Abstract
Purpose
Transcatheter aortic valve replacement (TAVR) is the standard of care in a large population of patients with severe symptomatic aortic valve stenosis. The sizing of TAVR devices is done from ECG-gated CT angiographic image volumes. The most crucial step of the analysis is the determination of the aortic valve annular plane. In this paper, we present a fully tridimensional recursive multiresolution convolutional neural network (CNN) to infer the location and orientation of the aortic valve annular plane.
Methods
We manually labeled 1007 ECG-gated CT volumes from 94 patients with severe degenerative aortic valve stenosis. The algorithm was implemented and trained using the TensorFlow framework (Google LLC, USA). We performed K-fold cross-validation with K = 9 groups such that CT volumes from a given patient are assigned to only one group.
Results
We achieved an average out-of-plane localization error of (0.7 ± 0.6) mm for the training dataset and of (0.9 ± 0.8) mm for the evaluation dataset, which is on par with other published methods and clinically insignificant. The angular orientation error was (3.9 ± 2.3)° for the training dataset and (6.4 ± 4.0)° for the evaluation dataset. For the evaluation dataset, 84.6% of evaluation image volumes had a better than 10° angular error, which is similar to expert-level accuracy. When measured in the inferred annular plane, the relative measurement error was (4.73 ± 5.32)% for the annular area and (2.46 ± 2.94)% for the annular perimeter.
Conclusions
The proposed algorithm is the first application of CNN to aortic valve planimetry and achieves an accuracy on par with proposed automated methods for localization and approaches an expert-level accuracy for orientation. The method relies on no heuristic specific to the aortic valve and may be generalizable to other anatomical features.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
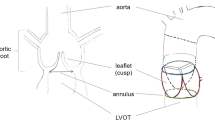
Transcatheter aortic valve replacement (TAVR) has become therapy of choice for a large proportion of patients with severe symptomatic aortic valve stenosis [1]. Multiple sizes of devices are available to accommodate different anatomies. The standard of care to perform the sizing of TAVR devices is based on CT aortic annulus planimetry [2,3,4]. The methodology requires the use of a multiplanar reconstruction of the aortic root to measure the area and perimeter of the aortic valve annulus. The aortic valve annulus is defined as the virtual endocardial ring of tissue measured in a plane defined by the nadir of the three cusps of the valve [5]. Given that the aortic root plane lies at an oblique angle, the operator typically uses a software package capable of 3D multiplanar reconstruction to sequentially rotate and translate reference planes to achieve an appropriate view through the valve [2, 6]. The process is time-consuming and requires significant training to achieve a reproducible annular plane. Given that the field is rapidly expanding, institutional-level expertise in CT analysis may be lacking. Automated image analysis may deliver a much-needed service for an efficient workflow and for quality assurance. Several manufacturers have developed software packages to streamline the annotation of CT dataset for TAVR planning.
Convolutional neural networks (CNN) have recently garnered much interest in the field of computer vision [7,8,9]. A significant research effort has been directed to their application in medical imaging [10,11,12]. They have been used successfully as both classification and regression models. Most computer vision algorithms suggested in the literature are aimed at 2D images. In the case of medical images, however, the information contained within 3D datasets is important when analyzing anatomical features [13]. Indeed, a feature may be readily identified only within its 3D environment. This is the case for the aortic valve annulus.
Herein, we propose an automated fully tridimensional recursive multiresolution CNN regression model to determine the location and orientation of the aortic valve plane in a volumetric cardiac CT dataset.
Related work
The automated landmarking and analysis of the aortic valve and its surrounding structures has been the topic of multiple studies. A statistical analysis of 2D region-growing algorithm applied to coronary artery segmentation on CT images was described in [14]. Model-based method for aortic root evaluation provided a landmark position error of 0.4 mm to 2.28 mm [14,15,16,17]. These methods rely on the availability of a 3D geometric model of the heart structure, which is highly nontrivial to generate. Similar methods have also been applied to C-arm CT images to provide optimal implantation fluoroscopic angulations [18]. Classic image segmentation techniques such as 3D normalized cuts have been applied to automatically extract the aortic root annulus [19, 20].
Machine learning has been applied to mitral valve analysis on echocardiographic images to automatically measure the mitral valve anteroposterior and intercommissural diameters in a small number of patients [21]. Machine learning has also been applied to modeling of the aortic valve with a mesh mean discrepancy of 1.57 mm in a small number of patients [22]. Extraction of aortic valve landmarks using colonial walk, a computationally efficient regression tree-based machine learning algorithm was recently published [23]. The number of CT image volumes used in that study was limited to 71. A recent article addressed landmarking of several aortic root features using neural networks trained on 198 image volumes and achieved an accuracy level of approximately 2.0 mm for several structures [24]. Multiscale deep learning approaches have been successfully applied to 3D landmarking of CT datasets [25].
Classical, non-machine learning methods/techniques rely generally on extensive a priori knowledge of the aortic valve geometry. In contrast, machine learning approaches, including the work presented here, rely on large amounts of data and virtually no a priori anatomical knowledge. The use of no heuristics specific to the aortic valve allows them to be readily adaptable to different anatomical structures if enough labeled data are available. Furthermore, the availability of high-performance open-source neural network libraries reduces the implementation complexity of many machine learning methods.
Methods
The aortic valve planimetry problem
The first and most crucial step in the analysis of a cardiac CT scan for the purpose of TAVR prosthesis sizing is to obtain an oblique multiplanar reconstruction at the level of the aortic valve annulus. In practice, the operator translates the volume in a visualization software until the valve in centered; they then successively rotate the multiplanar plane until a view that displays the nadirs of the three coronary cusps is obtained. This process takes on average 1–5 min depending on the expertise of the operator. Figure 1 demonstrates three multiplanar reconstructions parallel to the aortic valve annular plane at different levels. The figure pane (d) is at the level of the aortic valve annulus, which has been traced in yellow. The accuracy of the plane selection is important for the measurement of the aortic valve annulus area, perimeter and diameter, which are crucial for TAVR device sizing. Excessive oversizing may result in annular rupture and coronary ostial obstruction, while undersizing may result in device embolization and paravalvular leakage. These complications have been linked to increased mortality [26]. Furthermore, planimetry of the aortic valve annulus provides optimal fluoroscopic implantation views used during the procedure [27].
The aortic valve annular plane is fully described by a plane unit normal vector \( \left\{ {\varvec{n} \in R^{3} : \left\| n \right\| = 1} \right\} \) and the position of a point on the plane \( \left\{ {\varvec{x}_{0} \in R^{3} } \right\} \). The plane is thus defined as the set \( P = \left\{ {\varvec{x} \in R^{3} : \varvec{n} \cdot \left( {\varvec{x} - \varvec{x}_{0} } \right) = 0} \right\} \). We use the aortic valve centroid as point \( \varvec{x}_{0} \).
The concept of recursive multiresolution CNN for feature localization
Fundamentally, a CNN formulates a regression or classification task as a deep neural network of convolutions. Most layers of a CNN correspond to sets of kernel filters. During inference, each filter is convolved with the activations of the previous layer. As the depth of the CNN increases, more and more complex features are described. This framework is particularly powerful when applied to spatial datasets such as images.
For the specific problem of 3D feature localization, one can use the information obtained from a low-resolution CNN as a provisional feature position. This provisional position is expected to be inaccurate, but it can be used to generate a recentered sampling of the 3D dataset with an equal dimension but a lesser voxel spacing and thus, a lesser field of view. If the feature is contained within the new field of view, this process can be applied recursively to achieve arbitrarily high position accuracy. This is what we refer to as recursive multiresolution CNN.
Formally, we define the recurrence relation for the provisional position \( \hat{x}_{i} \in R^{3} \) at step i as
where \( C_{i} :R^{n \times n \times n} \to R^{3} \) represents the CNN induction at step i; \( S_{i} :R^{l \times m \times o} \to R^{n \times n \times n} \) is the trilinear interpolation of image volume \( \varvec{\mu}\in R^{l \times m \times o} \) over a regular n × n×n × grid with isotropic voxel spacing \( s_{i} \in R \) and centered at image position \( \hat{x}_{i - 1} \in R^{3} . \) For simplicity, we omit the formal definition of \( C_{i} \) and \( S_{i} \).
The choice of the initial provisional position \( \hat{\varvec{x}}_{0} \) is set to the center of the image volume \( \varvec{\mu} \), and \( s_{0} \) is set such that the field of view of the \( S_{0} \) interpolation covers the entire image volume to ensure that the feature of interest is contained. The field of view at step i is defined as the region
The sequence of voxel spacings \( s_{i} \) determines the rate of reduction in the size of the field of view. In order for the accuracy to improve at each step, we generally require
i.e., that the field of view decreases and that the interpolated image has a greater resolution at each step. Furthermore, the reduction in the voxel spacing must not be so great as to exclude defining regions of the image for the feature of interest. Specifically, \( \varvec{x}_{0} \in F_{i} \) must hold true at all steps, where \( x_{0} \) is the actual position of the feature of interest.
In the work presented here, we used \( s_{i} = rs_{i - 1} \) with \( r = \frac{1}{2} \). In other words, the field of view and voxel spacing are reduced by half as each recursive step. We also selected the total number of steps such that the final voxel spacing has the least difference with the original image volume voxel spacing. We selected a final voxel size of \( s_{4} = 0.5\, {\text{mm}} \), a maximum number of recursive steps of i = 4, an interpolation grid of 64 × 64 × 64 \( (n = 64) \). The algorithm defined by these design choices is depicted graphically in Fig. 2.
An important point to note regarding this architecture is that it enables efficient serving of the trained CNNs for inference over the network. Indeed, the interpolation and recentering step \( S_{i} \) can be performed on the client side, while the inference step \( C_{i} \) can be performed on the server side as depicted in Fig. 2.
This framework does not specify the architecture of the set of CNNs \( \left\{ {Ci} \right\} \). In the current work, we used a convolutional neural network defined in Fig. 3 to perform a regression. An identical neural network architecture is used at all resolution steps.
Generic regression CNN used at each step of the recursive multiresolution architecture of the localization algorithm. Node tensors are shown as rectangles. The same CNN is also used for the orientation neural network. Each layer called CONV is a 3D convolution operation with kernel size 3 × 3×3 and uses a rectified linear unit (ReLu) activation function. The layers labeled as FC are fully connected dense layers with 512 nodes for FC 512 and three nodes for FC 3. Output tensor dimensions are indicated after each operation
CNN for feature orientation
The framework described in the previous section focuses on the localization of the aortic valve annulus centroid. We also require the orientation of the valve annular plane. We formulated a single CNN with an identical architecture to that described in Fig. 3 to infer the three components of the normal vector. Note that inference of the valve orientation is not amenable to a recursive CNN formulation as is the case of the localization problem. Our implementation used a voxel spacing of 0.75 mm and an interpolation grid of 64 × 64 × 64. It uses the valve position inferred at the last step of the localization algorithm from the previous section as the center of the recentered and resampled image volume.
Tomographic data
We used multiphase ECG-gated datasets from 94 patients with severe symptomatic degenerative aortic stenosis who were referred to the McGill University Health Centre for TAVR candidacy evaluation. The Research Ethics Board approved this study. Clinical characteristics are described in Table 1. The dataset is representative of patients undergoing CT-based sizing of a TAVR device. We excluded patient with a prior surgical valve replacement. The CT scanner used for data collection was a Discovery CT750 HD scanner (GE Healthcare, Chicago, USA). The slice thickness was 0.625 mm and the spacing between slices was 0.625 mm. The in-plane pixel spacing was 0.88 mm. Retrospective ECG-gating was applied to yield five to ten image volumes at cardiac phases from 5% to 95% R–R. An additional volume at 35% is also produced by our TAVR protocol. An ungated volume is also acquired in order to plan vascular access, which we also included in analysis if it covered the aortic root. The volumetric datasets were anonymized to remove all identifying information prior to image analysis.
A single operator experienced with aortic valve planimetry manually segmented the aortic valve annulus with a closed 3D cubic spline for all 1007 CT image volumes. The operator processed images from different phases individually in ascending order. The oblique multiplanar reconstruction method was applied starting from standard axial, sagittal and coronal views for each image volume. The technique used was previously described in detail in [2]. A visualization software developed in-house and capable of 3D multiplanar reconstruction was used for the labeling process.
Implementation
The convolutional neural networks were implemented using TensorFlow (Google, USA) version 1.13 using the Python 3.7.2 API and a NVIDIA CUDA 10.1 backend. The Python code defining the generic regression CNN from Fig. 3 as a TensorFlow Estimator including specification of the training operations comprises 100 lines of code. Training and evaluation were performed using a workstation with dual Nvidia GTX 1080Ti graphic cards (NVIDIA, USA) each with 11 GB of memory. When running inference locally, each CNN produces predictions in 195 ms. The entire localization recursive algorithm can be executed in 780 ms.
Training and data augmentation
We used K-fold cross-validation for training and evaluation with K = 9. The image volumes were thus separated randomly into nine separate groups each with ten or 11 patients. Image volumes from any one patient were assigned to only one group. Nine separate training datasets were created by assembling eight groups, leaving in each case one of the groups as an evaluation dataset. Nine separate CNNs were trained at each resolution level of the localization CNN in addition to nine separate CNNs for the orientation CNN. A total of 45 CNNs were trained.
Training was performed using the TensorFlow (Google, USA) framework version 1.13. The pre-built Adam optimizer was used, and batch normalization was applied after each convolution layer and prior to the rectified linear unit (ReLu) activation function. The learning rate hyperparameter was optimized manually.
A batch size of 18 was used since it was the maximum allowed given our GPU memory restriction. Each batch of data was generated by randomly selecting 18 distinct image volumes, applying data augmentation and resampling the image on a \( 64 \times 64 \times 64 \) isometric grid. Data augmentation was applied on a total of 7 random degrees of freedom:
3D translation within range 32 times the target voxel spacing (3 degrees of freedom)
3D rotation about 3 Euler angles for the full angular range, i.e., from − 90° to 90° in cranio-caudal direction, − 180° to 180° in right- and left-anterior oblique and − 180° to 180° about the axial direction (3 degrees of freedom)
Scaling of voxel spacing by a factor from 0.8 to 1.2 (1 degree of freedom)
A total of 3500 epochs of training were applied. Each CNN was trained in approximately 12.4 h. K-fold cross-validation required a total of 558 h of training.
Evaluation metrics
The accuracy of the four localization CNNs was evaluated using two metrics: 3D error and out-of-place error. The 3D error \( e_{3D} \) for estimated position \( \hat{x} \in R^{3} \) is defined as:
where \( x_{0} \in R^{3} \) is the labeled position of the aortic valve annulus centroid. The out-of-plane error \( e_{\text{OOP}} \) is defined as:
where \( n \in R^{3} \) is the unit normal vector of the labeled aortic valve annular plane. These metrics are expressed in millimeters.
The accuracy of the orientation CNN was evaluated using the 3D angular error \( e_{\text{angular}} \) defined as:
where \( \hat{n} \in R^{3} \) is the estimated unit normal vector. The 3D angular error is expressed in degrees.
The error metrics were evaluated for each CNN for both the evaluation and training data to assess the presence of overfitting and assess the model generalizability. The error metrics were also correlated with the aortic valve area and the cardiac phase of the image volume to evaluate the algorithm robustness relative to these variables. Evaluating the dependence on the aortic valve area is important because at each recursive step of the multiresolution algorithm, the field of view of the resampled volume decreases by half. It is important for defining features of the structure of interest to remain within the volume. Should this not be the case, this may lead to a dependence of accuracy metrics on valve size. Evaluating the dependence on the cardiac phase is also crucial because the aortic valve changes configuration significantly between systole and diastole. All phases may be relevant for TAVR planning [2].
The ultimate goal of the proposed method is to simplify measurements of the aortic valve annulus dimensions. Errors in the plane position and orientation are expected to generate inaccuracies in the measurement of annulus area and perimeter. To study this effect, we randomly selected an image volume for each patient (n = 94). We generated oblique multiplanar reconstructions at the location and angulation inferred by the algorithm for which the patient was in the evaluation set. We traced the annulus manually using a 3D cubic spline to assess the error in the measured annulus area and perimeter.
Statistical analysis
The statistical analysis was performed using SciPy Library (SciPy Developers) version 1.2.1. The error values are expressed as mean ± standard deviation. The Pearson correlation coefficient is used. Statistical tests used p ≤ 0.05 as a significance level.
Results and discussion
Aortic valve annulus centroid localization
After training was completed at each of the resolution steps, we applied the recursive localization CNNs to the entire training and evaluation datasets without data augmentation. Figure 4 illustrates the evaluation metrics in a representative example with localization errors in the 50th percentile.
Orthogonal slices through the resampled datasets at each resolution used for the localization algorithm. The first row (a–d) presents axial slices; the second row (e–h), coronal slices; and the third row (i–l), sagittal slices. Images in each column have a different voxel spacing: 4 mm in the first column, 2 mm in the second column, 1 mm in the third column and 0.5 mm in the fourth column. The blue point is the labeled aortic valve annulus centroid and the dotted line the aortic valve annular plane. The yellow point is the provisional centroid position inferred by the CNN at that resolution level. The red line is the 3D error and the green line is the out-of-plane error
Table 2 reports the mean 3D and out-of-plane errors at each resolution step. Note that both metrics improve at each step of the algorithm. The error in both metrics is slightly greater for the evaluation datasets. This is expected given that the neural nets were not trained on the evaluation datasets. For unseen image volumes in patients with severe aortic stenosis, the error in the centroid 3D localization is (1.4 ± 0.8) mm and the error in the out-of-plane localization (0.9 ± 0.8) mm. Note that the localization error is approximately 5% of the annulus systolic excursion measured at (32.1 ± 35.2) mm in 3D and (16.5 ± 15.9) mm out-of-plane.
Histograms of the absolute error metrics at each resolution step for both the training and evaluation datasets are presented in Fig. 5. We observe that the distribution is different for the 3D and out-of-plane errors; the peak of the distribution is near a value of zero for the out-of-plane error, while this is not the case of the 3D error. This is interesting since it means that the 3D error is biased away from zero, while the out-of-plane error is unbiased.
Histogram of the absolute inference error achieved with the localization recursive CNN applied to the training and evaluation datasets. Each column corresponds to a different resolution step from 4 mm on the left to 0.5 mm on the right. The evaluation dataset is plotted in blue, and the training dataset in orange
Aortic valve annular plane orientation
After training of the orientation CNN, we applied it to the entire training and evaluation datasets without data augmentation. We used the inferred aortic valve annulus centroid from the recursive localization CNN as the center of the image volume. Figure 6 illustrates the difference in the angulation and position of the image centroid in three representative patients near the 5th, 50th and 95th percentiles of angular error.
Representative CT slices in three patients at the 5th percentile of angulation error (a)–(e), 50th percentile (f)–(j) and 95th percentile (k)–(o). The first column is an axial slice; the second column is a coronal slice; the third column is a sagittal slice, the forth column is an oblique multiplanar reconstruction at the level of the manually labeled aortic valve annulus (cyan), while the last column is at the level of the inferred aortic valve annulus (yellow)
The mean angular orientation error is presented in Table 3. For unseen image volumes in patients with severe aortic stenosis, the angular error was measured at (6.4 ± 4.0)°, which is approximately 53% of the systolic annular plane deflection (12.1 ± 5.0)°.
Figure 7 presents histograms of the angular error for both the training and evaluation datasets. The 95% percentile error was 8.2° for the training dataset and 13.4° for the evaluation dataset. For the evaluation dataset, we observe that in 84.6% of cases, the angular error was within 10° of the labeled angular plane. The gold standard used here is a trained expert human observer. A published study reported that trained expert human observers disagreed by less than 10° in 84.3% of cases [6]. While we cannot reach a strong conclusion given that our datasets are different, we believe that the accuracy of our algorithm approaches published expert-level accuracy.
Correlation of error with cardiac phase and aortic valve annulus area
We stratified image volumes based on the cardiac phase during which they were acquired. This observation did not translate into a statistically significant linear correlation as shown in Table 4.
We measured the projected area of the 3D cubic spline manually traced at the level of the aortic valve annulus during the labeling of each image volume. We studied the dependence of the error metrics relative to the labeled aortic valve annulus area. The gold standard used here is thus a trained expert human observer. We noted a trend toward higher errors at greater aortic valve areas. This observation is supported by the positive, statistically significant correlation between the 3D centroid error and the aortic valve area as reported in Table 4. This is a consequence of the progressively smaller field of view \( F_{i} \) of the resampled image volume at each resolution step of the algorithm. Note that despite the slightly greater inaccuracy at larger aortic valve annuli for the CNN at 0.5 mm voxel spacing, the accuracy in the inferred centroid position improved by more than 50% from the voxel size of 1 mm to 0.5 mm as shown in Table 2.
The angular error of the orientation CNN did not demonstrate a significant correlation with the aortic valve annulus area as reported in Table 4 and Fig. 7. This is likely a consequence of the voxel size selected at 0.75 mm, corresponding to a field-of-view dimension of 48 mm, which covers the entire aortic valve annulus even in larger anatomies.
Impact of inferred aortic valve annulus plane on dimensions measurements
The errors in the measured aortic valve annulus area and perimeter when measured in the inferred plane location and orientation are presented in Table 5. Note that on average the relative error in the valve area was (4.73 ± 5.32)% and the relative error in the valve perimeter was (2.46 ± 2.94)%. We noted a positive linear correlation between the error in plane position and orientation and the error in measured area and perimeter (Fig. 8).
Errors in the annulus area and perimeter measured in the plane predicted by the proposed algorithm as a function of the error in the angular orientation and 3D centroid distance of the predicted plane. The red line represents a linear regression relative to these quantities. Each data point corresponds to a different patient (n = 94)
Limitations and future directions
The dataset included only 94 patients for a total of 1007 CT image volumes, which remains limited. The patients included in the study had degenerative aortic valve stenosis. We excluded patient with prior surgical valve replacement. We also excluded congenitally abnormal valves such as bicuspid or quadricuspid valves. It would be an interesting study to extend our dataset to include cases with these pathologies. It would also be interesting to study how image quality affects the results.
The method presented in this manuscript does not directly provide a measurement of the aortic valve annular size. By using the method presented herein, one may generate a multiplanar reconstruction at the level of the aortic valve annulus. The sizing problem reduces to a 2D image segmentation, which has been addressed extensively in the literature [28]. In a recently published manuscript, the area and perimeter of the aortic valve annulus were measured accurately by a convolutional 2D neural network when applied to 2D MPR at the level of the aortic valve annulus [29]. An alternative approach would be to formulate the problem directly as a regression in order to yield the area and perimeter directly from a CNN. We aim to study these methods in follow-up studies.
Another important aspect is that the design of the method does not rely on a priori information specific to the aortic valve annulus but on a large amount of labeled CT datasets. We hypothesize that the method could be readily generalized to other anatomical structures, cardiac or otherwise.
Conclusion
Herein, we provided implementation details of a recursive multiresolution CNN algorithm to infer the location and orientation of the aortic valve annular plane in CT image volumes. Despite using fully tridimensional convolutions, this algorithm remains computationally efficient. We trained and evaluated the algorithm in 1007 ECG-gated CT image volumes. We observed a performance that approached expert-level accuracy. The algorithm includes no heuristics specific to the aortic valve and may thus be generalized to other anatomical features in the future.
References
Matiasz R, Rigolin VH (2018) 2017 Focused update for management of patients with valvular heart disease: summary of new recommendations. J Am Heart Assoc 7(1):e007596
Blanke P, Weir-McCall J, Achenbach S, Delgado V, Hausleiter J, Jilaihawi H, Marwan M, Norgaard BL, Piazza N, Schoenhagen P, Leipsic JA (2019) Computed tomography imaging in the context of transcatheter aortic valve implantation (TAVI)/transcatheter aortic valve replacement (TAVR): an expert consensus document of the society of cardiovascular computed tomography. J Cardiovasc Comput Tomogr 13(1):1–20. https://doi.org/10.1016/j.jcct.2018.11.008
Vaquerizo B, Spaziano M, Alali J, Mylote D, Theriault-Lauzier P, Alfagih R, Martucci G, Buithieu J, Piazza N (2015) Three-dimensional echocardiography vs. computed tomography for transcatheter aortic valve replacement sizing. Eur Heart J Cardiovasc Imag 17(1):15–23
Mylotte D, Dorfmeister M, Elhmidi Y, Mazzitelli D, Bleiziffer S, Wagner A, Noterdaeme T, Lange R, Piazza N (2014) Erroneous measurement of the aortic annular diameter using 2-dimensional echocardiography resulting in inappropriate corevalve size selection. JACC Cardiovasc Intervent 7(6):652–661
Piazza N, de Jaegere P, Schultz C, Becker AE, Serruys PW, Anderson RH (2008) Anatomy of the aortic valvar complex and its implications for transcatheter implantation of the aortic valve. Circ Cardiovasc Interv 1(1):74–81
Schuhbaeck A, Achenbach S, Pflederer T, Marwan M, Schmid J, Nef H, Rixe J, Hecker F, Schneider C, Lell M, Uder M, Arnold M (2014) Reproducibility of aortic annulus measurements by computed tomography. Eur Radiol 24(8):1878–1888. https://doi.org/10.1007/s00330-014-3199-5
Krizhevsky A, Sutskever I, Hinton GE (2017) Imagenet classification with deep convolutional neural networks. Commun ACM 60(6):84–90. https://doi.org/10.1145/3065386
LeCun Y, Boser B, Denker JS, Henderson D, Howard RE, Hubbard W, Jackel LD (1989) Backpropagation Applied to Handwritten Zip Code Recognition. Neural Comput 1(4):541–551
Lecun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proc IEEE 86(11):2278–2324
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: Medical image computing and computer-assisted intervention—MICCAI 2015, pp 234–241
Zhang W, Doi K, Giger ML, Wu Y, Nishikawa RM, Schmidt RA (1994) Computerized detection of clustered microcalcifications in digital mammograms using a shift-invariant artificial neural network. Med Phys 21(4):517–524. https://doi.org/10.1118/1.597177
Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, Liang J (2016) Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging 35(5):1299–1312
Milletari F, Navab N, Ahmadi S (2016) V-net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 fourth international conference on 3D vision (3DV), pp 565–571
Hennemuth A, Boskamp T, Fritz D, Kühnel C, Bock S, Rinck D, Scheuering M, Peitgen H (2005) One-click coronary tree segmentation in CT angiographic images. Int Congr Ser 1281:317–321
Ecabert O, Peters J, Schramm H, Lorenz C, von Berg J, Walker MJ, Vembar M, Olszewski ME, Subramanyan K, Lavi G, Weese J (2008) Automatic model-based segmentation of the heart in CT images. IEEE Trans Med Imaging 27(9):1189–1201
Ionasec RI, Voigt I, Georgescu B, Wang Y, Houle H, Vega-Higuera F, Navab N, Comaniciu D (2010) Patient-specific modeling and quantification of the aortic and mitral valves from 4-D cardiac CT and TEE. IEEE Trans Med Imaging 29(9):1636–1651
Waechter I, Kneser R, Korosoglou G, Peters J, Bakker NH, van der Boomen R, Weese J (2010) Patient specific models for planning and guidance of minimally invasive aortic valve implantation. Med Image Comput Comput Assist Interv 13(Pt 1):526–533
Zheng Y, John M, Liao R, Nottling A, Boese J, Kempfert J, Walther T, Brockmann G, Comaniciu D (2012) Automatic aorta segmentation and valve landmark detection in C-Arm CT for transcatheter aortic valve implantation. IEEE Trans Med Imaging 31(12):2307–2321
Elattar MA, Wiegerinck EM, Planken RN, vanbavel E, van Assen HC, Baan J, Marquering HA (2014) Automatic segmentation of the aortic root in CT angiography of candidate patients for transcatheter aortic valve implantation. Med Biol Eng Comput 52(7):611–618. https://doi.org/10.1007/s11517-014-1165-7
Elattar M, Wiegerinck E, van Kesteren F, Dubois L, Planken N, Vanbavel E, Baan J, Marquering H (2016) Automatic aortic root landmark detection in CTA images for preprocedural planning of transcatheter aortic valve implantation. Int J Cardiovasc Imaging 32(3):501–511
Jeganathan J, Knio Z, Amador Y, Hai T, Khamooshian A, Matyal R, Khabbaz KR, Mahmood F (2017) Artificial intelligence in mitral valve analysis. Ann Cardiac Anaesth 20(2):129–134
Liang L, Kong F, Martin C, Pham T, Wang Q, Duncan J, Sun W (2017) Machine learning-based 3-D geometry reconstruction and modeling of aortic valve deformation using 3-D computed tomography images. Int J Numer Method Biomed Eng. https://doi.org/10.1002/cnm.2827
Al WA, Jung HY, Yun ID, Jang Y, Park H, Chang H (2018) Automatic aortic valve landmark localization in coronary CT angiography using colonial walk. PLoS ONE 13(7):e0200317. https://doi.org/10.1371/journal.pone.0200317
Noothout JM, de Vos BD, Wolterink JM, Leiner T, Išgum I (2018) CNN-based landmark detection in cardiac CTA Scans. arXiv Preprint arXiv:1804.04963
Zheng Y, Liu D, Georgescu B, Nguyen H, Comaniciu D (2015) 3D deep learning for efficient and robust landmark detection in volumetric data. In: International conference on medical image computing and computer-assisted intervention, 2015, pp 565–572
Binder RK, Webb JG, Willson AB, Urena M, Hansson NC, Norgaard BL, Pibarot P, Barbanti M, Larose E, Freeman M, Dumont E, Thompson C, Wheeler M, Moss RR, Yang T, Pasian S, Hague CJ, Nguyen G, Raju R, Toggweiler S, Min JK, Wood DA, Rodés-Cabau J, Leipsic J (2013) The Impact of integration of a multidetector computed tomography annulus area sizing algorithm on outcomes of transcatheter aortic valve replacement: a prospective, multicenter, controlled trial. J Am Coll Cardiol 62(5):431–438
Spaziano M, Thériault-Lauzier P, Meti N, Vaquerizo B, Blanke P, Deli-Hussein J, Chetrit M, Galatas C, Buithieu J, Lange R, Martucci G, Leipsic J, Piazza N (2016) Optimal fluoroscopic viewing angles of left-sided heart structures in patients with aortic stenosis and mitral regurgitation based on multislice computed tomography. J Cardiovasc Comput Tomogr 10(2):162–172
Hesamian MH, Jia W, He X, Kennedy P (2019) Deep learning techniques for medical image segmentation: achievements and challenges. J Digit Imag 32(4):582–596. https://doi.org/10.1007/s10278-019-00227-x
Astudillo P, Mortier P, Bosmans J, De Backer O, de Jaegere P, De Beule M, Dambre J (2019) Enabling automated device size selection for transcatheter aortic valve implantation. J Interv Cardiol 2019:3591314. https://doi.org/10.1155/2019/3591314
Funding
This study received no external funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Pascal Theriault-Lauzier is a consultant for Circle CVI and receives royalties from sales of their software. Nicolo Piazza is a consultant and proctor for Medtronic and receives royalties from sales of Circle CVI software. Giuseppe Martucci is a proctor for Medtronic. All other authors have no conflict of interest to declare.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Human and animal rights
This article does not contain any studies involving animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
P. Theriault-Lauzier formally at the Division of Cardiology, McGill University, Montreal, QC, Canada.
Rights and permissions
About this article
Cite this article
Theriault-Lauzier, P., Alsosaimi, H., Mousavi, N. et al. Recursive multiresolution convolutional neural networks for 3D aortic valve annulus planimetry. Int J CARS 15, 577–588 (2020). https://doi.org/10.1007/s11548-020-02131-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-020-02131-0