Abstract
Purpose
The size information of detected polyps is an essential factor for diagnosis in colon cancer screening. For example, adenomas and sessile serrated polyps that are \(\ge 10\) mm are considered advanced, and shorter surveillance intervals are recommended for smaller polyps. However, sometimes the subjective estimations of endoscopists are incorrect and overestimate the sizes. To circumvent these difficulties, we developed a method for automatic binary polyp-size classification between two polyp sizes: from 1 to 9 mm and \(\ge 10\) mm.
Method
We introduce a binary polyp-size classification method that estimates a polyp’s three-dimensional spatial information. This estimation is comprised of polyp localisation and depth estimation. The combination of location and depth information expresses a polyp’s three-dimensional shape. In experiments, we quantitatively and qualitatively evaluate the proposed method using 787 polyps of both protruded and flat types.
Results
The proposed method’s best classification accuracy outperformed the fine-tuned state-of-the-art image classification methods. Post-processing of sequential voting increased the classification accuracy and achieved classification accuracy of 0.81 and 0.88 for polyps ranging from 1 to 9 mm and others that are \(\ge 10\) mm. Qualitative analysis revealed the importance of polyp localisation even in polyp-size classification.
Conclusions
We developed a binary polyp-size classification method by utilising the estimated three-dimensional shape of a polyp. Experiments demonstrated accurate classification for both protruded- and flat-type polyps, even though the flat type have ambiguous boundary between a polyp and colon wall.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The size information of detected polyps is an essential factor for diagnosis in colon cancer screening [1]. The American [2] and European [3] guidelines for colonoscopy surveillance define what treatments should be chosen after a physician finds polyps with respect to their size estimations.
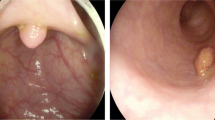
Difficulty of polyp-size estimation from a colonoscopic image: a Loss of three-dimensional information by projection to an image plane: Appearance size highly depends on distance from image plane \(\pi \) to an object. In (a), \({\varvec{c}}\) denotes optical centre. b Examples of appearance sizes of polyps: Polyps of different sizes (2, 6, and 10 mm) can appear to be almost identical. c Gross morphology of polyps
In both definitions, the size determination of whether a polyp size is equalling or exceeding 10 mm, or not is critical. Adenomas and sessile serrated polyps of 10 mm are considered advanced, and shorter surveillance intervals are recommended for smaller polyps [1, 2]. The determination of polyp size is based on subjective estimation by endoscopists rather than objective measurements by pathologists. However, subjective estimation tends to be overestimation, where small polyps are mistakenly considered 10 mm or larger [4].
Subjective estimations are essentially an ill-posed problem, as depicted in Figs. 1a and b, since the two-dimensional endoscopic view fails to completely capture all of the three-dimensional information. Furthermore, several kinds of gross morphology shown in Fig. 1c complicate estimations. To circumvent this difficulty, subjective observations with a device have been proposed [5,6,7]. Rex et al. measured polyp size by comparing the size of open forceps [5]. Hyun et al. [6] and Kaz et al. [7] developed graduated measuring devices, which have scale marks of 5-mm intervals to measure a polyp in vivo. Hyun et al. achieved classification accuracy of 0.93, 0.16 and 0.58 with their graduated device for polyps ranging 0–5 mm, 6–9 mm and \(\ge \)10 mm, respectively [7]. Even if endoscopists use a device, there is a terminal digit preference bias for aesthetically pleasing numbers, where endoscopists are apt to determine in such increments as 5, 10, 15, 20, \(\dots \) mm [8]. As a device-free method, Itoh et al. proposed image-based binary polyp-size classification using convolutional neural networks (CNNs), and achieved classification accuracy of 0.79 for 50 protruded polyps [9].
In this paper, instead of polyp-size estimation, we tackle binary polyp-size classification between polyps of 1-9 mm and those of \(\ge 10\) mm. This approach bases on the following two reasons. The first reason is that the determination of whether a polyp size is equalling and exceeding 10 mm, or not is critical for diagnosis in colon cancer screening. The second one is that precise polyp-size estimation under the ill-posed condition is still too challenging at this point. Therefore, we propose a new polyp-size classification method as a natural extension of our previous method [9]. In this extended method, we introduce new techniques: model-based precise depth estimation and polyp localisation. We used the unsupervised estimation of relative depth in previous work, which has no physical length. Instead of relative depth, we adopted a depth with a physical length obtained from a model-based RGBD-GAN that achieved accurate depth estimation with an average error less than 1 mm [10]. We combine the depth and polyp location information to express the three-dimensional shape of a polyp with physical length for the classification.
Methods
Single-shot classification
This paper assumes that a three-channel colonoscopic image \({\mathcal {X}} \in {\mathbb {R}}^{H \times W \times 3}\) with height H and width W expresses only one polyp, and refer to \({\mathcal {X}}\) as a polyp image.
The straightforward estimation of polyp size s from an image appearance of \(\mathcal {X}\) is expressed with a parameter vector \({\varvec{\theta }}\) as a function \(f({\mathcal {X}}; {\varvec{\theta }})=s\). As the regression problem for pairs of polyp image \({\mathcal {X}}_i\) and ground-truth size \(s^{*}_i\) of each polyp in \({\mathcal {X}}_i\) for index \( i=1,2,\dots , N \), we obtain the optimised parameter vector \(\hat{{\varvec{\theta }}}\) by
where \(\Vert \cdot \Vert _2 \) is the Euclidean norm. However, an obtained function \(f({\mathcal {X}}; \hat{{\varvec{\theta }}})\) does not work for an unseen image \({\mathcal {X}} \notin \{ {\mathcal {X}}_i \}_{i=1}^N\) due to the ill-posed condition as shown in Figs. 1a and b.
To circumvent the ill-posed condition, depth information is essential. Previous work [9] estimated a depth image \({\varvec{D}} \in {\mathbb {R}}^{H \times W}\) from \({\mathcal {X}}\) by an unsupervised method. By expressing all the parameters for depth estimation as \({\varvec{\theta }}_{{D}}\), we have a depth-estimation function
Furthermore, we relaxed polyp-size estimation to the binary polyp-size classification. In this classification, we use category labels 0 and 1 to express the 1–9 mm and \(\ge 10\) mm polyps, respectively. By using binary cross-entropy \({\mathscr {H}}\), optimised depth estimation \(g({\mathcal {X}}_i ; \hat{{\varvec{\theta }}}_D)={\varvec{D}}_i \), and ground truth \(t_i^{*}\), we define a parameter optimisation as
By thresholding a probability \(f({\mathcal {X}}, {\varvec{D}};\hat{{\varvec{\theta }}})\) with \(\xi \), we obtain a category label for input . To solve Eq. (4), we used RGB-D CNN [9]. However, this CNN must learn how to detect polyp locations from training images without annotations of polyp locations.
In our method, we estimate a polyp-location map \({\varvec{W}} \in \{ 0, 1 \}^{H \times W}\), whose elements of 0 and 1 express polyp’s in-existence and existence, respectively. By expressing all the parameters for a polyp detection as \({\varvec{\theta }}_{{L}}\), we have a polyp detector \(h({\mathcal {X}}; {\varvec{\theta }}_{{L}})\) that outputs \({\varvec{W}}\) and a likelihood \(p \in [0, 1]\) for a polyp existence. Using a detector with an optmised parameter vector \(\hat{{\varvec{\theta }}}_L\), we define polyp localisation by
where \(\tau \) is a criterion. We then integrate \({\varvec{W}}\) to \({\varvec{D}}\), since the combination of the depth and location information expresses a polyp’s three-dimensional shape. By expressing a localised-depth image \({\varvec{D}}_i \odot {\varvec{W}}_i\) with the Hadamard product \(\odot \) for \({\mathcal {X}}_i\), we search the solution \(\hat{{\varvec{\theta }}}\) of
We then defined binary classification with a criterion \(\xi \) as
Figure 2a illustrates the proposed method. For the practical construction of \(g({\mathcal {X}}; \hat{{\varvec{\theta }}}_D), h({\mathcal {X}}; \hat{{\varvec{\theta }}}_L), f({\varvec{D}} \odot {\varvec{W}}; \hat{{\varvec{\theta }}})\) , we optimise RGBD-GAN [10], YOLOv3 [11], and a three-layer CNN, respectively. We explain how to optimise them in Sect. 4: Optimisations.
Sequential-voting classification
We propose a voting-based classification shown in Fig. 2b for a short colonoscopic video. Single-shot classification often suffers from illumination changes, motions, colour blurs, and so on. To achieve a robust polyp-size classification, we use sequential polyp images \({\mathcal {X}}_i, i=1,2, \dots , M\) extracted from a short colonoscopic video. Using \(\hat{{\varvec{\theta }}}\) obtained from Eq. (7), we obtain a set \(\{ p_i \}_{i=1}^M\) of probabilities \(p_i = f( {\varvec{D}}_i \odot {\varvec{W}}_i; \hat{{\varvec{\theta }}})\), \(i=1,2, \dots , M\) for each \({\mathcal {X}}_i\). Let \({\varvec{v}} \in \{0, 1\}^{M}\) be a binary vector defined by
With \({\varvec{v}}\) and \(L_0\) norm \(\Vert \cdot \Vert _0\), we classify an input video as
Dataset construction
Dataset for depth estimation
To optimise RGBD-GAN, we select the real-and-virtual colonoscopy dataset [10]. This dataset offers 9189 colonoscopic images and 8085 RGB-D virtual colonoscopic images. These colonoscopic images are extracted from colonoscopic videos of 30 patients at five frames per second. In the video collection, we used a high-definition colonoscope (CF-HQ290ZI; Olympus). The virtual colonoscopic images are generated from CT colonography data of seven patients with CT colonography software [12]. For the generation of the virtual depth image, pixel values are set to a range of [0, 255] for distances in the range of [0, 10] cm.
Dataset for polyp localisation
To optimise YOLOv3, we selected 67,824 polyp images and 1028 non-polyp images collected at five hospitals [13]. Each polyp image includes one polyp of protruded or flat type, which has a bounding box annotation of its location. Each non-polyp image includes one of eight typical hard negative elements, such as bubbles, halation, droplets, ..., with a bounding box annotation.
To evaluate localisation accuracy, we used publicly available SUN database [14]. This database offers 49,136 images of 100 different polyps, which are protruded- and flat-type polyps, and 109,554 images of no-polyp scenes with their location annotation.
Dataset for polyp-size classification
We constructed a polyp-size dataset. First, we collected colonoscopic videos during typical colonoscopies at two hospitals using a colonoscope (CF-HQ290ZI; Olympus) and a video recorder (IMH-10; Olympus) with Institutional Review Board approval. Expert endoscopists with experience of over 5000 colonoscopy cases measured polyps by comparing the tip of an inserted sheath and the polyp diameters. We used sheaths of polypectomy snares (snare master; Olympus, captivator; Boston Scientific), whose tips have diameters of 2.6 and 2.4 mm. To reduce personal bias in their measurements, two or three expert endoscopists checked a polyp’s size and reached a consensus. Endoscopists annotated the polyp-existing frames with labels of gross morphology (Ip, Isp, Is, and IIa) and size (1, 2, 3, \(\dots \), 14, 15 mm) with an annotation tool [15].
Next, we divided these videos into training, validation, and test data without duplication of polyp cases. We extracted still images from these data at 30 frames per second (fps). We removed such inappropriate images as motion blurred, colour blurred, bubbles, out-of-focus, and successive images with a similar appearance from only the training data. The test dataset has 84 polyps, each of which has an average of 185 successive images.
Finally, we generated depth and localised-depth images for the all images by using RGBD-GAN and YOLOv3. Table 1 summarises the number of images in the constructed dataset. Figure 3 shows examples of the training data. Figure 4 shows the image-number-based distributions of size and gross morphology in our dataset.
Architecture for depth estimation and polyp localisation: a RGBD-GAN [10] for depth estimation; b YOLOv3 [11] for polyp detection; In (a) and (b), numbers above and below square boxes express the spatial resolutions and channel sizes, respectively. As the exception, a number below a residual unit in (b) expresses the number of residual units
Optimisations
\({\varvec{\theta }}_{{\mathcal {D}}}\) for depth estimation
For the domains \({\mathcal {R}}_{\mathrm {RGB}}\) and \({\mathcal {V}}_{\mathrm {RGBD}}\) of RGB colonoscopic and RGB-D virtual colonoscopic images, respectively, we define \(F: {\mathcal {R}}_{\mathrm {RGB}} \rightarrow {\mathcal {V}}_{\mathrm {RGBD}}\) and \(G: {\mathcal {V}}_{\mathrm {RGBD}} \rightarrow {\mathcal {R}}_{\mathrm {RGB}}\). For the generation of RGB channel of these virtual images, we assume the Lambertian-reflection model. For given images \({\mathcal {X}}_i \in {\mathcal {R}}_{\mathrm {RGB}}\) and \({\mathcal {Y}}_j \in {\mathcal {V}}_{\mathrm {RGBD}}\) for \(i=1, 2, \dots , n_{{\mathcal {R}}}\) and \(j=1, 2, \dots , n_{{\mathcal {V}}}\), we introduce discriminators \(D_{{\mathcal {V}}}\) and \(D_{{\mathcal {R}}}\), which distinguish \({\mathcal {Y}}_j\) from \(\{ F({\mathcal {X}}_i) \}_{i=1}^{n_{{\mathcal {R}}}}\), and \({\mathcal {X}}_i\) from \(\{ G({\mathcal {Y}}_j) \}_{i=1}^{n_{{\mathcal {V}}}}\), respectively. We then obtain an object function
where \(\lambda \) controls the importance of adversarial losses \({\mathscr {L}}_{\mathrm {GAN}}\) and a cycle-consistency loss \({\mathscr {L}}_{\mathrm {cyc}}\) [10]. By expressing the all parameters of F and G as \({\varvec{\theta }}_D\) and \({\varvec{\theta }}_R\), respectively, we search \(\hat{{\varvec{\theta }}}_D\) by solving the min–max problem
With the obtained \(\hat{{\varvec{\theta }}}_D\), we estimate depth image \({\varvec{D}}\) as the fourth channel of \(\hat{{\mathcal {Y}}} = F({\mathcal {X}})\) [10].
Figure 5a shows the architecture of F for depth estimation. By using training data described in Sect. 3.1: Dataset of depth estimation, we optimised RGBD-GAN with Adam of base learning \(lr=0.002\), minibatch size of 64, leaky ReLU coefficient \(\alpha =0.2\), \(\lambda =10\), and data argumentation of random flips for 300 epochs.
\({\varvec{\theta }}_{{L}}\) for polyp localisation
We briefly summarise the optimisation of YOLOv3 [11], since YOLO has a few versions and partial updates presented by different papers.
As shown in Fig. 5b, YOLOv3 comprises two parts: backbone and head. The backbone part is Darknet-53 which extracts image features from input. The head part outputs predictions for three scales. In the i-th scale, YOLOv3 divides an input image \({\mathcal {X}}\) into \(S_i \times S_i\) regions as cells and predicts B candidates of an object’s location for each cell.
YOLOv3 uses bounding box priors. The k-th bounding box prior of the i-th scale has predefined width \(A_{w_{ik}}\) and height \(A_{w_{ik}}\). Furthermore, YOLOv3 adopts gird-cell coordinate \((c_{x_{ij}}, c_{y_{ij}})\) in the divided \({\mathcal {X}}\). The origin in grid-cell coordinate is the left-top corner of \({\mathcal {X}}\), and \((c_{x_{ij}}, c_{y_{ij}})\) represents grid-corner indices for the j-th cell in the i-th scale. YOLOv3 predicts \(t_{x_{ijk}}, t_{y_{ijk}}, t_{w_{ijk}}, t_{h_{ijk}}\), and outputs a centre \( (b_{x_{ijk}}, b_{y_{ijk}})\), width \(b_{w_{ijk}}\) and height \( b_{h_{ijk}}\) of a location for an object by
where \(\sigma (\cdot )\) is a sigmoid function. Furthermore, YOLOv3 predicts an objectness score \(\sigma (t_{o_{ijk}})\) and category scores \(\sigma (t_{c_{ijk1}})\), \(\sigma (t_{c_{ijk2}})\), \(\dots \sigma (t_{c_{ijkC}})\) for the k-th bounding box of the j-th cell in the i-th scale. Therefore, for the i-th scale, YOLOv3 outputs a tensor \({\mathcal {T}}_{i} \in {\mathbb {R}}^{S_i \times S_i \times (B(4+1+C)))}\). As the outputs of \(h({\mathcal {X}}; {\varvec{\theta }}_L)\), we obtain \({\varvec{W}}\) by Eqs. (16)-(19) and \(p=\sigma (t_{c_{ijk1}})\).
YOLOv3 ignores low-confident predictions by thresholding object scores with \(\eta \). For each object, YOLOv3 finds the best localisation by computing IoU (Intersection over Union) \(S \cap S^{*} / S \cup S^{*}\) between a predicted region S and ground truth \(S^{*}\). We set weights \(\mathbbm {1}^{\mathrm {obj}}_{ijk}=1\) and \(\mathbbm {1}^{\mathrm {obj}}_{ijk} =0\) for the best localisation and the others, respectively, for each object. Furthermore, we set \(\mathbbm {1}^{\mathrm {non}}_{ijk}=1 - \mathbbm {1}^{\mathrm {obj}}_{ijk}\). By using ground truth \(t_{x_{ijk}}^{*}\), \(t_{y_{ijk}}^{*}\), \(t_{w_{ijk}}^{*}\), \(t_{h_{ijk}}^{*}\), \(t_{c_{ijk1}}^{*}\), \(t_{c_{ijk2}}^{*}\), \(\dots t_{c_{ijkC}}^{*}\), for \({\mathcal {X}}\), we have a loss functional by
By using a training set \(\{ {\mathcal {X}}_m \}_{m=1}^N\), we search \(\hat{{\varvec{\theta }}}_{L}\) by solving
To optimise \({\varvec{\theta }}_{{L}}\), we used 68,852 images described in Sect. 3.2: Dataset for polyp localisation. We set \(C=9\) and \(\eta =0.6\). As shown in Fig. 6b, we used \(S_1=6, S_2=12, S_3=24\) and \(B=3\). We used the same bounding-box priors as the original [11]. Setting \(\alpha =0.1\) for all leaky ReLU and \(\lambda _{\mathrm {box}}=1.0\) for Eq. (17), we applied fine-tuning to the weights of the pre-trained backbone [11] with the stochastic gradient descent and the same data augmentations of Ref. [13]. With a minibatch size of 64, we used a burn-in of 1000 iterations and obtained a learning rate of \(lr = 1.0 \times 10^{-4}\). We then started a model training of 70,000 iterations where lr was multiplied with 0.10 at 56,000 and 63,000 iterations.
\({\varvec{\theta }}\) for polyp-size classification
To solve Eq. (7), we optimised the three-convolution CNN shown in Fig. 2a with the training data described in Sect. 3.3: Dataset for polyp-size classification. Setting leaky ReLU coefficient \(\alpha =0.3\) and dropout ratio 0.2, we used He’s weight initialisation, Adam of a learning rate \(lr=1.0 \times 10^{-5}\), and weighted binary cross-entropy loss for 150 epochs. As the data argumentation, we used random shift, rotation, horizontal and vertical flips, and a small perturbation of the pixel values. In this optimisation, we multiplied 0.1 to lr at 15 epochs. Based on the evaluations with the validation data, we selected the early stopping model at 25 epochs.
Experimental evaluations
Evaluation of localisation
First, we evaluated the accuracy of polyp detection and localisation, by using the SUN database described in Sect. 3.2: Dataset for polyp localisation. We detected polyps in these images using YOLOv3. Although it outputs several predictions for input, we selected only one output of the highest likelihood for input and applied thresholding in Eq. (5). Furthermore, we added IoU thresholding in this evaluation. If IoU is less than criterion \(\rho \in [0, 1]\), we reject the detection results. We defined sensitivity and specificity as the ratios of the correctly detected polyp images and the correctly undetected non-polyp images, respectively. Figure 6a shows the ROC curves for the criteria of likelihood \(\tau = 0.025, 0.050, \dots , 0.975\) and IoU \(\rho =0.0, 0.1, 0.2, \dots , 0.5\), where area under curve is 0.98 for \(\rho =0.0\). Figure 6b shows the mean IoU.
Evaluation of single-shot classification
Next, we evaluated the accuracy of the single-shot classification method by comparing the proposed and CNN architectures. As comparison methods, we adopted NASNet-A [16], InceptionResNet-v2 [17], and Xception [18], which achieved the best-, second-, and third-ranked top-1 image classification accuracies in the ImageNet dataset. As the depth-based method, we added to this comparison our previous work: BseNet [9]. In this evaluation, we used images described in Sect. 3.3: Dataset for polyp-size classification.
For NASNet-A, InceptionResNet-v2, and Xception, we applied fine-tuning to their pre-trained models of ImageNet using the colonoscopic images of our training data. Just for InceptionResNet-v2, we applied fine-tuning using the depth images in our training data. In such fine-tuning, we used only the feature extraction part of each architecture and added a classification part, which consists of global average pooling and a fully connected layer. For the optimisation of BseNet, we used the colonoscopic and depth images in our training data.
Distributions of probabilities in correctly and incorrectly classified cases in sequential-voting classification: a and b are distributions for correct and incorrect classified cases in Fig. 10
We classified the test data by the six trained models. We defined sensitivity and specificity of the ratios of the correctly classified images of polyps over 10 mm and those in the 1 to 9 mm range, respectively. Figure 7a summarises the ROC curves with the criteria of likelihood \(\xi = 0.05, 0.10, \dots , 0.95\). Figures 8a and b show the classification accuracy in each size for the InceptionResNet-v2 and the proposed method. In Figs. 8a and b, we set \(\xi \) to 0.4 and 0.7, respectively. Figure 9 illustrates examples of the correctly classified inputs by the proposed method.
Evaluation of sequential-voting classification
Finally, we evaluated our sequential-voting classification with the test dataset described in Sect. 3.3: Dataset for polyp-size classification. Figure 7b summarises the classification results of the sequential-voting and single-shot classifications. We defined accuracy as the ratio of correctly classified scenes. Figure 8c shows the accuracy in each size of the sequential-voting classification. Figure 10 shows a typical comparison between correctly and incorrectly classified scenes. Figure 11a and b show the distributions of the likelihoods for the correctly and incorrectly classified scenes in Figs. 10a and b.
Discussion
In Fig. 7a, the proposed method achieved the best result among the six methods, demonstrating the importance of combining a polyp’s location and depth information. BseNet with RGB-D images achieved almost the same performance as Xception, even though this method used RGBD-GAN for depth estimation. These results emphasise the importance of location information.
InceptionResNet-v2 with colonoscopic images achieved the second-best classification accuracy, even though it achieved meaningless random classification with depth images in Fig. 7a. These results imply that deep learning might not identify polyps just using depth images, since there is no texture information of a polyp.
Figure 8a shows that InceptionResNet-v2 with colonoscopic images only correctly classified polyps from 2 to 9 mm. On the other hand, our proposed method correctly classified the polyps of every size except 9 and 11 mm in Fig. 8b. These two sizes are difficult queries, since they exist around the border of the binary categories. Figure 9 shows the correct classification for both small- and large-appearance polyps. These results suggest that the proposed method captured the three-dimensional information of polyps.
In Figs. 7b and 8c, we improved the classification accuracy by sequential voting. The classifications of the polyps from 1 to 9 mm and those of \(\ge \) 10 mm achieved accuracies of 0.81 and 0.88. Figures 11a and b show the probability distributions for the correctly and incorrectly classified polyps in Fig. 10. In the former case, the proposed method captured the correct location of a polyp, whereas miss-localisations exist in many frames in the latter case. These miss-localisations can happen in the images when a flat polyp exists far from a colonoscope.
From Fig. 4b, we confirmed a less-biased size distribution over our dataset than subject estimation results [4, 8]. In subjective estimation, including size determination with measurement devices, estimated sizes tend to concentrate on only specific sizes such as 5, 10, 15, 20 mm [8]. In Ref. [4], among 99 polyps determined as 10 mm in subjective estimation, 71 polyps were actually <10 mm. On the other hand, the polyp-size distribution of our dataset is a smooth continuum in Fig. 4b. Therefore, the distribution of our dataset implies that our polyp-size measurement achieved the reduction in the terminal digit bias. Furthermore, the distribution of our dataset has a similar size distribution as the one of pathological measurement [4]. These observations imply that our measurements reduced the overestimation, too. The expert endoscopists in our group reached a consensus that a possible measurement error might be less than 1 mm.
Even though our localisation is imperfect for the SUN database as shown in Fig. 6, we can handle possible localisation errors. Since our purpose is polyp-size classification, we can get close enough to a polyp to correctly capture its location. The possible errors both in the ground truth data and depth estimation results [10] might cause lower classification accuracy for polyp sizes from 8 to 12 mm than the other sizes in Figs. 8b and c. However, our method achieved 88% classification accuracy for large polyps, whereas only 58% of 34 endoscopists correctly recognised large polyps in subjective-estimation tests with artificial polyps and the graduated device [7]. Compared with the subjective estimation [7], our method has an advantage in the correct recognition of large polyps.
Furthermore, our binary polyp-size classification has an advantage compared with the polyp-size regression. As shown in Fig. 4, the distribution of polyp sizes is imbalanced. If we adopt the regression, a trained model might overfit the dominant sizes of the distribution such as 2, 3, \(\dots \), 7 mm polyps. However, by splitting sizes into 1–9 mm and \(\ge 10\) mm as binary polyp-size categories, we can prevent the overfitting to the dominant sizes.
Conclusions
We developed a new method for binary polyp-size classification by integrating the deep-learning-based estimation of a polyp’s spatial information, since spatial information is essential for classifying object sizes in a three-dimensional space. In this method, precise polyp localisation and depth estimation with physical length contributed to estimating the three-dimensional shape of a polyp. Furthermore, the binary classification is not suffered from the imbalanced distribution of polyp sizes. Experimental results demonstrated accurate classification for both protruded- and flat-type polyps. Our method achieved 88% classification accuracy for large polyps, whereas only 58% of 34 endoscopists correctly recognised large polyps in the subjective estimation [7]. Moreover, the implementation of our method is feasible, since the pipeline in our method bases on deep-learning frameworks. Therefore, in the future, we can improve our method by integrating more precise depth-estimation and localisation methods into the pipeline.
References
Hassan C, Repici A, Rex D (2016) Addressing bias in polyp size measurement. Endoscopy 48(10):881–883
Lieberman DA, Rex DK, Winawer SJ, Giardiello FM, Johnson DA, Levin TR (2012) Guidelines for colonoscopy surveillance after screening and polypectomy: a consensus update by the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology 143(3):844–857
Hassan C, Quintero E, Dumonceau J-M, Regula J, Brandão C, Chaussade S, Dekker E, Dinis-Ribeiro M, Ferlitsch M, Gimeno-García A, Hazewinkel Y, Jover R, Kalager M, Loberg L, Pox C, Rembacken B, Lieberman D (2013) Post-polypectomy colonoscopy surveillance: European Society of Gastrointestinal Endoscopy (ESGE) Guideline. Endoscopy 45(10):842–864
Anderson B, Smyrk T, Anderson K, Mahoney D, Dovens M, Sweetser S, Kisiel J, Ahlquist D (2015) Endoscopic overestimation of colorectal polyp size. Gastrointestinal Endoscopy 83(1):201–208
Rex DK, Rabinovitz R (2014) Variable interpretation of polyp size by using open forceps by experienced colonoscopists. Gastrointestinal Endoscopy 79(3):402–407
Hyun YS, Han DS, Bae JH, Park HS, Eun CS (2011) Graduated injection needles and snares for polypectomy are useful for measuring colorectal polyp size. Digestive and Liver Disease 43(5):391–394
Kaz AM, Anwar A, O’Neill DR, Dominitz JA (2016) Use of a novel polyp “ruler snare’’ improves estimation of colon polyp size. Gastrointest Endoscopy 83(4):812–816
Plumb A, Nickerson C, Wooldrage K, Bassett P, Taylor S, Altman D, Atkin W, Halligan S (2016) Terminal digit preference biases polyp size measurements at endoscopy, computed tomographic colonography, and histopathology. Endoscopy 48:899–908
Itoh H, Roth HR, Lu L, Oda M, Misawa M, Mori Y, Kudo S-E, Mori K (2018) Towards Automated Colonoscopy Diagnosis: Binary Polyp Size Estimation via Unsupervised Depth Learning. Proc. Medical Image Computing and Computer Assisted Intervention LNCS 11071:611–619
Itoh Oda M, Mori Y, Misawa M, Kudo S-E, Imai K, Ito S, Hotta K, Takabatake H, Mori M, Natori H, Mori K (2021) Unsupervised Colonoscopic Depth Estimation with a Lambertian-Reflection Keeping Auxiliary Task. International Journal of Computer Assisted Radiology and Surgery 16:989–1001
Redmon J, Farhadi A (2018) YOLOv3: an incremental improvement. CoRR arXiv:1804.02767https://pjreddie.com/darknet/yolo/
Mori K, Suenaga Y, Toriwaki J (2003) Fast Software-based Volume Rendering Using Multimedia Instructions on PC Platforms and Its Application to Virtual Endoscopy. Proc SPIE Medical Imaging 5031:111–122
Misawa M, Kudo S-E, Mori Y, Hotta K, Ohtsuka K, Matsuda T, Saito S, Kudo T, BaBa T, Ishida F, Itoh H, Oda M, Mori K (2021) Development of a computer-aided detection system for colonoscopy and a publicly accessible large colonoscopy video database (with video). Gastrointestinal Endoscopy 93(4):960–967
Itoh H, Misawa M, Mori Y, Oda M, Shin-Ei Kudo, Mori K (2020) SUN Colonoscopy Video Database. http://amed8k.sundatabase.org/
Lausberg H, Sloetjes H (2009) Coding gestural behavior with the NEUROGES-ELAN system. Behav Res Methods 41:841–849, https://tla.mpi.nl/tools/tla-tools/elan/
Zoph B, Vasudevan V, Shlens J, Le QV (2018) Learning transferable architectures for scalable image recognition. In: Proceedings of IEEE international conference on computer vision, pp 8697–8710
Szegedy C, Ioffe S, Vanhoucke V, Alemi A (2017) Inception-v4, Inception-ResNet and the impact of residual connections on learning. In: Proceedings of thirty-first AAAI conference on artificial intelligence, pp 4278–4284
Chollet F (2017) Xception: deep learning with depthwise separable convolutions. In: Proceedings of IEEE international conference on computer vision, pp 1800–1807
Acknowledgements
This study was funded by Grants from AMED 445 (19hs0110006h0003), JSPS MEXT KAKENHI (26108006, 17H00867, 446 17K20099), JST CREST (JPMJCR20D5), and the JSPS Bilateral Joint Research Project. 447.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Kudo SE, Misawa M, and Imai K received lecture fees from Olympus. Mori Y and Hotta K received consultant and lecture fees from Olympus. Mori K is supported by Cybernet Systems and Olympus (research grant) in this work and by NTT outside of the submitted work. The other authors have no conflicts of interest.
Ethical approval
All the procedures performed in studies involving human participants were in accordance with the ethical committee of Nagoya University (No. 357), the Shizuoka Cancer Center (No. T30-45-30-1-5), and the 1964 Helsinki declaration and subsequent amendments or comparable ethical standards. Informed consent was obtained by an opt-out procedure from all individual participants in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Itoh, H., Oda, M., Jiang, K. et al. Binary polyp-size classification based on deep-learned spatial information. Int J CARS 16, 1817–1828 (2021). https://doi.org/10.1007/s11548-021-02477-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-021-02477-z