Mairal A, Chibon F, Rousselet A, Couturier J, Terrier P, Aurias A (2000) Establishment of a human malignant fibrous histiocytoma cell line, COMA. Characterization by conventional cytogenetics, comparative genomic hybridization, and multiplex fluorescence in situ hybridization. Cancer Genet Cytogenet 121(2):117–123. https://doi.org/10.1016/S0165-4608(00)00261-2
Nakatani T, Marui T, Yamamoto T, Kurosaka M, Akisue T, Matsumoto K (2001) Establishment and characterization of cell line TNMY1 derived from human malignant fibrous histiocytoma. Pathol Int 51(8):595–602. https://doi.org/10.1046/j.1440-1827.2001.01253.x
Nishio J, Iwasaki H, Nabeshima K, Ishiguro M, Isayama T, Naito M (2010) Establishment of a new human pleomorphic malignant fibrous histiocytoma cell line, FU-MFH-2: molecular cytogenetic characterization by multicolor fluorescence in situ hybridization and comparative genomic hybridization. J Exp Clin Cancer Res 29(1):153. https://doi.org/10.1186/1756-9966-29-153
Oyama R, Takahashi M, Yoshida A, Sakumoto M, Takai Y, Kito F, Shiozawa K, Qiao Z, Arai Y, Shibata T, Araki Y, Endo M, Kawai A, Kondo T (2017) Generation of novel patient-derived CIC- DUX4 sarcoma xenografts and cell lines. Sci Rep 7(1):4712. https://doi.org/10.1038/s41598-017-04967-0
Pan X, Yoshida A, Kawai A, Kondo T (2016) Current status of publicly available sarcoma cell lines for use in proteomic studies. Expert Rev Proteomics 13(2):227–240. https://doi.org/10.1586/14789450.2016.1132166
The International Cell Line Authentication Committee (2018) Available via http://standards.atcc.org/kwspub/home/the_international_cell_line_authentication_committee-iclac_/
Acknowledgments
We thank Drs. M. Endo, Y. Minami, K. Shimizu, T. Mori, T. Uehara M. Sugawara, Y. Araki, and Ms. R. Nakano, Division of Musculoskeletal Oncology, National Cancer Center Hospital, for sampling tumor tissue specimens from surgically resected materials. We would like to thank Editage (www.editage.jp) for English language editing and constructive comments on the manuscript.
Funding
This research was supported by the National Cancer Center Research and Development Fund (26-A-3 and 26-A-9).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This study was approved by the ethics committee of the National Cancer Center, and written informed consent was obtained from the patient.
Additional information
Editor: Tetsuji Okamoto
Electronic supplementary material
Supplementary Table 1
(XLSX 4516 kb)
Supplementary Table 2
(XLSX 7035 kb)
Supplementary Table 3
(XLSX 12 kb)
Rights and permissions
About this article
Cite this article
Oyama, R., Kito, F., Sakumoto, M. et al. Establishment and proteomic characterization of a novel cell line, NCC-UPS2-C1, derived from a patient with undifferentiated pleomorphic sarcoma. In Vitro Cell.Dev.Biol.-Animal 54, 257–263 (2018). https://doi.org/10.1007/s11626-018-0229-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11626-018-0229-7