Abstract
Background
The most common complications after colorectal surgery, postoperative ileus, surgical site infections, and anastomotic leaks continue to occur despite advances in surgical technique and enhanced recovery pathways. Preclinical studies have documented that intestinal bacteria play a role in the development of these complication, yet human data is lacking. Here we hypothesized that patients that develop ileus, surgical site infection, and/or anastomotic leak following colorectal surgery harbor a specific preoperative gut microbiome.
Methods
We performed a prospective cohort study on 101 patients undergoing colon or rectal resection at the Mayo Clinic. Rectal samples were collected preoperatively and on the ward on postoperative day two. The bacterial community from each sample was characterized by 16S rRNA and associated with the development of complications.
Results
The rectal microbiome collected from patients in the operating room (p = .003) and on postoperative day two (p = .001) was significantly difference in patients whom later developed postoperative ileus compared with patients that had a normal return of bowel function. Patients whom developed ileus showed increased abundance of Bacteroides spp., Parabacteroides spp., and Ruminococcus spp., bacteria that are associated with promoting intestinal inflammation. There were no differences in the microbiome in patients that developed surgical site infections or anastomotic leaks.
Conclusions
In this pilot study, patients that develop postoperative ileus harbor a specific gut microbiome during the perioperative period. These findings demonstrate that the preoperative bacterial composition may predispose patients to the development of ileus and that perioperative manipulation of the gut bacteria may provide a novel method to promote normal return of bowel function.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colon or rectal resection with anastomosis continues to have an overall morbidity rate of nearly 35%.1,2 The most common complications driving this morbidity are postoperative ileus (POI), surgical site infection (SSI), and anastomotic leak (AL). Despite preoperative optimization of comorbidities, use of minimal invasive surgery, and utilization of enhanced recovery pathways, these complications continue to cause hospital readmissions, increased inpatient length of stay, and $1.5 billion in healthcare costs annually in the USA.3,4
The primary reason that POI, SSI, and AL continue to cause major postoperative morbidity is because the mechanism by which they develop remains elusive.5 Yet, the explosion of microbiome research over the last decade has clearly demonstrated that the gut microbiota is critical to maintaining normal health and is responsible for an ever expanding list of processes such as nutrient metabolism, barrier function, and immune cell modulation.6,7 Building upon this foundation, there is now emerging preclinical evidence from animal studies that demonstrate that perturbations of the gut microbiota promote the development of POI, SSI, and AL.8,9,10,11,12 Furthermore, human-based publications have shown that preoperative intestinal decontamination with oral and intravenous antibiotics significantly reduces the incidence of POI, SSI, and AL, lending concrete evidence that their pathogenesis is, at least in part, microbial driven.13,14
While the current strategy of bowel prep is effective in some, why some patients appear to be immune to the protective effect of preoperative decontamination remain unknown. Risk factors common to POI, SSI, and AL (i.e., smoking, obesity, steroids, previous surgery) have independent effects on the microbiome, yet, how these variables coalesce to result in a preoperative microbiota that influences the development of postoperative complications is essentially unstudied.15,16,17 Here we hypothesize that patients whom develop POI, SSI, and AL harbor a specific perioperative microbiome. To investigate this hypothesis, we performed a pilot study and analyzed the perioperative composition of the gut-associated microbiota in patients undergoing colorectal surgery to determine if a specific bacterial community structure associates with the development of postoperative complications.
Methods
Study Design and Participants
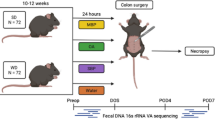
This prospective cohort study of patients undergoing colon or rectal resection at the Mayo Clinic Rochester between 3/1/2017 and 7/31/2017 was approved by the Mayo Clinic Institutional Review Board. All patients ≥ 18 years of age undergoing colon or rectal resection by a colorectal surgeon were eligible for inclusion. Patients with an existing ileostomy were excluded. To keep rectal swab sampling consistent, patients whom were given an end ileostomy during the index surgery were excluded. Patients whom were given an end colostomy or a diverting loop ileostomy with retention of the anus/rectum were included. Preoperative bowel preparation consisted of Golytely and oral neomycin and Flagyl on the day before surgery. At the time of incision, each patient was given intravenous cefazolin.
Procedures
Sample Collection
Prior to sample collection, all patients underwent written consent. Rectal samples were collected from patients intraoperatively (POD0) and on postoperative day two (POD2). POD0 samples were collected after induction of anesthesia and prior to any application of antiseptics; POD2 samples were collected in the hospital room. Anal samples were collected in all patients whom had an anus (even if they had a diverting loop ileostomy) by placing a swab (Copan FLOQ Swab) just passed the anus and rubbed for 10 s. If the patient had postoperative colostomy, the sample was taken by inserting the swab 3 cm into the colostomy and swabbing for 10 s. All samples were immediately snap-frozen in isopentane and stored at − 80 °C.
Data Collection
At the time of consent, each patient completed a questionnaire documenting current and/or previous 90-day use of steroids, antibiotics, and tobacco. Clinical and 30-day outcomes data was collected from the electronic medical record. If they did not follow-up locally, patients were contacted on postoperative day 30 and using a script, queried on the development of complications. SSI was defined using the NSQIP criteria as an infection of the skin with purulent drainage, localized edema, erythema, or diagnosis made by the attending physician. AL was defined by clinical and radiographically evidence of anastomotic leakage. A pelvic fluid collection was defined as a fluid collection adjacent to a newly formed anastomosis; because fluid collections adjacent to an anastomosis likely represent anastomotic leakage, for analysis purposes, these two entities were combined. POI was defined by the presence of ≥ 2 of the following over a 24-h period occurring on or after the fourth postoperative day: ≥ 2 episodes of vomiting, inability to tolerate diet, absence of flatus, abdominal distension.18 As per routine, no patient had a nasogastric tube placed prophylactically postoperatively. Placement of a postoperative nasogastric tube for the treatment of POI was determined by the primary surgeon.
DNA Extraction, MiSeq Preparation, and Sequencing
To avoid batch effect, DNA extraction and MiSeq sequencing were performed for all samples in a single batch. Total DNA was extracted using the PowerSolid Kit (#12888-100). The V3 to V4 region of the 16s rRNA gene was amplified by PCR. Paired R1 and R2 sequence reads were processed via the hybrid-denovo bioinformatics pipeline, which clustered a mixture of good quality paired-end and single-end reads into operational taxonomic units (OTUs) at 97% similarity level.19 OTUs were assigned taxonomy using the RDP classifier trained on the GreenGenes database (v13.5). A phylogenetic tree based on FastTree algorithm was constructed based on the OTU representative sequences. Singleton OTUs and samples with > 1000 reads were removed as a quality control (QC) step.20
Statistical Analysis
This study was designed as a pilot study to associate the perioperative microbiome with the development of complications. Alpha (within sample) and beta (between sample) diversity were analyzed. Four alpha diversity measures were calculated on the rarefied OTU data: observed number of OTUs, Chao1 estimator, Shannon index, and inverse Simpson index (R “phyloseq” package, v1.19.1).21 Beta diversity reflects the shared diversity between microbial communities in terms of ecological distances and quantifies the overall compositional difference between samples, and different beta diversity measures provide distinctive views of the community structure. Three phylogenetic beta diversity measures, unweighted, generalized, and weighted UniFrac distances, were calculated using the OTU table and a phylogenetic tree (R “GUniFrac” package, v1.0). The unweighted UniFrac reflects differences in community membership, whereas the weighted UniFrac prioritizes differences in the abundance. Generalized UniFrac reduces the weight on abundance reveals community differences in less abundant lineages. The Bray-Curtis distance, which does not depend on the phylogenetic tree, was calculated (R package “vegan”, v2.4.3) to capture potential non-phylogenetically related community changes. Rarefaction was performed on the OTU table for the diversity analyses.
A multiple linear regression model was used for testing the association with alpha diversity. F test/t test was used for assessing statistical significance. Beta diversity was tested using PERMANOVA. An omnibus test was used to combine multiple sources of association provided by different beta diversity measures and an overall association p value was provided (“PermanovaG” in the R “GUniFrac” package v1.0). Ordination plots were generated using principal coordinate analysis (“cmdscale” in R) for visualizing the association of covariates with the beta diversity. Taxa-level association analyses were performed at the phylum, class, order, family, and genus levels. Taxa with prevalence < 10% or with a maximum proportion < 0.2% were excluded from testing to reduce multiple testing burden. The count data was normalized into relative abundances by dividing by the GMPR size factor.22 A permutation test (1000 permutations) was used to identify differentially abundant taxa based on the F-statistic of a linear regression model with the square-root transformed taxa relative abundance as the response variable.23 False discovery rate (Benjamini-Hochberg) was used to correct for multiple testing on each taxonomic level, and FDR-adjusted p values or q values < 0.05 were considered significant. All analyses adjusted the Illumina flow-cell ID. All statistical analyses were performed in R 3.3.2.
Results
Demographics and Samples
A total of 101 patients undergoing colon or rectal resection at the Mayo Clinic Rochester between 3/1/2017 and 7/31/2017 were included in the study (Table 1). The mean age of the cohort was 53.4 years, and 56% of patients were male. The most common surgical indication was malignancy or an unresectable polyp (n = 52, 51.4%), inflammatory bowel disease (n = 27, 26.7%), diverticular disease (n = 20, 19.8%), and other (n = 2, 1.9%). The most common operation was segmental resection (n = 45, 45%) followed by proctectomy (n = 36, 35.6%), abdominal perineal resection (n = 13, 12.8%), total abdominal colectomy (n = 5, 4.9%), and other (n = 2, 1.9%). Seventy-six percent of surgeries were performed in a minimally invasive fashion. Thirty-day follow-up was successful in 95 patients (94%).
A total of 155 microbial samples were collected, passed quality control, and were analyzed. POD0 samples were collected in all 101 patients, whereas POD2 samples were collected in 54 patients. The most common reason for a missed POD2 sample was patient refusal or early discharge.
Overall Microbial Composition
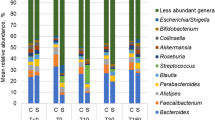
The mean relative abundance of the entire cohort is presented in Supplemental Fig. 1. Analysis of the cohort as whole revealed that on POD2, there was a significantly increased abundance of the phylum Bacteroidetes and decreased abundance of phyla Actinobacteria and Firmicutes compared with samples collected on POD0.
Association of the Compositional Changes of the Microbiota with Surgical Outcomes
By comparing bacterial communities from patients who developed complications to those that did not, we next assessed for associations between specific bacterial compositions and surgical outcomes (Table 2). We found no significant differences in the rectal-associated bacterial communities recovered on POD0 or POD2 in patients who developed SSI’s, deep pelvic infections, or anastomotic leaks. However, we found significant differences in the composition of the rectal-associated bacterial communities recovered on both POD0 and on POD2 between those who developed POI compared with those that did not. Clinically, patients who developed POI were more likely to be a current or past smoker and showed a significantly increased length of stay compared with those patients that did not develop POI (Table 3). All patients whom were diagnosed with POI had a nasogastric tube placed for treatment.
Bacterial Changes in Patients Who Developed POI
We next wanted to understand the differences in the bacterial communities associated with the development of POI. Analysis of the alpha diversity, representing the number (richness) and distribution (evenness) between taxa within a single population, trended to be increased in patients who developed POI, but was only significant in the inverse Simpson index (Supplemental Fig. 1). Beta diversity analysis was used to compare the compositional difference between bacterial populations associated with patients who developed POI compared with those that did not (Figs. 1 and 2). We found that in all four of the tested beta diversity measures, there were significant differences in the bacterial community composition recovered on both POD0 and POD2 in patients that developed POI compared with those that did not. Strikingly, the magnitude of increased beta diversity in patients who developed POI was significantly more profound in the bacterial communities recovered on POD2 than on POD0. To account for the potential differences between rectal-associated microbiota and ostomy-associated microbiota, this analysis was repeated excluding patients who underwent a postoperative diversion. Similarly, we found a significant difference in the rectal-associated microbiota in patients with POI (Supplemental Fig. 2).
Patients whom develop postoperative ileus have a different rectal-associated bacterial composition on POD0 and POD2. Principal coordinate analysis (based on beta diversity measures) of rectal-associated bacterial communities showing that the bacterial composition recovered on POD0 (a) and on POD2 (b) was significantly different in patients whom developed POI to those that had normal return of bowel function (POD, postoperative day; POI, postoperative ileus, p < .05 considered significant)
The phyla of rectal-associated microbiota in patients whom develop postoperative ileus is significantly different on POD0 and POD2. Mean compositional phyla analysis comparing rectal-associated microbiome samples collected in the operating room (POD0) and on the hospital ward (POD2). Patients whom developed POI showed significantly increased abundance of Proteobacteria and Bacteroidetes and decreased Actinobacteria and Firmicutes (*p < 0.05)
The major taxonomic differences accounting for the diversity changes in patients with POI. The rectal-associated microbiota from POD0 and POD2 in patients that later developed POI showed increased Proteobacteria and Bacteroidetes and decreased Actinobacteria and Firmicutes. We found that at the family and genus level, the most significant changes where increased abundance of Bacteroides spp., Parabacteroides spp., and Ruminococcus spp., (Table 4).
Drivers of the Bacterial Compositional Changes Associated with Postoperative Ileus
To understand which preoperative factors contribute to the development of the communities associated with POI, we investigated what preoperative variables significantly altered the microbiome on samples recovered from the rectum on POD0. This analysis was performed by comparing the bacterial communities in patients with that variable compared with those that did not have that variable. We found that patients > 65 years of age, surgical indication of ulcerative colitis or malignancy, previous intestinal surgery, current steroid use, and preoperative chemotherapy/radiation were associated with significant alterations in the microbiota compared with patients that did not undergo those interventions (PERMANOVA, p < 0.01; Supplemental Table 1). On post hoc analysis, the variables that were significantly associated with alterations of the microbiome on POD0 were not significantly associated with the development of POI, concluding that the ileus-microbiota association was likely not driven by these confounding variables.
Discussion
The human microbiome is a vast ecosystem encompassing 100 trillion bacterial cells and 20 million unique microbial genes that colonize the human.24 While the role of the human microbiome on the pathogenesis of diseases such as inflammatory bowel disease, obesity, and malignancy has matured over the last decade, our understanding on how microbiota alterations cause complications following colorectal surgery is in its infancy. Given the now, well-known role of microorganisms in maintaining gut homeostasis, it is logical to hypothesize that preoperative dysbiosis predisposes to postoperative complications. The primary aim of this study was to determine if patients whom develop postoperative complications harbor a common and specific microbiome. Our results show that the rectal-associated microbiota recovered on both POD0 and POD2 was significantly different in patients that subsequently developed POI compared with those that did not. For the first time, we show that in patients undergoing colorectal surgery, perturbations of the gut microbiome may predispose them towards a postoperative delay in return of bowel function.
POI affects 10–25% of patients undergoing colorectal surgery and is the leading cause of readmissions.25,26,27,28,29 The implementation of enhanced recovery protocols (ERP) has been effective in promoting quicker return of bowel function and reducing length of stay.30 However, due to the development of POI, up to 50% of patients continue to have hospital stays that are longer than 5 days even using ERP and/or minimally invasive surgery.31 Based on the existing literature and known risk factors for the development of POI, it remains very difficult to predict which patient will quickly recover bowel motility versus those whom will have an extended hospital stay because of intolerance to diet.
The results of our current study suggest that certain bacterial populations may drive the development of POI in patients undergoing colorectal surgery. While POI has traditionally been framed as a consequence of excessive inflammation due to tissue trauma inherent during any intestinal operation, recent animal studies have provided compelling evidence that the gut microbiome is the initiating trigger for this inflammatory cascade.32,33,34,35 Alterations of the colon microbiome with antimicrobials in a murine model significantly influenced gut motility. Further, POI was prevented in mice lacking toll-like reports (the key component of microbial sensing), strongly suggesting a microbial-centered process.36
Which precise taxa influence this inflammatory cascade resulting in POI remains unknown. We demonstrated that the patients whom developed POI had a significant increase in taxa associated with inflammation. For instance, enhanced abundance of Bacteroidetes Odoribacteraceae has been shown in a murine model of colitis and in human acute appendicitis samples, increased colonization of Bacteroidetes Rikenellaceae is associated with increasing systemic lupus erythematosus severity and with inflammatory colorectal cancer, and Ruminococcus spp. have been associated with inflamed mucosa in human patents.37,38,39,40,41 Conversely, reduced amounts of Actinobacteria and Bifidobactor spp., as seen in our patients whom developed POI, was associated with worse inflammation in a model of pancreatitis and ulceration in a model of peptic ulcer disease.42,43,44 Finally, the observation of a 30-fold enhanced proliferation of the genus Bacteroides in POI patients, particularly on POD2, is intriguing. Via induction of the anti-inflammatory cytokine IL-10, Bacteroides has consistently been shown to proliferate to limit intestinal inflammation.45,46,47 Therefore, the enhancement of Bacteroides on POD2 in patients with POI may represent a compensatory mechanism to limit intestinal inflammation. Taken together, our data suggest that a bacterial community structure enriched with taxa associated with intestinal inflammation and devoid of anti-inflammatory taxa may drive POI.
The potential that a microbial signature exists for the development of POI carries important clinical implications. Over the last decade, the cost of 16S rRNA microbiome analysis has considerably decreased and certain centers have successfully used it in patient management.48 Along with clinical data, microbiome analysis was used to generate a predictive model of individualized glycemic response to food.49 It is therefore imaginable that patients could be preoperatively screened to determine if they harbor the “ileus-associated microbiome” and consequently at high risk of developing a POI. Similarly, because the community structure on POD2 demonstrated a greater degree of dysbiosis, it is conceivable that microbiome analysis could be used in deciding discharge criteria. Rather than just a predictive tool, equally intriguing is the potential to introduce clinical interventions, such as an individualized bowel prep or fecal transplant, to promote colonization of the intestinal bacteria towards a gut motility phenotype.
In this study, we relied upon rectal swabs as a proxy for the composition of the intestinal microbiome. This method has now been well validated and analysis of the microbial profiles from a rectal swab is reproducibly similar to expelled stool.50,51 Yet, how well-expelled stool represents the entirety of the gut microbiome is largely unknown. Clearly, there are differences in the bacterial composition in the colon versus the small intestine, and even within the geography of the colon.52 Similarly, luminal contents (i.e., stool) and mucosal samples can have widely different microbiomes.53 Thus, because POI is largely thought of as a disease of the small intestine, it remains unclear as to how a microbial signature recovered from the distal GI tract predicts small bowel paralysis. Similar to other inflammatory biomarkers that have been associated with POI, we hypothesize that the microbial signature in the distal rectum represents global intestinal inflammation.54 While it is technically challenging to acquire proximal samples from human patients, further work needs to be performed to determine if the bacterial composition from the proximal GI tract (small bowel, stomach, oropharynx) are different in patients with POI and validate our results.
In our cohort, we did not find any association of the microbiota with the development of AL or SSI. Animal models have implicated specific organisms in driving AL.8,55 van Praagh et al. analyzed the microbiota associated with the “donuts” in patients undergoing a stapled anastomosis and found that patients with AL had a microbiome that was less diverse and enriched with Lactonospiraceae and Bacteroidaceae compared with those that did not leak.56 While we did not observe a similar pattern in our study, methodological differences likely account for the divergent results. van Praagh analyzed the bacterial composition at the site of the anastomosis, while we recovered rectal-associated bacteria even in patients with the right-sided colon resections. This suggests that the bacterial composition localized to the healing anastomotic tissue is a better predictor of AL than the bacteria in expelled stool. Conversely, because POI affects the entire length of the GI tract, the rectal-associated microbiota may better represent entire gut composition.
There are several limitations to our study. Importantly, this was designed as a pilot study to screen patients for an association of certain bacterial communities with the development of complications. Because of the pilot study design, our patient population was purposely heterogeneous and included patients with various indications, approaches, and comorbidities. Because we observed that other demographic variables (i.e., age, steroid use) were associated with certain bacterial communities our study does not indicate causality. Our limited sample size may have resulted in statistical error and thus not demonstrating that these perioperative variables influence the microbiome in POI patients. We are currently recruiting patients such that we reach appropriate statistical power based upon a simulated Bray-Curtis and Euclidean power analysis using date from this preliminary study.
We strategically designed this study to collect samples on POD2 to assess the progression of the microbiome postoperatively. While this will be an important aspect of follow-up studies, we encountered limitations with postoperative sampling. First, a fraction of patients had a postoperative diverting loop ileostomy at their index operation. While sampling in these patients occurred intra-anally to compare with the intraoperative anal swab, diversion of the fecal stream can influence the distal microbiome. For instance, it has been shown that the microbiome in the diverted segment has less diversity and reduced abundance of Clostridia and Streptococcus, compared with segments that are not diverted.57 Because exclusion of these patients did not have an effect on the overall analysis, we hypothesize that because the swabs were taken early in their postoperative course (POD2), these microbial differences were not yet apparent.
Second, POD2 samples were collected in 54% of the entire cohort. The taxa differences found on POD0 in patients whom developed POI were amplified on POD2. While this could be due to enrichment of the population with patients whom developed POI, within the boundaries of this limitation, our analysis of collected POD2 samples is intriguing. Whether the dramatic differences observed on POD2 in patients with POI are causative or consequence will need further study.
In conclusion, we have demonstrated that patients undergoing colorectal resection whom develop a POI harbor a specific rectal-associated bacterial composition in the operating room and on POD2, compared with patients with a normal return of bowel function. This observation provides a framework to better predict those patients at risk of a delay in bowel function and develop novel strategies to manipulate the microbiome in favor of a motility promoting microbiota.
Conclusions
In this pilot study, patients that developed postoperative ileus harbored a specific gut microbiome during the perioperative period. These findings demonstrate that the preoperative bacterial composition may predispose patients to the development of ileus and that perioperative manipulation of the gut bacteria may provide a novel method to promote normal return of bowel function.
References
Alves A, Panis Y, Mathieu P, et al. Postoperative mortality and morbidity in French patients undergoing colorectal surgery: results of a prospective multicenter study. Arch Surg. 2005;140:278–83, discussion 284.
Tevis SE, Kennedy GD. Postoperative Complications: Looking Forward to a Safer Future. Clin Colon Rectal Surg. 2016;29:246–52.
Cheadle WG. Risk factors for surgical site infection. Surg Infect (Larchmt). 2006;7 Suppl 1:S7-11.
Boccola MA, Buettner PG, Rozen WM, et al. Risk factors and outcomes for anastomotic leakage in colorectal surgery: a single-institution analysis of 1576 patients. World J Surg. 2011;35:186–95.
Shogan BD, Carlisle EM, Alverdy JC, et al. Do we really know why colorectal anastomoses leak? J Gastrointest Surg. 2013;17:1698–707.
Sanford JA, Gallo RL. Functions of the skin microbiota in health and disease. Semin Immunol. 2013;25:370–7.
Jandhyala SM, Talukdar R, Subramanyam C, et al. Role of the normal gut microbiota. World J Gastroenterol. 2015;21:8787–803.
Shogan BD, Belogortseva N, Luong PM, et al. Collagen degradation and MMP9 activation by Enterococcus faecalis contribute to intestinal anastomotic leak. Sci Transl Med. 2015;7:286ra68.
Zhang M, Jiang Z, Li D, et al. Oral antibiotic treatment induces skin microbiota dysbiosis and influences wound healing. Microb Ecol. 2015;69:415–21.
Scales BS, Huffnagle GB. The microbiome in wound repair and tissue fibrosis. J Pathol. 2013;229:323–31.
Pohl J-M, Gutweiler S, Thiebes S, et al. Irf4-dependent CD103+CD11b+ dendritic cells and the intestinal microbiome regulate monocyte and macrophage activation and intestinal peristalsis in postoperative ileus. Gut. 2017;66:2110–2120.
Farro G, Stakenborg M, Gomez-Pinilla PJ, et al. CCR2-dependent monocyte-derived macrophages resolve inflammation and restore gut motility in postoperative ileus. Gut. 2017;66:2098–2109.
Klinger AL, Green H, Monlezun DJ, et al. The Role of Bowel Preparation in Colorectal Surgery: Results of the 2012-2015 ACS-NSQIP Data. Ann Surg. . Epub ahead of print October 23, 2017. DOI: https://doi.org/10.1097/SLA.0000000000002568.
Kiran RP, Murray ACA, Chiuzan C, et al. Combined preoperative mechanical bowel preparation with oral antibiotics significantly reduces surgical site infection, anastomotic leak, and ileus after colorectal surgery. Ann Surg. 2015;262:416-25; discussion 423–5.
Savin Z, Kivity S, Yonath H, et al. Smoking and the intestinal microbiome. Arch Microbiol. 2018;200:677–684.
Parekh PJ, Balart LA, Johnson DA. The Influence of the Gut Microbiome on Obesity, Metabolic Syndrome and Gastrointestinal Disease. Clin Transl Gastroenterol. 2015;6:e91.
Huang EY, Inoue T, Leone VA, et al. Using corticosteroids to reshape the gut microbiome: implications for inflammatory bowel diseases. Inflamm Bowel Dis. 2015;21:963–972.
Vather R, Trivedi S, Bissett I. Defining postoperative ileus: results of a systematic review and global survey. J Gastrointest Surg. 2013;17:962–72.
Cole JR, Wang Q, Fish JA, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633-42.
Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–50.
McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217.
Chen L, Reeve J, Zhang L, et al. GMPR: A robust normalization method for zero-inflated count data with application to microbiome sequencing data. PeerJ. 2018;6:e4600.
Hale VL, Chen J, Johnson S, et al. Shifts in the Fecal Microbiota Associated with Adenomatous Polyps. Cancer Epidemiol Biomarkers Prev. 2017;26:85–94.
Proctor LM. The Human Microbiome Project in 2011 and beyond. Cell Host Microbe. 2011;10:287–91.
Wolthuis AM, Bislenghi G, Fieuws S, et al. Incidence of prolonged postoperative ileus after colorectal surgery: a systematic review and meta-analysis. Colorectal Dis. 2016;18:O1-9.
Asgeirsson T, El-Badawi KI, Mahmood A, et al. Postoperative ileus: it costs more than you expect. J Am Coll Surg. 2010;210:228–31.
Senagore AJ. Pathogenesis and clinical and economic consequences of postoperative ileus. Am J Health Syst Pharm. 2007;64:S3-7.
Schmitt SL, Cohen SM, Wexner SD, et al. Does laparoscopic-assisted ileal pouch anal anastomosis reduce the length of hospitalization? Int J Colorectal Dis. 1994;9:134–7.
Merkow RP, Ju MH, Chung JW, et al. Underlying reasons associated with hospital readmission following surgery in the United States. JAMA. 2015;313:483–95.
Thiele RH, Rea KM, Turrentine FE, et al. Standardization of care: impact of an enhanced recovery protocol on length of stay, complications, and direct costs after colorectal surgery. J Am Coll Surg. 2015;220:430–43.
Wood T, Aarts M-A, Okrainec A, et al. Emergency Room Visits and Readmissions Following Implementation of an Enhanced Recovery After Surgery (iERAS) Program. J Gastrointest Surg. 2018;22:259–266.
Stein K, Lysson M, Schumak B, et al. Leukocyte-Derived Interleukin-10 Aggravates Postoperative Ileus. Front Immunol. 2018;9:2599.
Schaefer N, Tahara K, Schmidt J, et al. Resident macrophages are involved in intestinal transplantation-associated inflammation and motoric dysfunction of the graft muscularis. Am J Transplant. 2007;7:1062–70.
Wehner S, Vilz TO, Stoffels B, et al. Immune mediators of postoperative ileus. Langenbeck’s Arch Surg. 2012;397:591–601.
Pohl J-M, Gutweiler S, Thiebes S, et al. Irf4-dependent CD103+CD11b+ dendritic cells and the intestinal microbiome regulate monocyte and macrophage activation and intestinal peristalsis in postoperative ileus. Gut. 2017;66:2110–2120.
Lin S-S, Zhang R-Q, Shen L, et al. Alterations in the gut barrier and involvement of Toll-like receptor 4 in murine postoperative ileus. Neurogastroenterol Motil. 2018;30:e13286.
Komanduri S, Gillevet PM, Sikaroodi M, et al. Dysbiosis in pouchitis: evidence of unique microfloral patterns in pouch inflammation. Clin Gastroenterol Hepatol. 2007;5:352–60.
Rogers MB, Brower-Sinning R, Firek B, et al. Acute Appendicitis in Children Is Associated With a Local Expansion of Fusobacteria. Clin Infect Dis. 2016;63:71–78.
Munyaka PM, Rabbi MF, Khafipour E, et al. Acute dextran sulfate sodium (DSS)-induced colitis promotes gut microbial dysbiosis in mice. J Basic Microbiol. 2016;56:986–98.
Johnson BM, Gaudreau M-C, Al-Gadban MM, et al. Impact of dietary deviation on disease progression and gut microbiome composition in lupus-prone SNF1 mice. Clin Exp Immunol. 2015;181:323–37.
Luo XM, Edwards MR, Mu Q, et al. Gut Microbiota in Human Systemic Lupus Erythematosus and a Mouse Model of Lupus. Appl Environ Microbiol.;84 . Epub ahead of print 2018. DOI: https://doi.org/10.1128/AEM.02288-17.
Ye C, Liu L, Ma X, et al. Obesity Aggravates Acute Pancreatitis via Damaging Intestinal Mucosal Barrier and Changing Microbiota Composition in Rats. Sci Rep. 2019;9:69.
Wallace JL, Syer S, Denou E, et al. Proton pump inhibitors exacerbate NSAID-induced small intestinal injury by inducing dysbiosis. Gastroenterology. 2011;141:1314–22, 1322.e1–5.
Sun T, Liu S, Zhou Y, et al. Evolutionary biologic changes of gut microbiota in an “adenoma-carcinoma sequence” mouse colorectal cancer model induced by 1, 2-Dimethylhydrazine. Oncotarget. 2017;8:444–457.
Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat Immunol. 2011;12:5–9.
Chang Y-C, Ching Y-H, Chiu C-C, et al. TLR2 and interleukin-10 are involved in Bacteroides fragilis-mediated prevention of DSS-induced colitis in gnotobiotic mice. PLoS One. 2017;12:e0180025.
Chiu C-C, Ching Y-H, Wang Y-C, et al. Monocolonization of germ-free mice with Bacteroides fragilis protects against dextran sulfate sodium-induced acute colitis. Biomed Res Int. 2014;2014:675786.
Akram A, Maley M, Gosbell I, et al. Utility of 16S rRNA PCR performed on clinical specimens in patient management. Int J Infect Dis. 2017;57:144–149.
Mendes-Soares H, Raveh-Sadka T, Azulay S, et al. Assessment of a Personalized Approach to Predicting Postprandial Glycemic Responses to Food Among Individuals Without Diabetes. JAMA Netw open. 2019;2:e188102.
Budding AE, Grasman ME, Eck A, et al. Rectal swabs for analysis of the intestinal microbiota. PLoS One. 2014;9:e101344.
Bassis CM, Moore NM, Lolans K, et al. Comparison of stool versus rectal swab samples and storage conditions on bacterial community profiles. BMC Microbiol. 2017;17:78.
Hillman ET, Lu H, Yao T, et al. Microbial Ecology along the Gastrointestinal Tract. Microbes Environ. 2017;32:300–313.
Lavelle A, Lennon G, O’Sullivan O, et al. Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut. 2015;64:1553–61.
Boersema GSA, Wu Z, Menon AG, et al. Systemic Inflammatory Cytokines Predict the Infectious Complications but Not Prolonged Postoperative Ileus after Colorectal Surgery. Mediators Inflamm. 2018;2018:7141342.
Olivas AD, Shogan BD, Valuckaite V, et al. Intestinal tissues induce an SNP mutation in Pseudomonas aeruginosa that enhances its virulence: possible role in anastomotic leak. PLoS One. 2012;7:e44326.
van Praagh JB, de Goffau MC, Bakker IS, et al. Mucus Microbiome of Anastomotic Tissue During Surgery Has Predictive Value for Colorectal Anastomotic Leakage. Ann Surg. . Epub ahead of print January 9, 2018. DOI: https://doi.org/10.1097/SLA.0000000000002651.
Beamish EL, Johnson J, Shaw EJ, et al. Loop ileostomy-mediated fecal stream diversion is associated with microbial dysbiosis. Gut Microbes. 2017;8:467–478.
Acknowledgments
We would like to thank Olga Zaborina Ph.D. for helping in preparing this manuscript and all of the consultants and operating room staff whom supported this manuscript.
Funding
Internal departmental funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This prospective cohort study of patients undergoing colon or rectal resection at the Mayo Clinic Rochester between 3/1/2017 and 7/31/2017 was approved by the Mayo Clinic Institutional Review Board.
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
ESM 1
(DOCX 1358 kb)
Rights and permissions
About this article
Cite this article
Shogan, B.D., Chen, J., Duchalais, E. et al. Alterations of the Rectal Microbiome Are Associated with the Development of Postoperative Ileus in Patients Undergoing Colorectal Surgery. J Gastrointest Surg 24, 1663–1672 (2020). https://doi.org/10.1007/s11605-020-04593-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11605-020-04593-8