Abstract
Introduction
Alterations in the microbiome contribute to the pathogenesis of many gastrointestinal diseases. However, the composition of the microbiome in gallbladder disease is not well described.
Methods
We aimed to characterize the biliary microbiome in cholecystectomy patients. Bile and biliary stones were collected at cholecystectomy for a variety of surgical indications between 2017 and 2019. DNA was extracted and metagenomic sequencing was performed with subsequent taxonomic classification using Kraken2. The fraction of bacterial to total DNA reads, relative abundance of bacterial species, and overall species diversity were compared between pathologies and demographics.
Results
A total of 74 samples were obtained from 49 patients: 46 bile and 28 stones, with matched pairs from 25 patients. The mean age was 48 years, 76% were female, 29% were Hispanic, and 29% of patients had acute cholecystitis. The most abundant species were Klebsiella pneumoniae, Staphylococcus aureus, and Streptococcus pasteurianus. The bacterial fraction in bile and stone samples was higher in acute cholecystitis compared to other non-infectious pathologies (p < 0.05). Neither the diversity nor differential prevalence of specific bacterial species varied significantly between infectious and other non-infectious gallbladder pathologies. Multivariate analysis of the non-infectious group revealed that patients over 40 years of age had increased bacterial fractions (p < 0.05).
Conclusions
Metagenomic sequencing permits characterization of the gallbladder microbiome in cholecystectomy patients. Although a higher prevalence of bacteria was seen in acute cholecystitis, species and diversity were similar regardless of surgical indication. Additional study is required to determine how the microbiome can contribute to the development of symptomatic gallbladder disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The human microbiome contributes to many systemic diseases, including gastrointestinal diseases. The importance of a diverse colonic microbiome is highlighted with the success of Clostridium difficile infections treated by fecal transplants.1 In inflammatory bowel disease, different fecal bacterial populations have been found in Crohn’s disease, ulcerative colitis, and healthy patients.2 The intestinal microbiota may even influence behavior and psychiatric disease.3 The distal bowel microbiome is more diverse than the proximal bowel, as the gastrointestinal microbiome is impacted by acidic gastric secretions and transit times.4 However, the association of the microbiome with biliary diseases—particularly gallstone formation—is not well studied.
Gallstone disease, the most common inpatient gastrointestinal diagnosis in the USA,5 is multifactorial. Cholesterol stones are the most common type of gallstones,6 while bacterial infections are known to cause brown pigment stones.7–9 Previously, normal bile in a healthy individual was considered sterile, but this assumption was based on data using less sensitive culture methods to detect the presence of bacteria,10 as well as known inhospitable culture aspects unique to bile.11 Recently, sequencing of bacterial ribosomal 16S RNA in the bile of healthy controls for liver transplant donations revealed the presence of bacteria.12 As such, bacteria may play a role in the pathogenesis of biliary disease in healthy individuals, as well as in patients with biliary stones.
Given the limited data of small cohorts using mostly 16S ribosomal sequencing, we aimed to characterize the microbiome of bile and biliary stones in patients referred for cholecystectomy using metagenomic sequencing. We hypothesized that the microbiome of the gallbladder will differ in patients depending on the type of surgical pathology. Secondarily, we looked for microbiome correlates with patient demographics, especially those associated with higher prevalence of cholelithiasis, such as age, gender, body mass index (BMI), and Hispanic ethnicity.13
Materials and Methods
Patient and Sample Characteristics
Human bile samples and biliary stones were obtained from the gallbladders of patients undergoing a cholecystectomy between 2017 and 2019 at two New York City hospitals—Jamaica Hospital Medical Center, an inner-city hospital that cares primarily for an indigent population, and New York-Presbyterian Hospital/Weill Cornell Medical Center, a tertiary referral center. Patients with the following post-operative diagnoses were included: acute cholecystitis, biliary dyskinesia, biliary colic, cholangitis, choledocholithiasis, cholelithiasis, and gallstone pancreatitis. Patients undergoing cholecystectomy for other indications, including malignancy, were excluded. Bile (1–10 mL) and gallstones (1–3 stones) were collected at the time of surgery from intact gallbladder specimens. Samples were then immediately stored at − 80 °C.
Patient demographic information, including age, sex, race, ethnicity, and nationality, was also collected. In addition, comorbidities [diabetes mellitus (DM), hypertension (HTN)], other clinical factors [BMI, smoking status, alcohol use, perioperative antibiotic use oral contraceptive (OCP) use], and any additional biliary procedures performed around the time of the cholecystectomy, such as endoscopic retrograde cholangiopancreatography (ERCP), sphincterotomy, stent placement, or cholecystostomy tube placement, were recorded from chart review. This protocol was approved by the institutional review boards at both institutions and preoperative informed consent was obtained from all patients.
DNA Extraction and Quantification
The research team collected bile and biliary stones aseptically to minimize cross-contamination, and specimen were stored in sterile Falcon 15-mL conical tubes (Thermo Fisher Scientific, Waltham, MA) in − 80° freezers until DNA extraction. Total DNA was extracted from 30 mg of homogenized biliary stones or 30μL of bile utilizing the QIAamp DNA Micro Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions with minor modifications. Briefly, 30 mg of biliary stone or 30μL of bile samples was lysed in 1.5 mL microcentrifuge tubes using 180μL of Buffer ATL and proteinase K. To ensure complete lysis, samples were placed in a thermomixer set to 56 °C for 12 h. DNA was then bound to the QIAamp MinElute column membranes by transferring the lysate to the columns and adding 200μL of Buffer AL followed by 200μL of 100% ethanol. Samples were then sequentially washed using wash buffers AW1 and AW2 as per the protocol. Elution of DNA was performed using 30μL of Buffer AE, centrifuging the samples at full speed (20,000 g) for 5 min. After the extraction was complete, DNA concentration was determined using a NanoDrop Microvolume Spectrophotometer (Thermo Fisher Scientific, Waltham, MA). The research team performed DNA extraction aseptically and changed gloves and pipette tips with each patient sample to minimize cross-contamination.
Metagenomic Sequencing
DNA samples were subsequently processed by the Weill Cornell Medicine Microbiome Core for metagenomic sequencing using the Illumina Nextera XT Preparation Kit (Illumina, San Diego, CA) according to the manufacturer’s instructions.
Shotgun Metagenomics Analysis
Raw sequence reads were filtered based on read quality and host reads were removed using kneadData software,14 which makes use of Trimmomatic15 and Bowtie2.16 Taxonomic classification of the resulting reads was performed using Kraken2.17 Species and genus relative abundance were then estimated using Bracken.18 The bacterial fraction of classified reads was determined by adding relative abundances of all bacterial species in the Bracken output, using Taxonkit19 to list bacterial species.
16S Library Generation, Library Verification, Quality Check, and Pooling
Library generation follows the protocol from Earth Microbiome Project.20 Amplicon libraries are washed using Beckman Coulter AMPure XP magnetic beads (Beckman Coulter, Pasadena, CA). Library quality and size verification was performed using PerkinElmer LabChip GXII instrument with DNA 1 K Reagent Kit (PerkinElmer, Waltham, MA). Library concentrations are quantified using Quant-iT dsDNA High Sensitivity Assay Kit using Promega GloMax plate reader on a microplate (Promega, Madison, WI). Library molarity is calculated based on library peak size and concentration. Libraries are normalized to 2 nM using the PerkinElmer Zephyr G3 NGS Workstation (PerkinElmer, Waltham, MA) and pooled together using the same volume across all normalized libraries into a 1.5-mL Eppendorf DNA tube (Eppendorf, Hamburg, Germany).
Sequencing
Pooled libraries are sequenced on the Illumina MiSeq instrument at loading concentration of 7 pM with 10% PhiX, paired-end 250 using MiSeq Reagent Kit v2, 500-cycles (Illumina, San Diego, CA).
Data Processing
Demultiplexed raw reads were processed to generate an operational taxonomic unit (OTU) table using USEARCH version 11.0.667.21 Specifically, forward and reverse reads were merged using a maximum of 5 mismatches in the overlap region, a minimum sequence identity in the overlap region of 90 percent, a minimum overlap length of 16 base pairs, and a minimum merged sequence length of 300 base pairs. PhiX contamination was then removed, followed by quality filtering based on FASTQ quality scores, with a maximum expected error number of 1.0. OTU clustering was performed using usearch -cluster_otus with default settings. Merged (pre-filter) reads were mapped to the OTU sequences to generate the OTU table. Taxonomic classification of OTU representative sequences was performed using usearch -sintax, an implementation of the SINTAX algorithm,22 using version 16 of the Ribosomal Database Project (RDP) Training Set.23 Alpha diversity estimation and principal coordinate analysis (PCoA) were performed using the phyloseq R package.24
Statistical Analysis
Statistical analysis of the microbiome data was performed using R 3.6.25 Differential prevalence was tested using Fisher’s exact tests. Shannon index of diversity and bacterial fractions were compared using the Wilcoxon rank test. Correlations of bacterial fraction in bile and stones were made using Pearson’s product-moment correlation. A multivariate linear regression for the bacterial relative abundance, including variables historically considered to contribute to gallstone disease, was performed in the non-infectious cohort. Variables included in the multivariate analysis were the type of sample (bile or stone), batch number, age, BMI, Hispanic ethnicity, and gender. BMI and age were included as dichotomous variables. BMI was characterized as above or below 30 kg/m2, and age was characterized as above or below 40 years of age. Additional demographic and clinicopathologic variables were compared via Fisher’s exact test for categorical variables or Student’s t-test for parametric continuous variables using Stata software, version 15.1 (Stata Corp. College Station, TX).
Results
Cohort Characteristics
In our surgical cohort of 49 cholecystectomy patients, 29% (14/49) had final pathology of acute cholecystitis while 71% (35/49) patients had non-infectious pathology—chronic cholecystitis, cholelithiasis, and normal gallbladder (patients with biliary dyskinesia) (Table 1). The two cohorts were similar in age, gender, and comorbidities. There were a variety of preoperative diagnoses in both cohorts, and as expected, more acute cholecystitis patients received preoperative antibiotics in addition to the standard perioperative single dose. Additionally, we had a subset (n = 25) of patients with matched bile and stone samples with characteristics representative of the entire cohort (Supplemental Table 1).
Genera and Species Were Similar in Paired Bile and Stone Samples
To further characterize the microbiome of our patients, we assessed genera represented in all patient samples. A total of 40 different genera were categorized in our cohort (Fig. 1), with variations dependent on pathologic diagnosis, ethnicity, and gender. Paired samples of bile and stone from the same patients (n = 25) had comparable genera. The most common genera included Streptococcus, Klebsiella, Burkholderia, Staphylococcus, and Escherichia. In addition to genera, we characterized our cohort by bacterial species (Fig. 2). The relative abundance of different bacteria species varied while paired bile and stone samples had analogous species (Fig. 2). The most common species in our cohort were Klebsiella pneumoniae, Staphylococcus aureus, and Streptococcus pasteurianus.
Genus heatmap showing all genera. Forty different genera are represented and varied based on pathologic diagnosis, ethnicity, and gender while paired bile and stone had similar genera. The most common genera included Streptococcus, Klebsiella, Burkholderia, Staphylococcus, and Escherichia. The 25 paired bile and stone samples (1–25) are denoted
Bile and Stones Phyla Bacterial Fractions and Shannon Index of Diversity
When looking more broadly at the composition of phyla, Proteobacteria had the highest relative abundance in both bile and stones followed by Firmicutes (Fig. 3A). In addition to similar phyla composition, the bacterial fraction in bile and paired stones was positively correlated (Fig. 3B). The relative abundance of bacterial fraction was also similar but tended to be higher in stones, although not statistically significant (Fig. 3C). Lastly, the median Shannon index of diversity between bile and paired stones were similar (Fig. 3D).
Representations of phyla from bile and stone. A The relative abundance of phyla was similar in bile and stone, with Proteobacteria as the most common for both bile and stone, making up 49% and 65% respectively. B The bacterial fraction in paired bile and stone samples correlated (p = 0.008). C Relative abundance analysis of bacterial fraction in paired samples of bile and stone showed similar relative abundance, and the fractions tended to be higher in stones although not statistically significant. D Shannon index comparing bile and stone phyla was similar. The central rectangle of each boxplot represents the interquartile range (IQR) while the inner line represents the median. The whisker lines represent the minimum and maximum values while the dot represents a suspected outlier, outside of 1.5 times the IQR
Differential Prevalence of Bacterial Species in Bile and Stones Across Pathologies
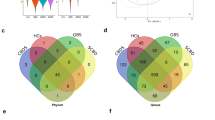
We next compared bacterial species in bile and stones based on final surgical pathology. There were 11 species found in bile and 12 species in stones (Fig. 4A and B). When looking at the prevalence of bacterial species in bile, there were similar rates in acute cholecystitis and the non-infectious pathologies, including chronic cholecystitis and non-inflammatory pathologies such as biliary dyskinesia (Fig. 4A). The prevalence was similar for most species, with an increased prevalence of Streptococcus pasteurianus in the non-infectious pathologies (chronic cholecystitis and non-inflammatory pathologies) compared to acute cholecystitis (0.46 vs 0.08, p = 0.027). However, the differential prevalence in stones varied across the pathologies (Fig. 4B). For example, Staphylococcus aureus had the highest differential prevalence in bile samples for all surgical pathologies, but stone samples had variable prevalence depending on pathology. The highest prevalence of Staphylococcus aureus in stones was in the non-inflammatory patients. Four different species had different prevalence in the stone cohort, with a statistically higher prevalence of Veillonella parvula, Streptococcus sanguinis, Kosakonia radicincitans, and Salmonella enterica in acute cholecystitis patients compared to the non-infectious pathologies.
Comparison of acute cholecystitis and non-infectious patholigies, including chronic cholecystitis and non-inflammatory pathologies. The differential prevalence varied in bile (A) and stone (B) depending on gallbladder pathology with non-infectious; however, there was a wide range of species across all pathologies. C The bacterial fraction was higher in acute cholecystitis bile (p = 0.024) and stones (p = 0.012) compared to non-infectious pathology D while the Shannon index of diversity was similar in all pathologies
The bacterial fraction was statistically higher in bile and stones in samples taken from patients with acute cholecystitis compared to non-infectious pathologies (p = 0.024, p = 0.012) (4C) while the Shannon index of diversity was similar (4D). Overall, there was a variety of bacterial species found in infectious and non-infectious bile and stones. On further sub-analysis of the non-infectious cohort, there was a similar variety of species in bile samples from patients with non-inflammatory pathology compared to those with acute and chronic cholecystitis. Three species (Citrobacter freundii, Streptococcis sanguinis, and Enterobacter roggenkampii) were found only in bile from patients with acute or chronic cholecystitis.
A Sub-analysis of the Non-infectious Cohort Based on Demographics
Finally, a multivariate linear regression analysis of the bacterial fraction of the non-infectious group revealed that the bacterial composition may be relatedS to age, BMI, and ethnicity. The overall regression was statistically significant (R2 = 0.476, F 6 = 6.65, p < 0.001). Age over 40 significantly predicted the relative bacterial abundance (β = 0.582, p = 0.049) while BMI over 40 (β = − 0.570, p = 0.064) and Hispanic ethnicity (β = 0.602, p = 0.074) approached significance. Based on our multivariate findings, we completed an additional analysis within the non-infectious cohort to look for differences between Hispanic and non-Hispanic patients. The relative abundance bacterial fraction (Fig. 5A) and the median Shannon index of diversity (Fig. 5B) were not significantly different in bile or stones of Hispanic patients compared to non-Hispanic patients. Similarly, the differential prevalence of genera varied by ethnicity for bile (Fig. 5C) and stones (Fig. 5D), but these differences were not statistically significant. Staphylococcus was the most prevalent genus in bile and stones regardless of ethnicity.
A sub-analysis of the non-infectious cohort showed differences in patients’ Hispanic ethnicity. A The relative abundance bacterial fraction was not significantly higher in stones (p = 0.06) or bile in Hispanic patients compared to non-Hispanic patients. B The Shannon index of diversity was also similar across ethnicities.. The differential prevalence of genera varied between Hispanic and non-Hispanic patients in bile (C) and stones (D), although these differences were not statistically significant
Discussion
In this analysis of bile and gallstones from patients undergoing cholecystectomy, we characterized the biliary microbiome in the largest cohort to date via metagenomic sequencing. We found a broad range of bacteria, with Proteobacteria the most common phyla in bile and gallstones, and the most abundant species were Klebsiella pneumoniae, Staphylococcus aureus, and Streptococcus pasteurianus. The biliary bacterial compositions were similar to previous small bowel microbiome analyses which found Proteobacteria the most common phyla26 and Streptococcus the most abundant genus.27 In paired samples of bile and stones, we found similar bacterial compositions while the species varied based on final surgical pathology. We found a higher prevalence of bacteria in acute cholecystitis compared to non-infectious pathologies while diversity was similar across all pathologies. Surprisingly, non-infectious pathologies often had a similar variety of bacteria to those found in patients with acute cholecystitis, including high prevalence of Staphylococcus aureus in patients with biliary dyskinesia.
Our study used the more robust shotgun metagenomic sequencing, in which all microorganisms in a sample are sequenced,28,29 to characterize the biliary microbiome. Most of the previous literature used 16S ribosomal RNA amplification, including one study that found “no significant bacterial signal” identified in the normal bile samples,30 suggesting 16S sequencing analyses of the biliary microbiome are not comprehensive. Moreover, shotgun metagenomic sequencing has been shown to yield more genera of bacteria compared to 16S ribosomal RNA amplification.29
Although all patients in our study had an indication for cholecystectomy, the reasons varied from acute cholecystitis to biliary dyskinesia. Four patients in our cohort (n = 2 acute cholecystitis, n = 2 non-infectious pathology) underwent endoscopic ERCP with sphincterotomy prior to collection of specimen. Although sphincterotomy is believed to lead to increased rates of ascending cholangitis due to reflux of bacteria from the duodenum into the biliary tree, recent studies suggest that the impact of sphincterotomy on bacterial contamination may be minimal.31,32 Unfortunately, our cohort was limited for further analysis due to sample size. Previous studies included the bile of healthy individuals, while our study only included patients referred for surgical resection in our analysis. Given the similar bacterial profiles identified in the paired bile and stone samples—also noted in a previous study using the 16S sequencing method33—similar inferences may be drawn from the samples in which we only have bile or stone. Finding a variety of diverse bacteria across surgical pathologies, including diseases not classically considered to be contaminated, highlights that bile and gallbladder stones are not sterile, even in the absence of acute cholecystitis, as has recently been suggested.12 When compared to healthy liver donors, the biliary microbiome of patients with cholelithiasis was comprised of different families of bacteria.12 Moreover, bacterial dysbiosis has also been associated with the development of cholangiocarcinoma.30,34 As dysbiosis of the microbiome contributes to diseases of the small bowel and colon, and bile has been shown to harbor bacteria, it is possible that biliary dysbiosis contributes to gallstone formation.
Age, gender, and obesity are considered risk factors for gallstone formation,35–37 as well as certain ethnicities.13,38 Previous data from the Hispanic Health and Nutrition Examination Survey have shown increased rates of gallstones among Hispanic patients, particularly Mexican American women, with rates of 44.1% in women aged 60 to 74 years old.13 This increased rate of gallstone diseases in Mexican American patients remained, even when other risk factors were controlled for in a logistic regression modeling.39 While the cohort in our study was small, there appears to be differences in the microbiomes of Hispanic patients, including higher bacterial fractions and increased Shannon index of diversity. These differences approached statistical significance and should be further explored on a larger scale.
Limitations to our study include a relatively small sample size of surgical patients. Nonetheless, this is the largest cohort of patients sequenced with shotgun metagenomics to our knowledge. Additionally, the bacterial diversity of our cohort may be limited by the threshold used for screening relevant bacteria, which was set at 0.02%. It is possible that we missed additional bacterial diversity because of this threshold; however, this was mitigated by our use of metagenomic sequencing instead of ribosomal 16S RNA sequencing. Our analysis may be impacted by the batch effect on our samples, as the samples were assessed in two batches and can be impacted by the other samples in the cohort. However, our statistical model accounts for the fact that samples were sequenced in two separate batches. Most of the limitations of our study are typical and inherent in microbiome analyses.
In conclusion, this study depicts the biliary microbiome in a variety of pathologies in patients undergoing cholecystectomy. This characterization highlights the large variety of bacterial species present in the biliary microbiome—including in patients with acute cholecystitis and non-infected pathologies—with similarities to previously characterized small bowel microbiomes. The diversity of the gallbladder’s microbiome and its potential impact on gallstone formation should be further studied on a larger scale.
References
Konturek PC, Koziel J, Dieterich W, Haziri D, Wirtz S, Glowczyk I et al. Successful therapy of Clostridium difficile infection with fecal microbiota transplantation. J Physiol Pharmacol. 2016;67(6):859-66.
Sokol H, Seksik P, Rigottier-Gois L, Lay C, Lepage P, Podglajen I et al. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm Bowel Dis. 2006;12(2):106-11. https://doi.org/10.1097/01.MIB.0000200323.38139.c6.
Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J et al. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology. 2011;141(2):599-609, .e1-3. https://doi.org/10.1053/j.gastro.2011.04.052.
Broido PW, Gorbach SL, Condon RE, Nyhus LM. Upper intestinal microfloral control. Effects of gastric acid and vagal denervation on bacterial concentrations. Arch Surg. 1973;106(1):90-3. https://doi.org/10.1001/archsurg.1973.01350130088020.
Russo MW, Wei JT, Thiny MT, Gangarosa LM, Brown A, Ringel Y et al. Digestive and liver diseases statistics, 2004. Gastroenterology. 2004;126(5):1448-53. https://doi.org/10.1053/j.gastro.2004.01.025.
Portincasa P, Moschetta A, Palasciano G. Cholesterol gallstone disease. Lancet. 2006;368(9531):230-9. https://doi.org/10.1016/s0140-6736(06)69044-2.
Maki T. Pathogenesis of calcium bilirubinate gallstone: role of E. coli, beta-glucuronidase and coagulation by inorganic ions, polyelectrolytes and agitation. Annals of surgery. 1966;164(1):90–100. https://doi.org/10.1097/00000658-196607000-00010.
Cetta FM. Bile infection documented as initial event in the pathogenesis of brown pigment biliary stones. Hepatology. 1986;6(3):482-9. https://doi.org/10.1002/hep.1840060327.
Trotman BW. Pigment gallstone disease. Gastroenterol Clin North Am. 1991;20(1):111-26.
Ikeda T, Yanaga K, Kusne S, Fung J, Higashi H, Starzl TE. Sterility of bile in multiple-organ donors. Transplantation. 1990;49(3):653-. https://doi.org/10.1097/00007890-199003000-00036.
Begley M, Gahan CG, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29(4):625-51. https://doi.org/10.1016/j.femsre.2004.09.003.
Molinero N, Ruiz L, Milani C, Gutiérrez-Díaz I, Sánchez B, Mangifesta M et al. The human gallbladder microbiome is related to the physiological state and the biliary metabolic profile. Microbiome. 2019;7(1):100. https://doi.org/10.1186/s40168-019-0712-8.
Maurer KR, Everhart JE, Ezzati TM, Johannes RS, Knowler WC, Larson DL et al. Prevalence of gallstone disease in Hispanic populations in the United States. Gastroenterology. 1989;96(2 Pt 1):487-92. https://doi.org/10.1016/0016-5085(89)91575-8.
KneadData. https://huttenhower.sph.harvard.edu/kneaddata/.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England). 2014;30(15):2114-20. https://doi.org/10.1093/bioinformatics/btu170.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9(4):357-9. https://doi.org/10.1038/nmeth.1923.
Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biology. 2019;20(1):257. https://doi.org/10.1186/s13059-019-1891-0.
Lu J BF, Thielen P, Salzberg SL. Bracken: estimating species abundance in metagenomics data. PeerJ Computer Science. 2017;3:e104. https://doi.org/10.7717/peerj-cs.104.
Shen W, Ren H. TaxonKit: A practical and efficient NCBI taxonomy toolkit. Journal of Genetics and Genomics. 2021;48(9):844-50. https://doi.org/10.1016/j.jgg.2021.03.006.
16S Illumina Amplicon Protocol. https://earthmicrobiome.org/protocols-and-standards/16s/.
Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460-1. https://doi.org/10.1093/bioinformatics/btq461.
Edgar RC. SINTAX: a simple non-Bayesian taxonomy classifier for 16S and ITS sequences. bioRxiv. 2016:074161. https://doi.org/10.1101/074161.
Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42(Database issue):D633-42. https://doi.org/10.1093/nar/gkt1244.
McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. https://doi.org/10.1371/journal.pone.0061217.
R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2019.
Chung CS, Chang PF, Liao CH, Lee TH, Chen Y, Lee YC et al. Differences of microbiota in small bowel and faeces between irritable bowel syndrome patients and healthy subjects. Scand J Gastroenterol. 2016;51(4):410-9. https://doi.org/10.3109/00365521.2015.1116107.
Dlugosz A, Winckler B, Lundin E, Zakikhany K, Sandström G, Ye W et al. No difference in small bowel microbiota between patients with irritable bowel syndrome and healthy controls. Scientific Reports. 2015;5(1):8508. https://doi.org/10.1038/srep08508.
Jovel J, Patterson J, Wang W, Hotte N, O’Keefe S, Mitchel T et al. Characterization of the Gut Microbiome Using 16S or Shotgun Metagenomics. Front Microbiol. 2016;7:459-. https://doi.org/10.3389/fmicb.2016.00459.
Brumfield KD, Huq A, Colwell RR, Olds JL, Leddy MB. Microbial resolution of whole genome shotgun and 16S amplicon metagenomic sequencing using publicly available NEON data. PLoS One. 2020;15(2):e0228899. https://doi.org/10.1371/journal.pone.0228899.
Choi SJ, Kim Y, Jeon J, Gwak HJ, Kim M, Kang K et al. Association of Microbial Dysbiosis with Gallbladder Diseases Identified by Bile Microbiome Profiling. J Korean Med Sci. 2021;36(28):e189. https://doi.org/10.3346/jkms.2021.36.e189.
Shen H, Ye F, Xie L, Yang J, Li Z, Xu P et al. Metagenomic sequencing of bile from gallstone patients to identify different microbial community patterns and novel biliary bacteria. Sci Rep. 2015;5:17450. https://doi.org/10.1038/srep17450.
Natsui M, Honma T, Genda T, Nakadaira H. Effects of endoscopic papillary balloon dilation and endoscopic sphincterotomy on bacterial contamination of the biliary tract. Eur J Gastroenterol Hepatol. 2011;23(9):818-24. https://doi.org/10.1097/MEG.0b013e328348c0bf.
Wu T, Zhang Z, Liu B, Hou D, Liang Y, Zhang J et al. Gut microbiota dysbiosis and bacterial community assembly associated with cholesterol gallstones in large-scale study. BMC Genomics. 2013;14:669. https://doi.org/10.1186/1471-2164-14-669.
de Martel C, Plummer M, Parsonnet J, van Doorn LJ, Franceschi S. Helicobacter species in cancers of the gallbladder and extrahepatic biliary tract. Br J Cancer. 2009;100(1):194-9. https://doi.org/10.1038/sj.bjc.6604780.
Barbara L, Sama C, Labate AMM, Taroni F, Rusticali AG, Festi D et al. A population study on the prevalence of gallstone disease: The sirmione study. Hepatology. 1987;7(5):913-7. https://doi.org/10.1002/hep.1840070520.
Friedman GD, Kannel WB, Dawber TR. The epidemiology of gallbladder disease: observations in the Framingham Study. Journal of chronic diseases. 1966;19(3):273-92.
Einarsson K, Nilsell K, Leijd B, Angelin B. Influence of age on secretion of cholesterol and synthesis of bile acids by the liver. New England Journal of Medicine. 1985;313(5):277-82.
Sampliner RE, Bennett PH, Comess LJ, Rose FA, Burch TA. Gallbladder disease in Pima Indians: demonstration of high prevalence and early onset by cholecystography. New England Journal of Medicine. 1970;283(25):1358-64.
Maurer KR, Everhart JE, Knowler WC, Shawker TH, Roth HP. Risk factors for gallstone disease in the Hispanic populations of the United States. Am J Epidemiol. 1990;131(5):836-44. https://doi.org/10.1093/oxfordjournals.aje.a115574.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This abstract was presented virtually at the Scientific Forum of the American College of Surgeons’ Clinical Congress October 2020.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Limberg, J., Egan, C.E., Mora, H.A. et al. Metagenomic Sequencing of the Gallbladder Microbiome: Bacterial Diversity Does Not Vary by Surgical Pathology. J Gastrointest Surg 26, 2282–2291 (2022). https://doi.org/10.1007/s11605-022-05418-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11605-022-05418-6