Abstract
Skin lesion is one of the severe diseases which in many cases endanger the lives of patients on a worldwide extent. Early detection of disease in dermoscopy images can significantly increase the survival rate. However, the accurate detection of disease is highly challenging due to the following reasons: e.g., visual similarity between different classes of disease (e.g., melanoma and non-melanoma lesions), low contrast between lesions and skin, background noise, and artifacts. Machine learning models based on convolutional neural networks (CNN) have been widely used for automatic recognition of lesion diseases with high accuracy in comparison to conventional machine learning methods. In this research, we proposed a new preprocessing technique in order to extract the region of interest (RoI) of skin lesion dataset. We compare the performance of the most state-of-the-art CNN classifiers with two datasets which contain (1) raw, and (2) RoI extracted images. Our experiment results show that training CNN models by RoI extracted dataset can improve the accuracy of the prediction (e.g., InceptionResNetV2, 2.18% improvement). Moreover, it significantly decreases the evaluation (inference) and training time of classifiers as well.

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
As the growing advance of deep learning, a numerous number of tasks have been solved by artifact intelligence (A.I). Especially, the demand of A.I for medical images has become emerging in recent years, since with the early detection of the disease, we are now able to provide a better treatment plans. However, the main issue related to medical image classification is due to the lack of enough number of sample images. Skin is the largest organ of the body that contains lots of information about the individual’s health condition and also their identity [1]. Skin lesion is a serious disease that, if not diagnose in a proper time, may lead to detrimental consequences. There are many sources for skin image datasets, among them, the International Skin Imaging Collaboration (ISIC) provides public datasets that are mainly used for skin lesion classification [2,3,4,5,6]. Image segmentation is one of the most important computer vision tasks, which is used to partition a given image into multiple segments. The main objective of segmentation is to locate objects of interest and its boundary in order for enabling more efficient and effective further analysis. Segmentation has been widely investigated and implemented in many works [7,8,9].
There are many machine learning methods for object segmentation. One of the well-known methods for object semantic segmentation tasks is U-Net [10]. In this method, the network can be trained on both the original and augmented dataset. This characteristic is primarily appropriate when the target datasets are from medical fields (mostly limited) since data augmentation enriches training samples. Likewise, [11]’s residual multitasking network achieved second place (among 28 teams) in ISBI 2016 Skin Lesion Analysis Towards Melanoma Detection Challenge segmentation task [3]. This model includes more than 50 layers with residual layers, separated into two sub-architectures for classification and segmentation.
Another promising method, namely fully convolutional network (FCNs), is introduced by [12]. This deep neural architect aims to localize the coarse approximation in the early learning stage; then, the exact approximation will be learned later. Besides, the author also introduced a fusion framework to facilitate their model’s performance. The final model achieved 90.66% in the PH2 dataset and 91.18% in ISIC 2016.
In [13], an end-to-end training procedure has been proposed that utilizes the Jaccard Distance loss. The model includes 19 layers which were trained thoroughly by their proposed loss function. Although the result is not outstanding for more challenging samples (involving hairs, badges, poor lightning condition, etc.), their approach outperforms the [3] and [12] within the same datasets.
The first attempt towards multi-class segmentation on ISBI 2016 was conducted by [14], enabling segmentation with classes’ information. The sequential learning method involved Faster-RCNN and U-Net in [15] also tackles the same segmentation task. In [16] a fully resolution convolutional network for learning visual representation from skin lesion images, reaching 77.11% Jaccard Index on ISIC 2017 private test set has been proposed.
In this study, we investigated the effectiveness of ROI extraction that comes after segmentation step to improve the performance of the classification task. We have experimented and evaluated recently developed methods of semantic segmentation so as to isolate and extract the RoI (lesion) of the images. It enables us to remove unwanted background image and artifacts such as hairs and badges before training CNN models.
2 Material and method
We started our study by a light-weight non-training-based segmentation method, then to have better result, we extended our study by implementing a complex training-based segmentation method. One of the non-training-based segmentation algorithm that we investigated is Otsu’s thresholding [17] which clusters the background (skin) and the foreground (lesions) based on the optimal threshold from the histogram of the pixel counts. Several previous works utilizing Otsu’s thresholding segmentation due to its simplicity, for example H&E staining images [18], MRI and CT scan images [19], and also melanoma lesions detection [20]. However, the main assumption of this segmentation method relies on the histogram of pixel counts, which is assumed to be bimodal distribution. Hence, the performance of Otsu’s segmentation on noisy images that possess badges, hairs, and black borders is unsatisfactory.
The second approach for non-training-based segmentation is K-means clustering based on the color spaces [21]. The unsupervised cluster took three inputs: (1) two components of three color spaces: Hue which is related to the color’s position on a color wheel, Cr and Cb which are the blue-difference and red-difference chroma components of an image, (2) pixel-based features, and (3) rough estimation of skin’s boundary gained from color-based classifier. In both segmentation algorithms, we have used Jaccard index which is used for evaluating segmentation performance as follows
where A corresponds to the ground truth binary segmentation mask, B represents the predicted binary segmentation mask, |A ∩ B| is area of overlap and |A ∪ B| is area of union [22]. We used ISIC-2017 which contains 2000 skin lesion samples with 2000 corresponding masks as a ground truth to evaluate the performance of these two algorithms [23]. Although K-means clustering method was well-performed than Otsu’s thresholding segmentation by having Jaccard Index of 76.2% in compare to 71.7%, it is still not able to eliminate artifacts efficiently. However, our experiments over skin dataset showed that both non-training-based approaches cannot segment images very well, since artifacts like badges are more often than not segmented as skin lesions. In addition, K-means approach does not consider the border of the region. We observed that ground truth mask of skin lesion are mostly solid closed-contour shape, however the K-means approach does not give solid shape. In other words, there are some small dark region inside a detected lesion contour. This issue is due to the fact that the algorithm tries to separate pixels into multiple clusters based on mean, and regardless of the position and value of near pixels. Figure 1 shows two samples of the ISIC-2017 that have been segmented based on K-means approach. The red circles inside lesion contour show the disability of this approach for detecting whole lesion part.
To address this issue, and regarding the availability of 2000 masks of ISIC-2017, we followed our investigation by evaluating training-based approaches. One of the most common training-based approaches for image segmentation is “U-net” convolutional neutral network [10]. U-net architecture is an evolution of traditional convolutional neutral network, which is so-called end-to-end fully convolutional network (FCN). The architecture includes two parts: (1) the contraction path (the encoder) and (2) the symmetric expanding path (the decoder). The encoder is basically a conventional CNN, which is a sequence of matrix operations (convolutional layers, max pooling, batch normalization, and so on). The main modification from conventional CNN is lied on the decoder - successive expanding path, where the upsampling operator is used instead of pooling operation. Thus, the resolution of outputs increase along as these layers. The features from the encoder are then combined with upsampled output in order to enable a precise localization. Since fully connected layers are absent from U-net architecture, the outputs of the network are segmentation maps in which represents the mask of lesion in corresponding image.
By using the train data (images and their corresponding masks), the FCN is able to segment the lesion without segmenting artifacts as a lesion part. However, the database for training segmentation task is rarely available since it requires expertise of related fields. Medical segmentation tasks often involve objects with varying size, ranging from cell nuclei [10], lung [24], retina vessels [25], and tumors [26]. Especially in dermoscopy images, the RoI often results in irregular shapes and varying sizes. Thus, the demand for a stable network which is robust to a wide scale of image sizes is necessary for further analysis. In this work, we have adopted the state-of-the-art MultiResUNet architecture which integrated the idea of Residual Inception blocks [27]. By utilizing multiple kernels with different size in parallel fashion, the MultiResUNet outperforms the conventional U-Net architecture by 5.065% in skin lesion boundary segmentation. From our experimental results, the MultiResUNet segmentation network overcomes both Otsu’s thresholding and K-means segmentation method due to its strong suit that is built on a expertise-involved training data. Within the scope of this paper, we used ISIC 2017 [23] database for skin lesion boundary segmentation. The MultiResUNet is trained by 2000 images along with corresponding masks produced by dermoscopic experts from ISIC 2017. We then selected the best segmentation model with Jaccard Index of 80.4 for segmenting 23331 remaining images of ISIC 2019.
2.1 RoI extraction
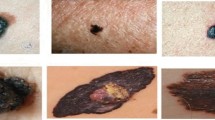
The image size of skin lesion dataset is various and large. Due to the computational limitations of CNN models, the input/output layers’ size of segmentation model is fixed and smaller than that of the raw image. For example, in Figs. 2 and 3 first and second rows, the raw image size is 682 × 1024 × 3, 1024 × 768 × 3, and 1024 × 682 × 3 pixels, respectively. But the size of input/output layers of segmentation model is fixed to 224 × 224 pixels. It means, regardless of the raw image’s size, the output segmented image’s size is 224 × 224 × 3 pixels. Moreover, by observing the segmented images in Fig. 3, it is obvious that many pixels are black (detected as background) and only a small number of pixels are related to the lesion part. If we directly input segmented image to a classifier, those black pixels do not contribute much to the classification tasks. Also, since the legion part is highly down-sampled, some critical information could be lost. To verify our assumptions, we trained and evaluated multiple CNN models based on segmented dataset, but their accuracy was much lower (up to 10%) than that of the raw dataset.
To overcome this issue, we developed an algorithm to extract the RoI from raw images based on derived masks. As illustrated in Fig. 2, the output mask is first resized to the same size of the raw image, then contour’s structural analysis was applied to detect locations of bounding rectangle box (green rectangle). After finding the rectangle of RoI, we extract this part of the raw image and input it to the classifier. In Fig. 3, two samples are depicted to show how our approach can focus on the most important part of the skin image. This approach can prevent to input useless pixels (black pixels) to CNN models, and the input lesion part would have more information.
In this study, the importance of extracting RoI on the performance of the CNN-based classifiers is investigated. While CNNs have been widely used for skin lesion classification [27,28,29,30], to the best of our knowledge, there is no work about the effect of RoI extraction before training CNN models. In next section, we discuss in more details about CNNs, and it followed by the experimental settings and results.
3 Classification
3.1 Transfer learning
Unlike traditional machine learning in which an expert needs to observe the target and extract good reliable features from it based on his knowledge, deep learning methods extract reliable high-quality features from large amounts of targets automatically which makes them more beneficial than traditional methods. Consequently, deep learning methods are highly dependent on mass data since they need a large amount of data to have a reliable comprehension of the patterns of the dataset. The more dataset a deep learning network has, the bigger it should be to extract well-behaved features. It means that for getting a really good performance from a deep neural network, it needs a large amount of data that requires a big network to be capable of understanding the patterns of them.
In deep neural networks, some of the final layers are responsible for making a decision related to the task and the rest of them can be used to extract high-level features. Lack of sufficient data is one of the main problems that researchers usually face when they want to train a model on specific data. In the biomedical image classification tasks, the problem is more severe since it is much harder to find a large amount of a high-quality dataset. Transfer learning is addressing this problem and is a solution. In traditional machine learning, we should consider the fact that training data must be independent and identically distributed with the test data. However, transfer learning assumes that training data and test data do not need to be independent and identically distributed entries. It means that for a specific task, the network is not required to train from scratch which has two specific benefits: first, it eliminates the requirement of accessing a big dataset and second, it reduces the time of training the network.
CNN pre-trained models usually trained on large image classification tasks. Convolutional layers are responsible for extracting high-level features from an image, while dense layers must decide on those features. There are two kinds of transfer learning: feature extraction and fine-tuning. In the former, the convolutional layer parameters are being frozen during back-propagation and are used to extract features on a new dataset and the new dense layer is added to fit the network for the new dataset. In the latter, after adding a new dense layer, relax back-propagation will be done on the whole parameters for tuning them with the new dataset. Pre-trained models by ImageNet have been widely used for skin lesion classification[31,32,33]. We also tested this property and found that if we initialize the models with ImageNet pre-traiend wights, the training process converges in much fewer number of epochs while maintaining the higher accuracy. Also, we used data augmentation by randomly rotating the training images up to 90 degrees, and flipping them horizontally.
3.2 Deep learning models
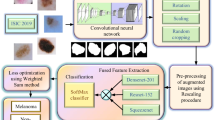
Since 2012, when Alex Krizhevsky et al. introduced the AlexNet [34], CNN models which were not able to absorb attention came back to the play and in the next few years, many researchers and experts tried to come up with new deep neural networks in the similar way to improve the accuracy on different tasks [35,36,37,38,39,40,41,42]. We have used some of these pre-trained networks via transfer learning in our work to evaluate the effectiveness of our proposed method. We have used InceptionResNetV2 [35], Xception [37], InceptionV3 [38], DenseNet [40], ResNet-152 [41], and VGG19 [42] which each of them has different architecture in the number of fully connected layers and convolutional layers.
All of these man-made models have tried to look at different problems and are designed by some experts since it requires a suitable selection of architectures that need a high-quality knowledge in machine learning as well as it is a tedious and time-consuming task. Moreover, for different targets, different architectures should be designed to get a better result. However, some works named the neural architecture search(NAS) are introduced recently that address this problem and try to find a good architecture for a certain target automatically which is logically suitable to use for different types of image classification. In our work, we also tried to use some of these networks which are trained well on ImageNet dataset to evaluate our work. We have used two networks EfficientNet [36] and NasNet [39] which both are automatically designed by Google brain team members.
4 Experiments and results
We used ISIC-2019 dataset that contains 25331 dermoscopic images belong to 8 classes: melanoma,melanocytic nevus,basal cell carcinoma,actinic keratosis,benign keratosis, dermatofibroma, vascular lesion, and squa-mous cell carcinoma. On the other hand, ISIC-2017 dataset contains 2000 samples with masks for skin lesion segmentation and classification. These 2000 samples of ISIC-2017 are exactly available in ISIC-2019 as well. We used those 2000 samples and their masks to train the MultiResUNet model, and we did not involve them for the evaluation. We set the input/output size of segmentation model to 224 × 224 pixels. After training MultiResUNet model for 50 epochs, we generated 23,331 segmented images (2000 samples out of 25,331 were excluded). The average required time for segmenting each image was 11 ms (Ms). Then, we applied our RoI extraction algorithm in order to focus on the main information of the image. After doing so, two sets of image were generated for our experiments: one contained 23,331 raw images and the other had 23,331 corresponding RoI extracted images. We randomly split the 23,331 samples into three sub-sets, training (18,890 samples), validation (2101 samples), and test (2340 samples). We applied same data augmentation from Keras framework over both training datasets by randomly rotating the images up to 90 degrees and horizontally flipping them.
Python was used as the programming language. We used Keras, a high-level neural networks API which is written in Python and is able to run on top of TensorFlow. All models were directly selected from Keras documanetation and the defualt input size of first layer was chosen according to the Keras documantation [43]. Keras framework provides models that can be converted to tensorflow lite (TFLite) format. TFLite models can be deployed over android operating system. We used a single GPU (Nvidia GeForce GTX 1080 Ti with 11 GB GDDR5X memory) for all of our experiments (training and evaluation).
We have done several experiments in order to demonstrate the effectiveness of the segmentation and RoI extraction before the classification. To have a fair comparison, all input and hyper parameters were set the same for each model while training raw or segmented datasets. We used learning rate 0.001, batch size 8 for the NasNet model, and batch size 16 for the rest of the models. Default input size of each model was used (refer to Table 1). Each model was trained for 50 epochs and 1000 steps per epoch.
In Table 1, we evaluated the accuracy of each trained model over test dataset. The results show that extracting the RoI prior to classification can improve the accuracy of skin lesion diagnosis. Table 1 only presents the results based on the default size of each network where we obtained the best accuracy of each model. However, we trained each model with smaller size as well. The smaller input size gave lower accuracy but higher improvement.
Sicne skin lesion contains 8 unbalanced classes, overall accuracy may not completely convey the effectiveness of our approach. Hence, to evaluate the impact of our approach over each class separately, we used F1-score which can yield a more realistic measure of the classifier’s performance. It avoids to be mislead by the average accuracy which can be wrongly obtained from a very poor precision or very high recall. Figure 4 shows the F1-score of two diseases, “melanoma” and “nevus,” and also weighted-average over all classes. It is obvious that proposed approach can improve the performance of almost all classifiers.
In Table 2, the required time for predicting the label of each input sample has been reported. It is evident that the inference time of segmented data is much lower than that of the raw data. To have stable results, we repeated our experiments 10 times over the test dataset and then the average of results is calculated. In Table 3, the required time for training each model has been evaluated for both datasets. It shows that segmented data can be trained in shorter period of time. Although training and generating segmented image take time, but in certain cases, we generate dataset one time and use it for training many models. For examples, in ensemble learning, many models are used to calculate the best accuracy [44]. If we use segmented dataset, it would significantly decrease the time of training, and as a result, saving more power and computational resources. In other words, by using segmented images, not only the required times for training all models would be decreased but also the required time for evaluation would be decreased accordingly. The size of raw dataset (23331 samples) is 9.4 GB (gigabyte) while the size of segmented dataset is 1.4 GB. It means on overage, the size of a segmented image is less than one-sixth of a raw sample. This property would be important if we need to send the data over a communication channel. For example, in a remote classification task, client may send the image to a remote server for doing more reliable classification. By doing a segmentation over the client-side (e.g., android device), we would be able to reduce the communication bandwidth.
5 Conclusion
In this study, we proposed a new preprocessing technique to separate the RoI section from unwanted background of skin lesion. To this end, we used one of the state-of-the-art segmentation algorithm, MultiResUnet, to firstly segment the skin lesion image. Then, we found the bounding box around the lesion and cropped that part of image. We applied this preprocessing over whole ISIC-2019 except 2000 samples of this dataset which had been used for training MultiresUnet model. This preprocessing enabled us to only input the most important part of image to the classifiers for both training and evaluation steps. Our investigation over different CNN models showed that if we train models with RoI extracted dataset, the accuracy of models would be increased, and the training and inference time would be dropped. This is intuitively due to the fact that we remove unwanted background and only input most important part of skin image to the classifiers. In this study, our results were based on Keras models, and we used ImageNet pre-retrained weights to train all classifiers. We trained the models based on their default input resolution size (based on Keras Documentaion). As the discussion above revealed, focusing on RoI part of image can improve the performance of skin lesion classification. Instead of segmentation technique, object detection algorithm can also be used to find the RoI of skin lesion. However, there is no available annotated dataset for training object detection algorithm. But, converting the masks labels of ISIC-2017 to annotated dataset would enable us to train an object detection algorithm. I can accelerate the RoI extraction phase, and potentially improving the final accuracy of classifiers.
References
Zeinali B, Ayatollahi A, Kakooei M (2014) A novel method of applying directional filter bank (dfb) for finger-knuckle-print (fkp) recognition. In: 2014 22nd Iranian conference on electrical engineering (ICEE), pp 500–504
Codella N, Rotemberg V, Tschandl P, Celebi ME, Dusza S, Gutman D, Helba B, Kalloo A, Liopyris K, Marchetti M et al (2019) Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (isic). arXiv:1902.03368
Gutman D, Codella NC, Celebi E, Helba B, Marchetti M, Mishra N, Halpern A (2016) Skin lesion analysis toward melanoma detection: a challenge at the international symposium on biomedical imaging (isbi) 2016, hosted by the international skin imaging collaboration (isic). arXiv:1605.01397
Tschandl P, Rosendahl C, Kittler H (2018) The ham10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci Data 5:180161
Codella NC, Gutman D, Celebi ME, Helba B, Marchetti MA, Dusza SW, Kalloo A, Liopyris K, Mishra N, Kittler H et al (2018) Skin lesion analysis toward melanoma detection: a challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In: 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018), IEEE, pp 168–172
Combalia M, Codella NC, Rotemberg V, Helba B, Vilaplana V, Reiter O, Halpern AC, Puig S, Malvehy J (2019) Bcn20000: dermoscopic lesions in the wild. arXiv:1908.02288
Isensee F, Kickingereder P, Wick W, Bendszus M, Maier-Hein KH (2017) Brain tumor segmentation and radiomics survival prediction: contribution to the brats 2017 challenge. In: International MICCAI brainlesion workshop. Springer, pp 287–297
Chen L-C, Papandreou G, Kokkinos I, Murphy K, Yuille AL (2017) Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans Pattern Anal Mach Intell 40(4):834–848
Ng H, Ong S, Foong K, Goh P, Nowinski W (2006) Medical image segmentation using k-means clustering and improved watershed algorithm. In: 2006 IEEE southwest symposium on image analysis and interpretation, IEEE, pp 61–65
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention, Springer, pp 234–241
Yu L, Chen H, Dou Q, Qin J, Heng P. -A. (2016) Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans Med Imag 36(4):994–1004
Bi L, Kim J, Ahn E, Kumar A, Fulham M, Feng D (2017) Dermoscopic image segmentation via multistage fully convolutional networks. IEEE Trans Biomed Eng 64(9):2065–2074
Yuan Y, Chao M, Lo Y-C (2017) Automatic skin lesion segmentation using deep fully convolutional networks with jaccard distance. IEEE Trans Med Imaging 36(9):1876–1886
Goyal M, Yap MH, Hassanpour S (2017) Multi-class semantic segmentation of skin lesions via fully convolutional networks. arXiv:1711.10449
Vesal S, Patil SM, Ravikumar N, Maier AK (2018) A multi-task framework for skin lesion detection and segmentation. In: OR 2.0 Context-aware operating theaters, computer assisted robotic endoscopy, clinical image-based procedures, and skin image analysis, Springer, pp 285–293
Soudani A, Barhoumi W (2019) An image-based segmentation recommender using crowdsourcing and transfer learning for skin lesion extraction. Expert Syst Appl 118:400–410
Zhang J, Hu J (2008) Image segmentation based on 2d otsu method with histogram analysis. In: 2008 International conference on computer science and software engineering, vol 6. IEEE, pp 105–108
Haggerty JM, Wang XN, Dickinson A, O’Malley CJ, Martin EB (2014) Segmentation of epidermal tissue with histopathological damage in images of haematoxylin and eosin stained human skin. BMC Med Imaging 14(1):7
Bindu CH, Prasad KS (2012) An efficient medical image segmentation using conventional otsu method. Int J Adv Sci Technol 38(1):67–74
Premaladha J, Ravichandran K (2016) Novel approaches for diagnosing melanoma skin lesions through supervised and deep learning algorithms. J Med Syst 40(4):96
Buza E, Akagic A, Omanovic S (2017) Skin detection based on image color segmentation with histogram and k-means clustering. In: 2017 10th International conference on electrical and electronics engineering (ELECO), pp 1181–1186
McGuinness K, O’Connor NE (2010) A comparative evaluation of interactive segmentation algorithms. Pattern Recogn 43(2):434–444. interactive Imaging and Vision. [Online]. Available: http://www.sciencedirect.com/science/article/pii/S0031320309000818
Berseth M (2017) Isic 2017-skin lesion analysis towards melanoma detection. arXiv:1703.00523
Zhao T, Gao D, Wang J, Tin Z (2018) Lung segmentation in ct images using a fully convolutional neural network with multi-instance and conditional adversary loss. In: 2018 IEEE 15th International symposium on biomedical imaging (ISBI 2018), IEEE, pp 505–509
Xiao X, Lian S, Luo Z, Li S (2018) Weighted res-unet for high-quality retina vessel segmentation. In: 2018 9th International conference on information technology in medicine and education (ITME), IEEE, pp 327–331
Li X, Chen H, Qi X, Dou Q, Fu C-W, Heng P-A (2018) H-denseunet: hybrid densely connected unet for liver and tumor segmentation from ct volumes. IEEE Trans Med Imaging 37(12):2663–2674
Ibtehaz N, Rahman MS (2020) Multiresunet: Rethinking the u-net architecture for multimodal biomedical image segmentation. Neural Netw 121:74–87
Jafari MH, Karimi N, Nasr-Esfahani E, Samavi S, Soroushmehr SMR, Ward K, Najarian K (2016) Skin lesion segmentation in clinical images using deep learning. In: 2016 23rd International conference on pattern recognition (ICPR), IEEE, pp 337–342
Kawahara J, Hamarneh G (2016) Multi-resolution-tract cnn with hybrid pretrained and skin-lesion trained layers. In: International workshop on machine learning in medical imaging, Springer, pp 164–171
Saba T, Khan MA, Rehman A, Marie-Sainte SL (2019) Region extraction and classification of skin cancer: a heterogeneous framework of deep cnn features fusion and reduction. J Med Syst 43(9):289
Mahbod A, Schaefer G, Wang C, Ecker R, Dorffner G, Ellinger I (2020) Investigating and exploiting image resolution for transfer learning-based skin lesion classification
Hosny KM, Kassem MA, Foaud MM (2018) Skin cancer classification using deep learning and transfer learning. In: 2018 9th Cairo international biomedical engineering conference (CIBEC), pp 90–93
Adegun AA, Viriri S (2020) Deep learning-based system for automatic melanoma detection. IEEE Access 8:7160–7172
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: NIPS
Szegedy C, Ioffe S, Vanhoucke V, Alemi AA (2017) Inception-v4, inception-resnet and the impact of residual connections on learning. In: Thirty-first AAAI conference on artificial intelligence
Tan M, Le QV (2019) Efficientnet: rethinking model scaling for convolutional neural networks. arXiv:1905.11946
Chollet F (2016) Xception: Deep learning with depthwise separable convolutions. arXiv:1610.02357
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2015) Rethinking the inception architecture for computer vision. arXiv:1512.00567
Zoph B, Vasudevan V, Shlens J, Le QV (2017) Learning transferable architectures for scalable image recognition. arXiv:1707.07012
Huang G, Liu Z, van der Maaten L, Weinberger KQ (2016) Densely connected convolutional networks. arXiv:1608.06993
He K, Zhang X, Ren S, Sun J (2015) Deep residual learning for image recognition. arXiv:1512.03385
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556
Chollet F, et al. (2015) Keras. https://keras.io, [accessed April 1 2020]
Liu Y, Yao X (1999) Ensemble learning via negative correlation. Neural Netw 12(10):1399–1404
Acknowledgements
Effort sponsored in whole or in part by United States Special Operations Command (USSOCOM), under Partnership Intermediary Agreement No. H92222-15-3-0001-01. The U.S. Government is authorized to reproduce and distribute reprints for Government purposes notwithstanding any copyright notation thereon.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
The authors used the data publicly available for their study and did not collect data from any human participant or animal.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Disclaimer
The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of the United States Special Operations Command
Rights and permissions
About this article
Cite this article
Zanddizari, H., Nguyen, N., Zeinali, B. et al. A new preprocessing approach to improve the performance of CNN-based skin lesion classification. Med Biol Eng Comput 59, 1123–1131 (2021). https://doi.org/10.1007/s11517-021-02355-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-021-02355-5