Abstract
Purpose
Navigation systems commonly used in neurosurgery suffer from two main drawbacks: (1) their accuracy degrades over the course of the operation and (2) they require the surgeon to mentally map images from the monitor to the patient. In this paper, we introduce the Intraoperative Brain Imaging System (IBIS), an open-source image-guided neurosurgery research platform that implements a novel workflow where navigation accuracy is improved using tracked intraoperative ultrasound (iUS) and the visualization of navigation information is facilitated through the use of augmented reality (AR).
Methods
The IBIS platform allows a surgeon to capture tracked iUS images and use them to automatically update preoperative patient models and plans through fast GPU-based reconstruction and registration methods. Navigation, resection and iUS-based brain shift correction can all be performed using an AR view. IBIS has an intuitive graphical user interface for the calibration of a US probe, a surgical pointer as well as video devices used for AR (e.g., a surgical microscope).
Results
The components of IBIS have been validated in the laboratory and evaluated in the operating room. Image-to-patient registration accuracy is on the order of \(3.72\pm 1.27\,\hbox {mm}\) and can be improved with iUS to a median target registration error of 2.54 mm. The accuracy of the US probe calibration is between 0.49 and 0.82 mm. The average reprojection error of the AR system is \(0.37\pm 0.19\,\hbox {mm}\). The system has been used in the operating room for various types of surgery, including brain tumor resection, vascular neurosurgery, spine surgery and DBS electrode implantation.
Conclusions
The IBIS platform is a validated system that allows researchers to quickly bring the results of their work into the operating room for evaluation. It is the first open-source navigation system to provide a complete solution for AR visualization.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
In image-guided neurosurgery (IGNS), surgical tools are tracked in real time and displayed on a navigation system in the correct location and orientation with respect to a virtual patient model. Commercial IGNS systems such as Medtronic’s StealthStation and BrainLab are routinely used in the operating room (OR) for various neurosurgical interventions. However, these systems have a number of shortcomings in terms of registration, visualization and surgeon–computer interaction. More importantly, they are not built to easily accommodate new types of imaging and devices.
There is a very active community of researchers constantly proposing improvements to IGNS techniques. But their efforts are too often limited to laboratory experiments on phantoms. In a recent review of visualization methods in image-guided surgery, Kersten-Oertel et al. [1] showed that only 16 % of the articles reviewed were evaluated in the OR and only 6 % measured the impact on clinical outcome of the patient. There is a significant gap between laboratory testing and clinical use of IGNS tools. The best solution to bridge this gap is to use open-source platforms that provide researchers with a reliable OR ready IGNS software package and an extensible API to integrate new contributions seamlessly.
In this paper, we introduce the Intraoperative Brain Imaging System (IBIS), an IGNS software platform that has been developed and used in the OR at the Montreal Neurological Institute and Hospital (MNI/H) over the past 14 years. The platform replicates the functionality of a commercial IGNS system while allowing components to be improved upon or replaced to accommodate research projects. In addition to standard IGNS features, IBIS implements and tightly integrates the following innovative functionality:
-
A simple user interface for tracked ultrasound (US) probe calibration.
-
Seamless acquisition and 3D reconstruction of intraoperative US (iUS) images.
-
Near real-time brain shift correction based on iUS.
-
A surgical microscope calibration module.
-
Augmented reality (AR) visualization in the surgical microscope.
This paper describes for the first time the architecture and complete functionality of the IBIS system and overviews the clinical applications for which it has been used. The original contributions of the paper are (1) an improved workflow for IGNS that takes advantage of IBIS functionality to maintain navigation accuracy throughout a neurosurgical procedure by registering preoperative models with iUS and providing more intuitive navigation through the use of AR, (2) a novel microscope calibration method that greatly facilitates the use of this type of device to produce AR images in the OR and (3) the first complete open-source implementation of AR neuronavigation software that includes all the tools necessary to calibrate and track instruments and to produce the augmented images.
Consistent with the idea that the publication of source code is an essential element of any scientific communication [2] and in support of a recent institute-wide open science initiative at the MNI [3, 4], the complete source code for the IBIS platform is now freely available at ibisneuronav.org and is distributed under the BSD 3-clauses license.
While not FDA nor CE approved, the IBIS platform can be used in the OR after going through an ethics research board approval process. Furthermore, the platform has been fine-tuned to run seamlessly alongside the FDA-approved Medtronics StealthStation™system to guarantee the safety of the procedures.
In the following sections, we review the existing ecosystem of open-source software for image-guided surgery, we describe the core of the IBIS system (basic features, interface and architecture), we describe the specific workflow implemented in IBIS, and finally we describe the various clinical applications where the platform has been used.
Related work
Several existing open-source platforms are available for developing image-guided intervention (IGI) applications. These platforms and toolkits can be characterized according to the level of system integration they provide. Toolkits that have low levels of integration require significant investment in software engineering, whereas high-level integration is ready to use ‘off the shelf.’ Existing platforms cover a full spectrum of integration levels, from very general component-based libraries that help in the creation of new applications to fully integrated systems that are ready for the OR (Fig. 1). Less integrated platforms have the advantage of supporting a wider range of potential applications, while more integrated systems sacrifice flexibility to focus on a particular procedure or group of procedures and to facilitate their use by non-technical OR personnel during clinical studies.
Existing open-source platforms dedicated to IGI and their dependencies. The platforms are laid out according to their level of integration: (1) Libraries of components required to build IGI applications. Packages at this level do not provide a usable application. (2) General imaging platforms. At this level, a functioning program is available, but additional components and customization are needed to use it in IGI. (3) Packages of IGI-specific components for the general platforms of level 2. The packages at this level are typically not geared toward a specific application and some customization is usually required to make the interface usable in the OR. (4) Fully integrated IGI platforms. These systems are ready for the OR and target specific procedures or groups of procedures
At the lowest level of integration (1), we find libraries of components required to build IGI applications. Packages at this level do not provide a usable application. Software development is required to build such application. IGSTK [5] offers tightly packaged modules that implement a state machine to manage specific aspects of an IGI application such as interfacing with tracking systems or visualizing medical images. The OpenIGTLink [6] package defines a protocol that allows communication between a wide variety of hardware devices typically used for IGI. The Plus Toolkit [7] provides support for hardware devices that do not implement the OpenIGTLink protocol. This toolkit, which acts as a translation layer between native hardware application programming interfaces (API) and OpenIGTLink clients, supports a wide variety of devices such as tracking systems, ultrasound machines, and commercial navigation systems.
Level 2 contains general imaging platforms such as the Medical Imaging Interaction Toolkit (MITK) [8] and 3D Slicer [9]. At this level, a functioning program is available, but additional components and customization are needed to use it in IGI. Neither MITK nor 3D Slicer is specifically geared toward IGI, but can be configured to act as navigation systems by installing plugins or external modules. 3D Slicer, a multi-million dollar project funded by the NIH, is supported by a large community of users and developers (50+ regular contributors, 500+ papers published, 1000+ downloads/month). Its modularity and the large number of extensions allow users to build new applications quickly without writing code. Furthermore, the simple user interface allows one to install extensions and download relevant datasets, thus facilitating the reproducibility of scientific results. MITK is supported by a community of 25+ regular contributors from several research groups and an active mailing list with 1000+ posts/year. The MITK platform uses a plugin system, and more than 30 plugins are available.
At the next level of integration (3), we find bundles of IGI-specific components that complement the general platforms of level 2. The packages at this level are typically not geared toward a specific clinical application, and some customization is required to make the interface usable in the OR. Such bundles include the SlicerIGT plugin package, which extends the 3D Slicer platform. Similarly, the MITK-IGT [10] package adds support for surgical tool tracking to MITK and the MITK-US package complements it with support for US imaging. An alternative to MITK-IGT/MITK-US is NifTK [11], also based on MITK. This toolkit favors the integration of modules running as separate applications (potentially on different machines) and communicating through NiftyLink, a wrapper around the OpenIGTLink protocol that facilitates the management of connections. Software packages on level 3 usually serve as a basis to build OR ready solutions where their graphical user interface is wrapped to hide the complexity of the system and facilitate usage in the OR. A good example of such application is the LumpNav system developed by Ungi et al. [12].
Overview of the main GUI components in IBIS, illustrated with a tumor resection case. a The main window shows the canonical 2D views (sagittal, coronal, transverse) of the patient’s gadolinium enhanced T1 MRI (in gray) with an overlaid intraoperative US volume (in green) and a 3D view with the volumetric rendering of the MRI showing the highlighted tumor (in red) and the live image from a tracked US probe. b The AR view can replace the 3D view. c Dual US-MRI view window: on the left, the live US image, on the right, the corresponding resliced MRI (gray) with overlaid US (green)
On level 4, we find fully integrated IGI platforms such as CustusX [13]. These systems are ready for the OR and target specific procedures or groups of procedures. The CustusX platform focuses on the use of iUS for patient registration, brain shift correction and guidance. The platform is already optimized for a certain number of procedures and can be used in the OR without the assistance of a technician. The CustusX platform has the additional advantage of having a long history of clinical use.
The IBIS platform we present in this paper lies at the fully integrated end of the spectrum (Level 4) alongside CustusX. It focuses on a very specific workflow that constitutes an improvement upon the traditional workflow of commercial navigation systems. While CustusX is probably the most advanced open platform in terms of iUS-based navigation, IBIS’s distinguishing feature is its complete augmented reality (AR) solution described below.
System description
The IBIS program supports most of the basic functionality found in other comparable platforms. It can read and write a wide variety of 3D medical image formats (Minc, Nifti, Vtk), polygonal model formats (Vtk, Obj, TAG) and 3D transform formats (XFM). It can display this data both in 2D, where the slices from each 3D volume are overlaid with outlines of polygonal models, and in 3D, where images can be displayed with different kinds of volumetric rendering techniques, and polygonal models are rendered using a traditional 3D rendering pipeline. Additionally, specialized data types can be loaded, visualized and interacted with by implementing custom external modules in the form of plugins. A good example of this functionality is found in the FiberNavigator plugin, which is based on the open-source FiberNavigatorFootnote 1 [14, 15] application. The plugin allows reading and displaying DTI fiber tracts and recomputes tracts in real-time based on raw diffusion data.
Graphical user interface
Figure 2 shows an overview of the graphical user interface (GUI) of IBIS.
The main window of IBIS (Fig. 2a) is divided into three distinct areas: the left panel, the graphical area and the right panel. The left panel shows the state of surgical tools tracked by the system, a list of objects managed by the system and the properties of the currently selected object. This panel is the researcher’s interface. It contains technical GUI elements used to build new experiments in the laboratory. It is typically collapsed during navigation sessions in the OR. The right panel is a container for GUI elements provided by the plugin system. In the OR, it typically displays GUI elements of a wizard that guides the operator through the steps of the operation.
The graphical area is made up of 4 views: a 3D representation of the scene and the three canonical 2D slice views (sagittal, coronal and transverse). Each of the 4 views can be selected and expanded to fill the whole graphical area. The AR features of the system can turn the 3D view into an AR view by overlaying images captured from a video device with the patient model (Fig. 2b). Alternatively, a copy of the 3D AR view can be injected into a surgical microscope, a head-mounted display or simply displayed on an external monitor. Furthermore, an additional window (Fig. 2c) can be opened to visualize data in US space with the option of showing the US images and their corresponding slice of the preoperative 3D data objects (e.g., MR and CT), either side-by-side or overlaid.
System architecture
Main modules
In Fig. 3, the basic components of the IBIS platform are shown. IBIS consists of a very thin main application that communicates with a list of plugins, as well as a module that controls hardware devices such as US machines, tracking systems or surgical microscopes. These three components sit on top of a main library called IbisLib that implements the core functionality of the platform and manages communication between modules. These high-level modules rely on basic libraries commonly used in the field: VTK 6.2 for visualization, ITK 4.5 for image processing, registration and image IO and OpenCV 3.0 for computer vision. The graphical user interface (GUI) is implemented with Qt5. The platform also contains a series of specialized ITK and VTK classes that implement extra low-level functionality specific to IBIS. These classes are bundled in the VTKExt and ITKExt libraries.
The IBIS platform can easily be compiled for all major operating systems (Linux, OSX, WindowsFootnote 2) using the CMake cross-platform build program. The required version of external dependencies like ITK, VTK and OpenCV is automatically downloaded and compiled as part of the build process.
Data structures
The core of the IBIS platform relies on the concept of a Scene, which is a data structure that represents everything that is known by the system about the OR. A scene contains an arbitrary number of Scene Objects (of different types), each of which is a hardware-independent abstraction for an entity needed for navigation. Scene Object types include volumetric images, 3D surfaces, point sets, as well as, models for tracked surgical tools such as pointers, US probes and camera objects. Each Scene Object type is able to graphically represent itself in both the 2D and 3D views. New types of Scene Objects can be added through the plugin system.
Objects in a scene are organized in a hierarchical structure, implementing the well-known composite design pattern [16]. Each of the nodes contains a transform that is automatically concatenated with parent transforms, thus enabling simultaneous registration of several objects by modifying their parent transforms.
Module communication
IBIS implements a simple module communication sequence that ensures synchronized updates of the objects in the scene. Figure 4 illustrates the sequence performed for each cycle of the main event loop of the application. The system periodically sends the update signal to the hardware module, which in turn pushes data captured from the hardware devices to the relevant Scene Object. After all of the information is up to date, the plugins are notified and can in turn update their internal data structures. For example, a plugin responsible for ultrasound volume reconstruction, upon update notification, would grab the new tracked video image from the US probe Scene Object and combine it to the reconstructed volume. Once all plugins have had a chance to react to the update, the application display is refreshed if needed.
IBIS workflow
As mentioned in the introduction, the main contribution of this paper is to propose a novel workflow for IGNS. In this section, we describe this workflow as well as each of the components provided by the IBIS platform. To simplify our description of the system, we base our discussion on the setup presented in Fig. 5, which is a typical setup for brain tumor resection operations and several vascular surgery procedures such as AVM obliteration.
A typical OR setup where the IBIS platform is used. The patient is rigidly attached to the operating table using a Mayfield clamp and the reference tool of an optical tracking system acts as the origin of IBIS’ frame of reference. Preoperative patient models and plans are registered to the real patient using a tracked surgical pointer based on anatomical landmarks. An ultrasound machine is connected to the navigation system to acquire US images. A tracker tool is attached to the US probe so that the position and orientation of every US image can be retrieved from the tracking system. The surgical microscope is also tracked. A live video feed is captured by the navigation system and overlaid on a 3D rendering of preoperative images in IBIS. The resulting AR image is reinjected into the microscope to be visualized by the surgeon
a Conventional workflow employed at the MNI while operating with the Medtronics StealthStation™ navigation system and where registration errors caused by brain shift must be accounted for by the surgeon. b Improved workflow implemented in the IBIS platform where intraoperative ultrasound is used to correct for brain shift
Figure 6 illustrates the difference between the conventional workflow employed by commercial navigation systems such as the Medtronics StealthStation™used at the MNI and the IBIS workflow. In the conventional intervention (Fig. 6a), the patient is registered to the preoperative anatomical images. After the initial preparation steps, the surgeon starts the surgery, pausing his work every now and then to use the navigation system. The surgeon then engages in two concurrent and cognitively demanding tasks: (1) to mentally map, with the use of the tracked pointer, preoperative images to the patient and (2) to estimate and account for the discrepancy between preoperative models and reality.
The IBIS workflow (Fig. 6b) seeks to facilitate these demanding tasks by providing the surgeon with a constantly updated model of the patient made available to the surgeon’s, in his/her field of view. Tracked intraoperative ultrasound (iUS) is used to reregister preoperative patient models to the patient, and AR is used to guide the acquisition of iUS, verify updates of the plans and navigate to the anatomy of interest without looking away from the operative field.
The goals of the IBIS platform are to:
-
1.
Provide a complete implementation of the workflow that is ready to be brought to the OR,
-
2.
Provide a mechanism that allows researchers to easily improve or replace any component of the workflow, and add new ones.
As such, IBIS will allow researchers to rapidly evaluate new methods in the OR without the need to develop the basic functionality of an IGNS system.
The implementation of the IBIS workflow requires a certain number of key components as illustrated in Fig. 7. Each of the components (red boxes) is implemented in at least one plugin distributed with the platform. In the following sections, we briefly describe the components that rely on standard methods (patient registration, US probe calibration and US volume reconstruction) in the interest of readers who would like to know what performance to expect from these components or in order to use them as a basis for a novel implementations. We describe in more detail the components that are unique to the system (GPU-based US-MRI registration) and original contributions of this paper (microscope calibration and open-source AR visualization).
IBIS workflow with key components (in red) involved in its implementation. Before the operation starts, the US probe and microscope need to be calibrated. Navigation and resection (red dotted box) are done under AR visualization. Specialized modules are needed to reconstruct US slices into a volume and register that volume to the preoperative scans to correct for brain shift
Patient registration
The IBIS platform implements a standard landmark-based patient registration method. The GUI lets users identify landmark points on preoperative images. In the OR, the surgeons capture the position of the corresponding landmark using a tracked pointer. Internally, the registration transform is computed using the method of Horn et al. [17], implemented in VTK as vtkLandmarkTransform.
In a recent study that used IBIS to investigate the influence of using different patterns of landmark points for patient-to-image registration, Gerard et al. [18] obtained a mean fiducial registration error (mFRE) between \(3.72\pm 1.27\,\hbox {mm}\) and \(3.88\pm 1.61\,\hbox {mm}\), depending on the pattern used. These numbers are comparable to others reported in the literature as reviewed by Stieglitz et al. [19].
AR visualization
We illustrate the method to generate AR images in IBIS using a surgical microscope, but the method is independent of the type of video device used. The AR images are created in 2 steps: (1) Render the patient model from the point of view of the microscope to create the virtual image and (2) combine the virtual image with live video captured from the microscope to produce the AR view. These steps are detailed below.
Virtual image
The model used to render the virtual image is illustrated in Fig. 8. The patient model is first transformed to the coordinate system of the microscope by combining the patient registration transform P, the tracking transform M and the extrinsic calibration transform E that maps the tracker tool attached to the microscope to the optical center of the device. Once the patient data are in a space aligned with the microscope’s optics, they are projected to the image plane using the camera model I. Both the extrinsic calibration transform, E, and the projection model I are estimated using the calibration procedure described below.
Typical OR setup for the integration of an optical device within an IGS system. a Overview of the OR with the patient rigidly attached to the operating table and the tracking system’s reference using a Mayfield clamp, the surgical microscope above with an attached tracker tool and the tracking system’s camera that computes the rigid transform between different tracker tools. b The different transforms involved in the computation of an AR view: I, the intrinsic microscope projection model; E, the extrinsic calibration matrix that transforms from the microscope-attached tracker to the optical center of the lens. M, the rigid transform between tracking reference and the microscope tracker; P, the rigid transform between the patient’s anatomy and the tracking reference
Microscope calibration
The novel calibration method relies on a simple device that is constructed by sticking a printed checkerboard pattern on a rigid planar surface with an attached tracker tool (Fig. 9). The calibration procedure consists of collecting a sequence of microscope images (and the corresponding transforms \(M_{i})\) of the device in different poses in the field of view of the microscope.
The projection model I is obtained by automatically detecting corners of the checkerboard pattern in the captured image and feeding their coordinates as well as their known 3D position to OpenCV’s implementation of Zhang’s camera calibration method [20]. This method also produces transform V which maps the space of the calibration grid to that of the microscope’s optical center. Both the extrinsic transform E and the grid tool transform G are unknown. However, these transforms remain constant for all images captured. We can express G as
A simple way to estimate the extrinsic calibration transform is then to find transform E that minimizes the standard deviation of grid points transformed by the right side of equation (1). The optimization is performed using the iterative Nelder–Mead algorithm [21].
The main advantage of our calibration method is the simplicity of building the calibration device. Most published methods assume G is known and thus must rely on a precisely manufactured calibration device or a manually measured transform G, which may introduce an additional source of error in the system. With our method, transform G needs not be known prior to calibration. The only requirement is that the grid and the tracker tool be rigidly attached.
a Main window of the calibration plugin with enlargement of a portion of the graphic view. The corners of the grid are automatically detected in the live video feed. b The AR view in IBIS during the calibration process with an area zoomed in. The real and virtual calibration grids are overlaid allowing users to visualize the discrepancy between real and virtual images during the calibration procedure (here the discrepancy is exaggerated for the figure)
The main window of the calibration plugin provided by IBIS is shown in Fig. 10a. The plugin uses OpenCV’s functionality to automatically track the checkerboard pattern of the calibration device in real time, helping the user position the device correctly. The plugin also updates both intrinsic and extrinsic calibration parameters in the program as images are acquired, allowing the users to visually assess the alignment of the real and virtual images of the calibration device. The reprojection error (mean distance between each 3D point of the grid and reprojection of its 2D image) is computed and displayed as more views are captured. These features allow users to obtain an accurate calibration very quickly (less than 2 min for approx. 20 views). The calibration procedure is usually performed once before bringing the microscope into the OR, and parameters remain fixed throughout the procedure.
To validate the calibration method, we captured 20 images of the calibration device using a Zeiss NC-31 surgical microscope tracked by a NDI Polaris optical tracking system. We ran a leave-one-out cross-validation procedure and obtained a reprojection error of \(0.37\pm 0.19\,\hbox {mm}\). This is an estimate of the reprojection error that would be obtained in the OR if the patient was perfectly registered with the system. During surgery, this error is compounded with the patient registration error and could lead to a larger discrepancy. This problem can be addressed by semiautomatically correcting the registration through the AR view as we have shown in [22].
Combining real and virtual images
Figure 11 illustrates AR visualization using a phantom (Fig. 11a) that was 3D printed from scans of a patient treated for an arteriovenous malformation (AVM). Figure 11b shows the virtual image rendered from the patient’s preoperative CTA using the method described above. The display of the real image is handled by a custom VTK class that uploads the image captured from the microscope (Fig. 11c) to the GPU, computes a transparency mask (Fig. 11d) and applies an image deformation filter to take into account the image center and lens distortion parameters of the projection model I before rendering it over the virtual image to produce the final AR view (Fig. 11e). The mask is computed by combining a user-defined point of interest that restricts transparency to a circular region with a GPU computed edge map of the input image. The edge map, which is computed using a Sobel filter, adds the occlusion cue and allows for better perception of the relative depth between real and virtual images.
a 3D printed phantom used to demonstrate the AR view. b Preoperative CTA of the patient rendered from the point of view of the microscope. c Live video feed captured from the microscope. d Opacity mask generated in the GPU. e Final AR view. f Enlargement of the AR view showing alignment of real and virtual superficial blood vessels. The dotted lines highlight a blood vessel that is present in both the real and virtual part of the image
a Illustration of an N-shaped wire intersecting the US image plane. b The Plexiglas phantom typically used in our laboratory with 4 N-shaped wires. c The main window of the calibration plugin in IBIS. d The 3D view of IBIS during the calibration that shows a virtual representation of the N-shaped wires intersecting the US image plane
US probe calibration
To enable the use of US images in a navigation system, we must first estimate the transform between a tracker tool attached to the probe and the US image pixels, a process called US probe calibration. For quality control reasons, this procedure must be performed before every operation and for each depth setting supported by the probe. During the operation, the surgeon can change the depth setting at any moment. The IBIS platform comes bundled with a plugin that estimates the calibration transform based on the position of N shaped wires.
The N-fiducial (also called Z-fiducial) calibration method, inspired by Brown’s method [23] for registering a stereotactic frame to CT, was first applied to US calibration by Comeau et al. [24]. The technique is based on a phantom that contains three or more wires arranged to form an N-shaped pattern. The intersection of the patterns with the US image defines the 3D position of a point in the US image and 3 or more patterns fully define the 3D pose of the US image and the calibration transform, provided the phantom has initially been registered with the tracking system.
Figure 12b shows an example of a 4 N-shaped wire phantom. The main window of the plugin (Fig. 12c) allows users to manually specify the intersection of the wires with a sequence of US images. The calibration is automatically recomputed after each interaction and a simple line representation of the N wires is displayed in the 3D view (Fig. 12d) to allow user to visually assess the accuracy of the calibration.
The precision, accuracy and bias of the calibration obtained with the N-shaped calibration phantom was evaluated by Mercier et al. in [25] using a second cross-wire phantom. Precision varied between 0.62 and 0.90 mm, depending on the depth setting of the probe. Similarly, accuracy varied between 0.49 and 0.82 mm. We refer the reader to [25] for more details.
US reconstruction
To minimize the computational overhead of integrating tracked 2D US in the operating room, a GPU-based 3D reconstruction algorithm was implemented as a plugin in IBIS. The 3D reconstruction algorithm takes as input a series of masked 2D US images with known location and orientation. It computes the bounding box of the slices and generates the corresponding volume with user-specified voxel spacing. The intensity of every voxel of the volume is determined by computing a distance (Gaussian) weighted average of the intensity of all US pixels lying within a user-defined distance (search radius) from the center of the voxel.
The speed of the plugin was tested on a PC equipped with an Intel Quad Core i7 processor, 32 Gb of RAM and an NVidia GeForce GTX 670 graphics card running on the Ubuntu 14.04 operating system. The plugin is able to reconstruct a volume within 5–10 s, depending on the number of US slices, the size of the volume spanned and the size of the Gaussian kernel. Table 1 shows an example of execution times to reconstruct 489 US slices with an image size of \(640\times 480\) within a \(173\times 107\times 181 (1\times 1\times 1\,\hbox {mm})\) volume.
US-MRI registration
During open brain surgery, a significant amount of tissue displacement and deformation can occur. This is the so-called brain shift. This phenomenon invalidates the initial registration of the patient with preoperative data and as such makes navigation systems less reliable. To correct for brain shift, IBIS can compute a transform that aligns the reconstructed US volume with the preoperative MRI scan. The platform comes with a plugin that automatically performs this rigid registration operation based on a gradient orientation alignment method developed by De Nigris et al. [26, 27].
A gradient orientation alignment similarity metric is used because it is robust even with non-homogeneous imaging modalities such as US. To take into account the noisy nature of the image gradient and improve the speed of the algorithm, the similarity metric is evaluated at a subset of locations in the image where uncertainty of the gradient orientation is low. This approach is asymmetric as the gradient uncertainty is only evaluated in the US image. An asymmetric method is used because the boundaries in the US image are more likely to have a counterpart in the MR image than the reverse. Therefore, the asymmetry increases the robustness of the algorithm. The optimization is performed using a covariance matrix adaptation evolution (CAE) strategy. We refer the reader to [27] for more details about the registration algorithm.
The validation of the registration plugin was done using US collected from the OR as described in more detail in the brain shift correction section bellow.
Clinical applications
Since its early phases of development in 2002, the IBIS platform has been brought regularly into the OR. This has enabled validation of new IGNS methods and progressive improvements of the system’s usability based on real evaluation in the operating room and clinician feedback. Here, we describe the clinical applications the platform has been used for.
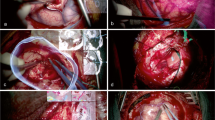
a Augmented reality is used for tailoring the craniotomy. The surgeon determined the location of important arteries (arising from the aneurysm) indicated as a green dot to plan the skin incision and craniotomy. b AR is used to differentiate between an arterialized vein (indicated with yellow arrow) and an artery (indicated with green arrow). c AR is used to plan a corridor of resection by placing a microlentine on the cortex above a deep feeding artery indicated with a pink dot (and blue arrow) in the AR view
Correction of brain shift during brain tumor resection
The first clinical application of the IBIS platform was the evaluation of brain shift compensation in tumor resections. The IBIS system was brought into the OR for 14 brain tumor resection cases, alongside a commercial navigation system. IBIS was found to be comparable to its commercial counterpart in terms of navigation accuracy after initial patient registration. US images were captured pre- and post-resection for each case. Between 19 and 40 corresponding landmarks were identified by 2 different imaging experts to evaluate registration. The resulting datasets (MRI, US and landmark coordinates) were made publicly available as the BITE database [28] and since then, several studies have used the data to investigate MRI-US registration methods that could be used to correct for the brain shift [29–34]. The US-MRI registration plugin described above was also validated on data from BITE [26, 27] (median mTRE: 2.54 mm). In our group, we have used these data to evaluate different objective functions [35, 36] (average mTRE: 2.35 mm), linear and nonlinear registration [37, 38] (average mTRE: linear registration, 2.7 mm, nonlinear registration, 1.5–1.7 mm), and for the development of a pseudo-US to MRI registration method [39] (mTRE: 2.97 mm).
AR in neurovascular surgery
We have explored the use of the augmented reality functionality of IBIS for the surgical treatment of arteriovenous malformations (AVMs), arteriovenous fistulas (AVFs) and aneurysms. In addition to the shortcoming of existing IGNS systems that place the burden of mapping preoperative patient models from the navigation system to the patient on the surgeon, there are shortcomings that are specific to neurovascular surgery. These include difficulty in localizing small and deep vessels and differentiating between arterialized veins and true veins. These tasks are made more difficult due to zoomed in microscope views (that do not provide contextual anatomy), a non-trivial mapping between the vascular images and the patient, and the inability to visualize the topology of the vessels below the exposed cortical surface.
To facilitate these tasks, we have explored: (1) the use of different perceptual visualization techniques (e.g., aerial perspective, chromadepth, edges) to improve the three-dimensional understanding of cerebral vascular volumes [40], (2) the merging of cerebral vascular volumes with live images of the surgical field to create an effective augmented reality visualization [41, 42], (3) the feasibility of using AR in the operating room in real clinical cases [43, 44] and (4) the usefulness of AR for particular surgical tasks [45]. Our results suggest that AR visualization may aid the surgeon in tailoring and planning the craniotomy (Fig. 13a), differentiating between arterialized veins and true veins (by using color coding in the AR view, Fig. 13b), and determining the optimal corridor of resection to the anatomy of interest (Fig. 13c).
AR + US in tumor surgery
In our most recent contribution, we leverage the full potential of the integrated workflow implemented in the IBIS platform by combining our two key technologies, iUS and AR, to improve the accuracy of AR visualizations during brain tumor resections [46]. In this work, the interpretation difficulties associated with US images are mitigated with detailed AR visualizations and the accuracy issues associated with AR are corrected through US registration (Fig. 14). The combination of these technologies allows for improved intraoperative, patient specific, planning by prolonging the reliable use of neuronavigation and improving the understanding of complex 3D medical imaging data so that different surgical strategies can be adapted when necessary.
a Surgical field of view (real image). b The segmented tumor and avatar showing location of tumor (virtual image). c Tumor boundary (green), manually segmented before the operation from the uncompensated gadolinium enhanced MRI overlaid on the brain shift compensated MRI. d The segmented tumor on pre-operative T1 gadolinium MRI after brain shift correction with ultrasound-MRI registration. e AR view prior to brain shift correction where the tumor seems to conform unnaturally to the surrounding tissue. f AR view after brain shift correction where the tumor visualization now lines up naturally with surrounding tissue and can be used for accurate intraoperative planning
Spine surgery
Several types of spine surgery require the implantation of screws into the pedicle of the vertebrae in order to stabilize parts of the spinal column. The accuracy of this procedure can greatly benefit from the use of navigation systems [47]. However, this type of navigation relies either on the extensive use of radiation or on a time-consuming procedure to register the vertebrae of interest with a preoperative CT scan.
IBIS was used to investigate new methods to obtain the CT-to-patient mapping from iUS acquisition followed by iUS-CT registration. This method has the potential to be faster and require less resection or retraction of patient tissues. Yan et al. [48] proposed a method to register 3D US reconstructed from a series of 2D iUS images with a preoperative CT of the spine. IBIS was employed to validate this method on porcine cadavers [49]. The evaluation procedure used 18 thoracic and lumbar vertebrae on 3 cadavers. For each case, iUS was acquired with 10 different sweep patterns. The optimal sweep pattern yielded a registration accuracy of 1.65 mm with 82.5 % of target registration errors \({<}2\) mm. This method was later improved to accelerate the registration procedure in the OR by performing the registration directly with the iUS slices without prior reconstruction of a volume [50].
Path planning for DBS and Epilepsy electrode implantation
Though the IBIS platform is mostly focused on the improvement of the AR-US workflow in neurosurgery, it can also be used for other types of procedures that take advantage of the machinery in place to start with an accurate patient registration and AR visualization. As such, the IBIS workflow has been applied to the planning of probe trajectories in Deep Brain Stimulation (DBS) and Stereoelectroencephalography (SEEG). Both DBS and SEEG are minimally invasive procedures that rely on visualization of preoperative MRI and precise tracking of surgical instruments, respectively, for pre-planning optimal electrode trajectories and executing the pre-planned implantations intraoperatively. In recent years, our group has focused on developing new automated planning software for minimizing risks of surgical complications [51–53] and improving therapeutic outcomes [54, 55].
The IBIS platform was used to visualize (concurrently) conventional T1-weighted and T2-weighted anatomical datasets, as well as 3D time-of-flight (TOF) angiography and susceptibility-weighed imaging (SWI) venography datasets co-registered to the anatomical data. Visualization features offered out-of-the-box in IBIS allowed researchers to create 2D min/max intensity projections and 3D cerebrovascular models using advanced SWI-TOF contrast toward enhancing visualization of fine cerebral vasculature, especially in comparison with standard angiography protocols based on injection of gadolinium contrast [52].
Furthermore, the IBIS plugin API enabled the creation of custom navigation applications dedicated to DBS and SEEG that were used by neurosurgeons to validate trajectories automatically predicted by the software [52, 55]. Most importantly, IBIS is an OR ready platform that can be used for conducting prospective experiments. Preliminary results on 8 DBS cases revealed that the use of IBIS can influence the neurosurgeon’s decision making [56]. For 7 out of 8 cases, the surgeon performed actual lead insertion (prospectively) according to surgical plans made with the experimental IBIS platform. Three of these plans differed by more than \(15^{\circ }\) in lead orientation from those originally planned using a commercial system.
Discussion and conclusion
In this paper, we have presented the IBIS platform as an essential component needed to bring the results of IGNS research from the lab to the OR. We have demonstrated that the platform includes all the components needed to implement an improved workflow for IGNS. Furthermore, as each of those components is implemented in the system as plugins, it is easy to replace them when new methods become available. We have shown that the patient can initially be linearly registered with an accuracy comparable with that obtained with commercial systems (\(3.72\pm 1.27\,\hbox {mm})\) and that an updated patient model can be obtained within less than 20 s by capturing tracked US images, reconstructing a 3D volume and using this volume to automatically realign preoperative plans. These steps can be done under constant AR visualization with a small reprojection error (\(0.37\pm 0.19\,\hbox {mm})\). Furthermore, we have shown the platform is flexible enough to accommodate new types of applications such as path planning for deep electrode implantation.
While general and flexible platforms such as MITK and 3D Slicer are essential for tech savvy researchers to rapidly implement bold new ideas, the IBIS platform sacrifices only some flexibility to provide a more streamlined workflow that is ready for the OR. IGI technologies, such as US-based navigation and AR, have been extensively investigated but seem to have a hard time finding their way to commercial navigation systems and routine use in the OR. A platform such as IBIS will facilitate this transition by providing an environment that is sufficiently integrated to be used routinely by clinical staff in long-term clinical studies while being flexible enough to allow researchers to fine-tune various aspects of the workflow such as image registration, visualization and interaction.
The CustusX platform provides a level of integration similar to IBIS, though with a different focus. While CustusX is dedicated to explore all the possibilities of intraoperative US for a wide range of probes and several type of surgeries, IBIS focuses specifically on neurosurgery and is mostly interested in leveraging the added value of combining iUS and AR. Because of this difference in focus, both initiatives will continue to coexist while trying to increase as much as possible their collaboration.
One of the main weaknesses of our system is the limited range of hardware devices that it supports. Currently, only NDI 3D tracking systems are supported and US is acquired through a video capture card without a possibility to acquire RF signal. Another limitation of the platform is the absence of a formal testing system. After having run for several hundred hours over consecutive OR visits without a crash, IBIS can now be considered stable. However, formal tests that run automatically every time a change is made to the code are still required to insure ongoing stability. Both of these limitations are part of the motivation to release the code as open source. A single laboratory does not have the resources to develop and maintain a complex testing infrastructure and support a wide range of hardware devices on its own. We hope that the availability of the code will bring more contributors and increase the flexibility and stability of the platform.
An important challenge for the IGI research community is to share an increasing amount of code while maintaining a sufficient level of diversity and solutions for researchers working at the different levels of integration described in this paper. It is not yet clear how this integration will happen; however, the release of the IBIS platform as open-source software simultaneously with the publication of this paper is an essential step to advance this integration. With this release, all systems that are part of the Image-Guided Systems Inter Operability (IGSIO)Footnote 3 initiative are now open source, which will facilitate code sharing and standardization.
Future work
Future development of the IBIS platform will focus more on the features that distinguish it from other platforms such as innovative AR visualization and the combination of AR and iUS and less on basic technical aspects of IGI systems such as interfacing with new hardware and software platforms. To achieve this, we will adopt the OpenIGTLink communication protocol as well as the meta-protocol that will result from the IGSIO initiative. By allowing all types of relevant data to easily flow between different open-source platforms, these protocols will allow developers of IBIS to stop maintaining certain technical parts of the code and rely on modules already implemented in other platforms. For example, in the near future, we will replace IBIS’ hardware module that handles optical tracking and video capture with the Plus Toolkit that supports a much wider range of devices and is maintained by a large community. In the long term, more functionality could similarly be offloaded to common platforms that already implement them and thus allow IBIS developers to focus on the most innovative aspects of their software.
From a user’s point of view, the IBIS platform will start to be employed for long-term clinical studies. Development of innovative techniques will take place mostly in new plugins, while existing validated and evaluated components will form the basis of a stable system that will demonstrate the value of iUS and AR on the outcome for patient and thus lead to a wider adoption of those techniques.
Notes
Presently, in May 2016, only Linux and OSX operating systems are actively maintained.
References
Kersten-Oertel M, Jannin P, Collins DL (2013) The state of the art of visualization in mixed reality image guided surgery. Comput Med Imaging Graph 37:98–112. doi:10.1016/j.compmedimag.2013.01.009
Ince DC, Hatton L, Graham-Cumming J (2012) The case for open computer programs. Nature 482:485–488. doi:10.1038/nature10836
Owens B (2016) Montreal institute going ‘open’ to accelerate science. Science 351:329–329. doi:10.1126/science.351.6271.329
Owens B (2016) Data sharing: access all areas. Nature 533:S71–S72
Enquobahrie A, Cheng P, Gary K, Ibanez L, Gobbi D, Lindseth F, Yaniv Z, Aylward S, Jomier J, Cleary K (2007) The image-guided surgery toolkit IGSTK: an open source C++ software toolkit. J Digit Imaging 20(Suppl 1):21–33. doi:10.1007/s10278-007-9054-3
Tokuda J, Gregory S, Yaniv Z, Cheng P, Blevins J, Golby AJ, Kapur T, Pieper S, Burdette EC, Fischer GS, Papademetris X, Ibanez L, Liu H, Arata J, Fichtinger G, Tempany CM, Hata N (2009) OpenIGTLink: an open network protocol for image-guided therapy environment. Int J Med Robot Comput Assist Surg 5:423–434. doi:10.1002/rcs.274
Lasso A, Heffter T, Rankin A, Pinter C, Ungi T, Fichtinger G (2014) PLUS: open-source toolkit for ultrasound-guided intervention systems. IEEE Trans Biomed Eng 61:2527–2537. doi:10.1109/TBME.2014.2322864
Nolden M, Zelzer S, Seitel A, Wald D, Müller M, Franz AM, Maleike D, Fangerau M, Baumhauer M, Maier-Hein L, Maier-Hein KH, Meinzer H-P, Wolf I (2013) The Medical Imaging Interaction Toolkit: challenges and advances?: 10 years of open-source development. Int J Comput Assist Radiol Surg 8:607–620. doi:10.1007/s11548-013-0840-8
Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin J-C, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward S, Miller JV, Pieper S, Kikinis R (2012) 3D slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging 30:1323–1341. doi:10.1016/j.mri.2012.05.001
Franz AM, Seitel A, Servatius M, Zöllner C, Gergel I, Wegner I, Neuhaus J, Zelzer S, Nolden M, Gaa J, Mercea P, Yung K, Sommer CM, Radeleff BA, Schlemmer H-P, Kauczor H-U, Meinzer H-P, Maier-Hein L (2012) Simplified development of image-guided therapy software with MITK-IGT. In: Holmes DR III, Wong KH (eds) SPIE Med. International Society for Optics and Photonics, Imaging, p 83162J
Clarkson MJ, Zombori G, Thompson S, Totz J, Song Y, Espak M, Johnsen S, Hawkes D, Ourselin S (2015) The NifTK software platform for image-guided interventions: platform overview and NiftyLink messaging. Int J Comput Assist Radiol Surg 10:301–316. doi:10.1007/s11548-014-1124-7
Ungi T, Gauvin G, Lasso A, Yeo CT, Pezeshki P, Vaughan T, Carter K, Rudan J, Engel CJ, Fichtinger G (2016) Navigated breast tumor excision using electromagnetically tracked ultrasound and surgical instruments. IEEE Trans Biomed Eng 63:600–606. doi:10.1109/TBME.2015.2466591
Askeland C, Solberg OV, Bakeng JBL, Reinertsen I, Tangen GA, Hofstad EF, Iversen DH, Våpenstad C, Selbekk T, Langø T, Hernes TAN, Olav Leira H, Unsgård G, Lindseth F (2015) CustusX: an open-source research platform for image-guided therapy. Int J Comput Assist Radiol Surg. doi:10.1007/s11548-015-1292-0
Chamberland M, Whittingstall K, Fortin D, Mathieu D, Descoteaux M (2014) Real-time multi-peak tractography for instantaneous connectivity display. Front Neuroinform 8:59. doi:10.3389/fninf.2014.00059
Chamberland M, Bernier M, Fortin D, Whittingstall K, Descoteaux M (2015) 3D interactive tractography-informed resting-state fMRI connectivity. Front Neurosci 9:1–15. doi:10.3389/fnins.2015.00275
Wolfgang P (1994) Design patterns for object-oriented software development. Addison-Wesley, Boston
Horn BKP (1987) Closed-form solution of absolute orientation using unit quaternions. J Opt Soc Am A 4:629. doi:10.1364/JOSAA.4.000629
Gerard IJ, Hall J, Mok K, Collins DL (2015) New protocol for skin landmark registration in image-guided neurosurgery. Neurosurgery. doi:10.1227/NEU.0000000000000868
Stieglitz LH, Fichtner J, Andres R, Schucht P, Krähenbühl A-K, Raabe A, Beck J (2013) The silent loss of neuronavigation accuracy: a systematic retrospective analysis of factors influencing the mismatch of frameless stereotactic systems in cranial neurosurgery. Neurosurgery 72:796–807. doi:10.1227/NEU.0b013e318287072d
Zhang Z, Member S (2000) A flexible new technique for camera calibration. IEEE Trans Pattern Anal Mach Intell 22:1330–1334. doi:10.1109/34.888718
Nelder JA, Mead R (1965) A simplex method for function minimization. Comput J 7:308–313. doi:10.1093/comjnl/7.4.308
Drouin S, Kersten-Oertel M, Collins DL (2015) Interaction-based registration correction for improved augmented reality overlay in neurosurgery. Lecture notes on comput science. Augment Environ Comput Interv 9365:21–29. doi:10.1007/978-3-319-24601-7_3
Brown RA (1979) A stereotactic head frame for use with CT body scanners. Invest Radiol 14:300–304
Comeau RM, Fenster A, Peters TM (1998) Integrated MR and ultrasound imaging for improved image guidance in neurosurgery. In: Hanson KM (ed) Med. Imaging ’98. International Society for Optics and Photonics, pp 747–754
Mercier L, Del Maestro RF, Petrecca K, Kochanowska A, Drouin S, Yan CXB, Janke AL, Chen SJ-S, Collins DL (2011) New prototype neuronavigation system based on preoperative imaging and intraoperative freehand ultrasound: system description and validation. Int J Comput Assist Radiol Surg 6:507–522
De Nigris D, Collins DL, Arbel T (2012) Multi-modal image registration based on gradient orientations of minimal uncertainty. IEEE Trans Med Imaging 31:2343–2354. doi:10.1109/TMI.2012.2218116
De Nigris D, Collins DL, Arbel T (2013) Fast rigid registration of pre-operative magnetic resonance images to intra-operative ultrasound for neurosurgery based on high confidence gradient orientations. Int J Comput Assist Radiol Surg 8:649–661. doi:10.1007/s11548-013-0826-6
Mercier L, Del Maestro RF, Petrecca K, Araujo D, Haegelen C, Collins DL (2012) Online database of clinical MR and ultrasound images of brain tumors. Med Phys 39:3253–3261. doi:10.1118/1.4709600
Heinrich MP, Jenkinson M, Papiez BW, Brady M, Schnabel JA (2013) Towards realtime multimodal fusion for image-guided interventions using self-similarities. In: International conference on medical image computing and computer-assisted intervention, pp 187–194
Wein W, Ladikos A, Fuerst B, Shah A, Sharma K, Navab N (2013) Global registration of ultrasound to MRI using the LC2 metric for enabling neurosurgical guidance. In: Medical image computing and computer intervention. Lecture notes on computer science, pp 34–41
Ferrante E, Paragios N (2013) Non-rigid 2D-3D medical image registration using markov random fields. In: Medical image computing and computer-assisted intervention, pp 163–70
Fuerst B, Wein W, Muller M, Navab N (2014) Automatic ultrasound-MRI registration for neurosurgery using the 2D and 3D LC2 metric. Med Image Anal 18:1312–1319. doi:10.1016/j.media.2014.04.008
Farnia P, Ahmadian A, Shabanian T, Serej ND, Alirezaie J (2015) Brain-shift compensation by non-rigid registration of intra-operative ultrasound images with preoperative MR images based on residual complexity. Int J Comput Assist Radiol Surg 10:555–562. doi:10.1007/s11548-014-1098-5
Ferrante E, Fecamp V, Paragios N (2015) Slice-to-volume deformable registration: efficient one-shot consensus between plane selection and in-plane deformation. Int J Comput Assist Radiol Surg 10:791–800. doi:10.1007/s11548-015-1205-2
Rivaz H, Karimaghaloo Z, Collins DL (2014) Self-similarity weighted mutual information: a new nonrigid image registration metric. Med Image Anal 18:343–358. doi:10.1016/j.media.2013.12.003
Rivaz H, Karimaghaloo Z, Fonov VS, Collins DL (2014) Nonrigid registration of ultrasound and MRI using contextual conditioned mutual information. IEEE Trans Med Imaging 33:708–725. doi:10.1109/TMI.2013.2294630
Mercier L, Araujo D, Haegelen C, Del Maestro RF, Petrecca K, Collins DL (2013) Registering pre- and postresection 3-dimensional ultrasound for improved visualization of residual brain tumor. Ultrasound Med Biol 39:16–29. doi:10.1016/j.ultrasmedbio.2012.08.004
Rivaz H, Collins DL (2015) Near real-time robust non-rigid registration of volumetric ultrasound images for neurosurgery. Ultrasound Med Biol 41:574–587. doi:10.1016/j.ultrasmedbio.2014.08.013
Mercier L, Fonov V, Haegelen C, Del Maestro RF, Petrecca K, Collins DL (2012) Comparing two approaches to rigid registration of three-dimensional ultrasound and magnetic resonance images for neurosurgery. Int J Comput Assist Radiol Surg 7:125–136. doi:10.1007/s11548-011-0620-2
Kersten-Oertel M, Chen SJ, Collins DL (2014) An evaluation of depth enhancing perceptual cues for vascular volume visualization in neurosurgery. IEEE Trans Vis Comput Graph 20:391–403. doi:10.1109/TVCG.2013.240
Drouin S, Kersten-oertel M, Chen SJ, Collins DL (2012) A realistic test and development environment for mixed reality in neurosurgery. In: Augmented environments for computer-assisted interventions, pp 13–23
Kersten-Oertel M, Chen SSJ, Drouin S, Sinclair DS, Collins DL (2012) Augmented reality visualization for guidance in neurovascular surgery. Stud Health Technol Inform 173:225–229. doi:10.3233/978-1-61499-022-2-225
Kersten-Oertel M, Gerard I, Drouin S, Mok K, Sirhan D, Sinclair DS, Collins DL (2015) Augmented reality in neurovascular surgery: feasibility and first uses in the operating room. Int J Comput Assist Radiol Surg 10:1823–1836. doi:10.1007/s11548-015-1163-8
Kersten-Oertel M, Gerard I, Drouin S, Mok K, Sirhan D, Sinclair D, Collins DL (2014) Augmented Reality in Neurovascular Surgery: First Experiences. In: Augmented environments for computer-assisted interventions. Lecture notes on computer science, vol 8678, pp 80–89
Kersten-oertel M, Gerard IJ, Drouin S, Mok K, Sirhan D, Sinclair D, Collins DL (2015) Augmented reality for specific neurovascular tasks. In: Augmented environments for computer-assisted interventions. Lecture notes on computer science, vol 9365, pp 92–103
Gerard IJ, Kersten-Oertel M, Drouin S, Hall J a., Petrecca K, De Nigris D, Arbel T, Collins DL (2015) Improving patient specific neurosurgical models with intraoperative ultrasound and augmented reality visualizations in a neuronavigation environment. In: Workshop on clinical image-based procedures: translational research in medical imaging. Lecture notes on computer science, pp 28–35
Langston TH, Kevin TF, Holly LT, Foley KT (2007) Image guidance in spine surgery. Orthop Clin North Am 38:451–461. doi:10.1016/j.ocl.2007.04.001
Yan CXB, Goulet B, Pelletier J, Chen SJ-S, Tampieri D, Collins DL (2011) Towards accurate, robust and practical ultrasound-CT registration of vertebrae for image-guided spine surgery. Int J Comput Assist Radiol Surg 6:523–537. doi:10.1007/s11548-010-0536-2
Yan CXB, Goulet B, Chen SJ-S, Tampieri D, Collins DL (2012) Validation of automated ultrasound-CT registration of vertebrae. Int J Comput Assist Radiol Surg 7:601–610. doi:10.1007/s11548-011-0666-1
Yan CXB, Goulet B, Tampieri D, Collins DL (2012) Ultrasound-CT registration of vertebrae without reconstruction. Int J Comput Assist Radiol Surg 7:901–909. doi:10.1007/s11548-012-0771-9
Bériault S, Al Subaie F, Collins DL, Sadikot AF, Pike GB (2012) A multi-modal approach to computer-assisted deep brain stimulation trajectory planning. Int J Comput Assist Radiol Surg 7:687–704. doi:10.1007/s11548-012-0768-4
Bériault S, Sadikot AF, Alsubaie F, Drouin S, Collins DL, Pike GB (2014) Neuronavigation using susceptibility-weighted venography: application to deep brain stimulation and comparison with gadolinium contrast. J Neurosurg 0:1–11. doi:10.3171/2014.3.JNS131860
Bériault S, Xiao Y, Collins D, Pike G (2015) Automatic SWI venography segmentation using conditional random fields. IEEE Trans Med Imaging 34:2478–2491. doi:10.1109/TMI.2015.2442236
Bériault S, Xiao Y, Bailey L, Collins DL, Sadikot AF, Pike GB (2012) Towards computer-assisted deep brain stimulation targeting with multiple active contacts. Med Image Comput Comput Assist Interv 15:487–494
Zelmann R, Beriault S, Marinho MM, Mok K, Hall JA, Guizard N, Haegelen C, Olivier A, Pike GB, Collins DL (2015) Improving recorded volume in mesial temporal lobe by optimizing stereotactic intracranial electrode implantation planning. Int J Comput Assist Radiol Surg 10:1599–1615. doi:10.1007/s11548-015-1165-6
Bériault S, Drouin S, Sadikot AF, Xiao Y, Collins DL, Pike GB (2013) A prospective evaluation of computer-assisted deep brain stimulation trajectory planning. In: Clinical image-based procedures from planning to intervention. Lecture notes on computer science, vol 7761, pp 42–49
Association WM (2013) World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA 310:2191–2194. doi:10.1001/jama.2013.281053
Acknowledgments
This work was financed by the Fonds Québécois de la recherche sur la nature et les technologies, the Canadian Institute of Health Research (MOP-97820) and the Natural Science and Engineering Research Council of Canada.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Simon Drouin, Anna Kochanowska, Marta Kersten-Oertel, Ian J. Gerard, Rina Zelmann, Dante De Nigris, Silvain Bériault, Tal Arbel, Denis Sirhan, Abbas F. Sadikot, Jeffery A. Hall, David S. Sinclair, Kevin Petrecca, Rolando F. DelMaestro and D. Louis Collins Collins declare that they have no conflict of interest.
Ethical standards
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008 [57].
Informed consent
Informed consent was obtained from all patients for being included in the study.
Rights and permissions
About this article
Cite this article
Drouin, S., Kochanowska, A., Kersten-Oertel, M. et al. IBIS: an OR ready open-source platform for image-guided neurosurgery. Int J CARS 12, 363–378 (2017). https://doi.org/10.1007/s11548-016-1478-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-016-1478-0