Abstract
Preclinical data show that autophagy delays age-related disease. It has been postulated that age-related disease is—at least in part—caused by an age-related decline in autophagy. However, autophagic flux has never been measured in humans across a spectrum of aging in a physiologically relevant context. To address this critical gap in knowledge, the objective of this cross-sectional observational study was to measure basal autophagic flux in whole blood taken from people at elevated risk of developing type 2 diabetes and correlate it with chronological age. During this study, 119 people were recruited and five people were excluded during sample analysis such that 114 people were included in the final analysis. Basal autophagic flux measured in blood and correlations with parameters such as age, body weight, fat mass, AUSDRISK score, blood pressure, glycated hemoglobin HbA1c, blood glucose and insulin, blood lipids, high-sensitivity C-reactive protein, plasma protein carbonylation, and plasma β-hexosaminidase activity were analysed. Despite general consensus in the literature that autophagy decreases with age, we found that basal autophagic flux increased with age in this human cohort. This is the first study to report measurement of basal autophagic flux in a human cohort and its correlation with age. This increase in basal autophagy could represent a stress response to age-related damage. These data are significant not only for their novelty but also because they will inform future clinical studies and show that measurement of basal autophagic flux in a human cohort is feasible.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The autophagic pathway is relevant to human age-related disease. Autophagy has two core cellular functions: (i) regenerating nutrients from macromolecules in response to nutrient deprivation; and (ii) acting as a key form of cell quality control by sequestering damaged cellular material and delivering it to the lysosome for degradation by powerful hydrolase enzymes. In this way, autophagy limits cell damage and slows biological aging [1], and autophagic dysfunction itself has recently been discussed as a hallmark of aging [2]. Reduction of basal autophagy limits tumor-free survival and lifespan in mice [3, 4], and increasing autophagy has the opposite effect [5]. Appropriate activation of autophagy is also important for maintaining metabolic health, including glucose tolerance and insulin resistance [6, 7] and so likely influences diabetes risk.
It is generally accepted in the research community that autophagic flux (acquisition of unwanted or damage intracellular cargo, its transport to, and destruction in the lysosome) decreases with age [8,9,10], which leads to the emergence of age-related diseases. However, a more detailed examination of the literature reveals a complex picture. It has long been known that lysosomal proteolytic capability decreases with age in the invertebrate model C elegans [11]. However, studies on whether autophagy changes with age in C. elegans have shown conflicting results [12, 13]. Research on autophagy in rodent aging has revealed further detail on how autophagy could change with age in humans. Basal autophagy measured by proteolytic release of valine in rodent hepatocytes changes relatively little, except in extreme old age when it decreases. This contrasts with the ability for proteolysis to increase in response to nutrient or pharmacological stress, which deteriorates dramatically with age from midlife in the rodent [14,15,16]. More recent research has shown autophagic flux decreases with rodent age in CD8+ T cells, liver, heart, kidney, and skeletal muscle [5, 17, 18], which could be due to age-related increases in expression of the autophagy inhibitor RUBCN (rubicon autophagy regulator) [19]. The exception to this is white adipose tissue, where RUBCN expression drops with age and autophagic flux increases [7]. Basal autophagic flux measured using autophagy adaptor SQSTM1/P62 (sequestosome 1) in mouse peripheral blood mononuclear cells (PBMCs) showed increased basal autophagic flux with age (21–22 week old compared with 105–106 week old mice) using a method that preserved physiological integrity of the sample. Interestingly, this effect was not observed in cardiomyocytes [20]. Taken together, regardless of consensus, basal autophagy appears to be less sensitive to aging than stimulated autophagy, and autophagy may increase or decrease with age in a tissue-dependent manner.
Research on autophagy and aging in human blood cells does show differences between young and old participants but is subject to methodological caveats. Bektas and colleagues [21] found no difference in the number of autolysosomes with age in CD4+ T lymphocytes; on the other hand, autophagosome numbers increased with aging in lymphocytes. They interpreted this as a decrease in clearance of autophagosomes but did not provide flux analyses to support this interpretation. Phadwal and colleagues [22] showed reduced overlap between MAP1LC3B/LC3B (microtubule associated protein 1 light chain 3 beta) and the acidic compartment in CD8+ T cells with aging. However, in the same experiment, lysosomal inhibition with protease inhibitors failed to increase overlap between LC3B and the acidic compartment in both young and old participants, rendering interpretation of this study difficult. McCormick and colleagues [23] found that PBMCs cultured in artificial media from older people had increased basal autophagic flux compared with younger people, however, PBMC responsiveness to stimulation of autophagy by rapamycin (a pro-autophagic mechanistic target of rapamycin complex 1 inhibitor) was diminished. This observed differential in sensitivity to aging between basal and stimulated autophagy is consistent with observations from rat studies [15]. Using a limited number of participants, Raz and colleagues [24] showed that autophagic flux in activated isolated CD4+ T cells decreased with age. Although images of LC3B puncta in resting CD4+ T cells were displayed in the paper, quantification of these cells was not presented.
Altogether, these data suggest that basal autophagy appears insensitive or at least less sensitive to age-related decline than stimulated autophagy. Stimulated autophagy (from nutrient restriction, pharmacological activation or cellular stimulation) appears to be very sensitive to age-related decline. Unsurprisingly, whereas high quality data from animals or animal-derived primary culture are available, data from humans has been generated in a way where flux is impossible to infer, and always with low numbers of participants. How basal autophagic flux changes with age in humans is consequently an important gap in knowledge that needs to be addressed if we are to understand how to change it to increase human healthspan, which currently ends on average in the seventh decade of human life [25]. We therefore adapted our recently devised whole blood assay [26] for autophagic flux for analysis with enzyme-linked immunosorbent assay (ELISA). The objective of this study was to measure basal autophagic flux in a cohort of humans to determine how autophagy changes with age. In so doing, we would demonstrate the feasibility of basal autophagic flux measurement and generate data that will be used in future power calculations. In a cross-sectional cohort of 114 people, we measured autophagic flux and determined that basal autophagic flux increased between the ages of 35–74 years. This increase in basal autophagy could represent a stress response to age-related damage.
Methods
Study design and setting
In this report, we present baseline characteristics of a sub-cohort of participants enrolled into an open-label, three-arm, parallel group sequential randomized controlled trial which was conducted between 26 September 2018 and 30 November 2021 [27, 28]. This sub-analysis included eligible people enrolled from 9 May 2019 to 6 March 2020. Samples from 119 participants were collected for this cross-sectional study. Study size was determined by our access to study participants over the dates specified.
The inclusion and criteria were: aged 35–75 years, weight-stable (< 5% fluctuation in their body weight for past 6 months at study entry); a score 12 or greater on the AUSDRISK calculator; HbA1c < 48 mmol/mol (measured at screening).
The Australian Type 2 Diabetes Risk Assessment Tool (AUSDRISK) requires demographic, lifestyle and anthropometric data. It assesses the risk of developing type 2 diabetes within 5 years for Australian adults [29]. A score of 12 or greater means a person is at high risk of developing type 2 diabetes (where one in 14 will develop diabetes in next 5 years) and it was used as an inclusion criterion for this study.
The exclusion criteria were: a personal history/diagnosis (self-reported) of diabetes (type 1 or 2); major psychiatric disorders (e.g. schizophrenia, major depressive disorder, bipolar disorder, and eating disorders); gastrointestinal disorders and previous gastric surgery (e.g. gastric band, gastric bypass) that may affect appetite; haematological disorders (e.g. thalassemia, iron-deficiency anaemia), insomnia, any other medical condition that is deemed unstable by the study physician; uncontrolled asthma, current fever, upper respiratory infections; current intake of > 140 g alcohol/week; current smokers of cigarettes/cigars/marijuana/e-cigarettes/vaporisers; current intake of any recreational drugs; taking medications that reduce weight or affect appetite or gut motility or lower blood glucose or antidiabetic medications (e.g. metformin, sulfonylureas, GLP-1 receptor agonists [e.g. exenatide], thiazolidinediones or dipeptidyl peptidase-IV (DPP-IV) inhibitors [e.g. ‘gliptins’]), orlistat, phentermine, topiramate, lorcaserin; donated blood within 3 months prior to commencement on trial; unable to comprehend study protocol due to English language or cognitive difficulties; unable to provide informed consent.
The detailed study protocol including detailed inclusion/exclusion criteria was reported previously [27, 28]. The study was registered on ClinicalTrials.gov, NCT03689608. Ethics approval was obtained from the Central Adelaide Local Health Network Human Research Ethics Committee (HREC/18/CALHN/171) and participants provided written informed consent. The study was performed at the South Australian Health and Medical Research Institute (SAHMRI) by researchers from The University of Adelaide and SAHMRI.
Participant involvement
Patients were not involved in the design or dissemination of this study.
Visits and measured outcomes
Variables measured in this study included basal autophagic flux, body weight, body mass index (BMI), waist circumference, body composition (fat mass and fat-free mass), blood pressure (systolic and diastolic), blood lipids, glycated hemoglobin (HbA1c), fasting blood glucose and insulin, plasma high-sensitivity C-reactive protein (hs-CRP), plasma protein carbonylation, and plasma β-hexosaminidase (βHex) activity.
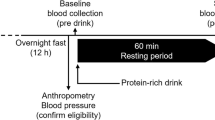
Participants arrived at 7:30am after an overnight 12-h fast and fasting blood samples were drawn. During the visit, body weight and waist circumference were measured in a gown after voiding. Blood pressure was measured after a 10-min seated rest. Whole body composition was measured by dual-energy x-ray absorptiometry (Lunar Prodigy; GE Healthcare, Madison, Wisconsin, USA).
Basal autophagic flux measurement by ELISA – rationale and experimental detail
Measurement of basal autophagic flux in the current study uses lysosomal inhibition and measurement of an autophagic cargo in a way that has been optimized for use in whole human blood [26]. The rationale behind using a lysosomal inhibitor to measure autophagic flux is as follows. A sample (in this case living whole human blood) is split into two tubes. One tube receives a lysosomal inhibitor (in this case chloroquine) and this stops the lysosomal degradation of incoming autophagic cargo. The other tube does not receive a lysosomal inhibitor. Both tubes are incubated at 37 °C for 1 h to allow time for incoming autophagic cargo to meet the lysosome. At this point in the process, we extract PBMCs for analysis of autophagic cargo.
One autophagic protein that is degraded at a relatively high rate is LC3B [26]. LC3B is present as a soluble cytosolic form (LC3B-I) or an autophagosome-bound form that is covalently conjugated to a membrane lipid (LC3B-II). The lipid bound LC3B-II is a lysosomal substrate, whereas the cytosolic LC3B-I form is not. To enrich samples for the lysosomal substrate LC3B-II, we gently permeabilize the plasma membrane with saponin and wash the soluble LC3B-I protein away. Western blot analysis for LC3B of cell pellets that have been washed with saponin are shown in Supplementary Fig. 1. Western blots were performed as described in Bensalem et al. [26]. LC3B-II is left behind in the cell pellet. It is this cell pellet that we analyse by ELISA for abundance of LC3B-II. If autophagy was occurring in the sample, then LC3B-II (being a lysosomal substrate) will be present in a higher abundance in the tube that was incubated with the lysosomal inhibitor than in the tube that was not incubated with a lysosomal inhibitor. In this paper, we use ELISA to quantitatively measure LC3B-II in cells extracted from both tubes. By subtracting the amount of LC3B-II measured from the uninhibited tube from the amount of LC3B-II measured in the inhibited tube, we have a quantitative measure for autophagic flux.
Specific experimental details are as follows. Fasted blood was collected in lithium heparin tubes (Greiner Bio-One, Vacuette tube 9 mL lithium heparin; 455,084) and processing was started within an hour. Blood samples were split into two 3 mL aliquots and were placed into 2 different 10 mL conical centrifuge tubes. One tube was treated with 150 µM chloroquine (CQ, chloroquine diphosphate; Sigma Aldrich; C6628) (3 µL of 50 mM CQ solution diluted in sterile water per mL of blood); the other remained untreated to act as a control. Both tubes were incubated at 37 °C for 60 min with rotation on a tube rotator (ThermoFisher Scientific Tube Revolver, 88881001) at 10 rpm. After incubation, blood samples were immediately put on ice. From there, samples were maintained cold on ice or at 4 °C during centrifugation, and cold buffers were used to stop or prevent vesicle trafficking during further processing. Samples were then diluted 1:1 with cold Dulbecco’s phosphate buffered saline (DPBS, GIBCO, ThermoFisher Scientific; 14190250). PBMCs were then isolated using Lymphoprep (4 mL, Stemcell Technologies; 07811) and tubes were centrifuged for 30 min at 800 × g, with brake off, at 4 °C. PBMCs were collected and washed in DPBS for a final volume of 5 mL and centrifuged at 600 × g for 10 min at 4 °C. The cell pellet was resuspended in 1 mL cold red blood cell lysis buffer (1X, BD Biosciences; 555899), incubated on ice for 2 min, then pelleted by centrifugation at 600 × g for 5 min at 4 °C. PBMC pellets were washed by resuspension in 5 mL of cold DPBS and pelleted again by centrifugation. The pelleted cells were resuspended in 1 mL of cold DPBS and transferred to a 1.5 mL microcentrifuge tube. This tube was centrifuged at 2,000 × g for 10 min at 4 °C. The supernatant was discarded and the pellet containing PBMCs was snap frozen on dry ice and stored at -80 °C until analysis.
PBMCs were thawed on ice and resuspended in 0.05% saponin (Sigma Aldrich, SAE0073) in DPBS containing protease and phosphatase inhibitors (0.1% sodium deoxycholate; 0.1% SDS; 140 mM NaCl; 2.5 mM sodium pyrophosphate; 1 mM sodium orthovanadate; 1 mM β-glycerophosphate; EDTA-free Protease Inhibitor Cocktail (Sigma-Aldrich; 4693132001))and centrifuged (20 °C, 13,000 × g, 5 min). The saponin wash gently permeabilized the cells to remove LC3B-I without affecting the membrane-associated LC3B-II [30]. Pellets were then washed in DPBS, centrifuged, and resuspended in a lysis buffer containing protease and phosphatase inhibitors (10 mM Tris, pH 7.0; 1 mM EDTA; 0.5 mM EGTA; 1% Triton X-100; 0.1% sodium deoxycholate; 0.1% SDS; 140 mM NaCl; 2.5 mM sodium pyrophosphate; 1 mM sodium orthovanadate; 1 mM β-glycerophosphate; EDTA-free Protease Inhibitor Cocktail (Sigma-Aldrich; 4693132001)). Cell suspensions were then sonicated (30 V, 20 s × 2 on ice). Cell lysates were centrifuged at 16,000 × g for 10 min at 4 °C and the supernatant collected. Total protein was measured using a micro BCA protein assay kit (ThermoFisher Scientific; 23235).
To determine LC3B-II sample concentration, samples were loaded in triplicate on a FastScan Total LC3B ELISA Kit (Cell Signaling Technology, 35172). A standard curve was made using recombinant human LC3B (Abcam, ab103506) prepared in the concentration range of 0 – 4 ng/mL in assay diluent. The absorbance of each well was measured at 450 nm using the Glomax plate reader (Promega, Wisconsin, USA). Standard curve results were constructed using sigmoidal fit and the unknown protein concentrations of each sample were interpolated and reported as ng LC3B-II/mg of total protein. Any sample with triplicate values giving a coefficient of variation (CV) > 10% was checked for outliers; obvious triplicate outliers were removed, and any final sample with a CV > 15% was excluded. Five samples (three controls and two CQ), out of 238 (119 participants ± CQ) were excluded for a final N = 114. To reduce bias the person excluding data based on these criteria was blind to the identity of these samples until after the data was excluded.
Basal autophagic flux for each participant was calculated as follows—ΔLC3B-II = (LC3B-II +CQ)—(LC3B-II -CQ) in ng of LC3B-II/mg of total protein/hour.
Other metabolic measurements
Blood glucose was assayed by the hexokinase method (Cobas Integra 400 Plus, Roche Diagnostics, Switzerland). Plasma insulin was measured by radioimmunoassay (HI-14 K, Millipore, Burlington, MA, USA). Whole blood HbA1c, hs-CRP, plasma triglycerides, high-density lipoprotein (HDL) and low-density lipoprotein (LDL) were measured using commercially available enzymatic kits on an automated clinical analyzer (Indiko Plus, Thermo Fisher Scientific Inc., Finland). Plasma β-hexosaminidase activity was measured as described previously [31, 32]. Results are reported as pmol/min/mg and calculated by comparing the average fluorescent readings of each sample to the average of the known standard according to the following equation: β-hexosaminidase activity [pmol/min/mg] = (fluorescence units (FU) sample – FU blank) / FU standard x standard amount [2,840 pmol] / incubation time [30 min] / volume of sample loaded [5 µL] x dilution factor [5] × 1000 / plasma protein concentration [mg/mL]. Total plasma protein was measured using a micro BCA protein assay kit (ThermoFisher Scientific; 23235).
Plasma protein carbonylation was measured using a protein carbonyl ELISA kit (Abcam; ab238536) according to the manufacturer’s instructions.
Statistical analysis
Baseline characteristics are presented as mean ± standard deviation (SD) for continuous measures and N (%) for discrete measures. Data normality was assessed using the Shapiro-Wilk test. Basal autophagic flux ΔLC3B-II did not follow a normal distribution and neither did most of the parameters assessed (including age), therefore these data were analyzed using non-parametric tests, i.e. Spearman correlation, Kruskal-Wallis test followed by Dunn’s multiple comparisons test or Mann-Whitney test. For normally distributed comparisons, unpaired t-test were performed. Missing data were not imputed (one missing value for hs-CRP, N = 113). The association between basal autophagic flux and age was assessed with linear regression models with sex, BMI, AUSDRISK score, and systolic blood pressure as confounders. Statistical analysis and graphs were performed using GRAPHPAD PRISM, version 9.4.1, for Windows (GraphPad Software, La Jolla, CA, USA) and a heatmap graph was generated using Microsoft Excel for Microsoft 365 MSO Version 2209 Build 16.0.15629.20200, 64-bit (Microsoft Corporation, Redmond, WA, USA). Figures were compiled using Microsoft PowerPoint for Microsoft 365 MSO Version 2209 Build 16.0.15629.20200, 64-bit (Microsoft Corporation, Redmond, WA, USA).
Results
Participant characteristics and sex differences
A total of 119 participant samples were collected and included in this analysis. Data from 5 participants were excluded due to high levels of variation within triplicate repeats due to sample processing error (defined above). Data exclusion was performed blind to sample identity. Participant characteristics are presented in Table 1 for all participants (N = 114) included in the final analysis set and compared between sex. We observed sex differences for most of the parameters measured, however, neither basal LC3B-II nor basal autophagic flux (ΔLC3B-II) displayed sex differences (p = 0.6795 and p = 0.3537, respectively) (Table 1).
Basal autophagic flux correlates with age, AUSDRISK, and systolic blood pressure
Basal autophagic flux was tested for correlation with participants’ demographic and metabolic characteristics (presented in Table 1). Spearman correlations showed that basal autophagic flux (ΔLC3B-II) positively correlated with age (p = 0.0015, r = 0.2943), AUSDRISK (p = 0.0406, r = 0.1921) and systolic blood pressure (p = 0.0177, r = 0.2219) (Fig. 1A, B). AUSDRISK, which has an age component in the scoring system, and systolic blood pressure both positively correlated with age (respectively, p < 0.0001, r = 0.3831; p = 0.0162, r = 0.2221) (Fig. 1A). Basal autophagic flux was analyzed per age quartiles which showed an increase with age (test for linear trend: p-trend = 0.0116), with the last quartile (66–74 years) driving most of the effect, older participants having higher basal autophagic flux, (Fig. 1C), as observed with the Spearman correlation (Fig. 1B). Multivariate analysis (Table 2) showed that age was associated with increased basal autophagic flux (model 1) after adjustment for sex and BMI (model 2) and after further adjustment for AUSDRISK score and systolic blood pressure (model 3). AUSDRISK score was not independently associated with increased basal autophagic flux after adjustment for sex and BMI (p = 0.4598) and after further adjustment for age and systolic blood pressure (p = 0.5883). Systolic blood pressure was associated with increased basal autophagic flux after adjustment for sex and BMI (p = 0.0310) but was no longer associated after further adjustment for age and AUSDRISK score (p = 0.0931). Summary data for variables described in Fig. 1 are presented in Supplementary Table 1.
Basal autophagic flux in human PBMCs positively correlates with age. A A heat map showing statistical significance for the correlation between variables in the cross-sectional cohort. Basal autophagic flux is shown at the bottom of the table and is highlighted in red. Statistical significance was calculated using Spearman correlations, N = 114 (one missing value for hs-CRP, N = 113). B A scatter plot showing a significant positive correlation between basal autophagic flux and age. Data points are shaded corresponding to age-related quartiles. Statistical significance was determined using Spearman correlation N = 114. C A histogram showing the same data presented in panel B across age split into quartiles where bars = mean ± SEM. Statistical significances denoted by stars were calculated using a Kruskal-Wallis test followed by a Dunn’s multiple comparisons test
Discussion
In this study we measured basal autophagic flux in PMBCs using lysosomal inhibition in whole blood to preserve a physiologically relevant environment. After adjustment for BMI, sex, AUSDRISK score, and systolic blood pressure, basal autophagy was correlated with chronological age. Although basal autophagy correlated with AUSDRISK score, after adjustment for sex and BMI, basal autophagy and AUSDRISK score were no longer correlated. Similarly, the correlation between basal autophagy and systolic blood pressure was no longer observed after adjustment for sex, BMI, AUSDRISK score, and age. Basal autophagic flux did not differ between sexes. This is interesting as sex-dependent differences in autophagy have been observed in animals [33, 34].
Observation of a small increase in basal autophagic flux with age is important for several reasons. Firstly, we need to understand why basal autophagy is increasing. We hypothesize that basal autophagy is increasing to cope with age-related macromolecular damage. While we did not demonstrate an increase in carbonylated (oxidized) protein with age in this cohort, or any correlation with autophagy, previous studies have demonstrated increasing macromolecular damage with aging [35,36,37]. Further, in this study we only assessed one marker of oxidative damage (protein carbonylation) and this does not exclude the accumulation of other markers of damage when basal autophagy is low. Other oxidative modifications of proteins could be analysed in future [38]. Secondly, the observation that autophagy increases with age is important because it demonstrates that people at the end of their healthspan in the seventh and eighth decades of life [25] have robust basal autophagic flux that is at least comparable or even superior to basal autophagic flux measured in younger people within the age range investigated in this study, and with a similar risk of developing diabetes. Based on our observations, it appears that basal autophagy does respond to aging, however, not in the direction we anticipated. This is not to say that basal autophagic flux is increased in age-related diseases: for example, ample preclinical and human data indicate that autophagic flux is not functional in the Alzheimer disease brain [39,40,41]. However, it is striking that basal autophagic flux measured in human blood in a physiologically relevant way does not appear to decrease during normal aging.
Of note, fasting insulin did not correlate with basal autophagy. This is noteworthy because insulin is known to suppress proteolysis in humans [42]. While it is still possible that healthy individuals possess a negative correlation between insulin and basal autophagy, we cannot assess that relationship in the current study where participants were obese on average. This is because age and obesity positively correlate with insulin resistance [43, 44], which is the inability for cells to respond to changes in insulin [45]. Taken together, this means that a simple negative correlation between insulin and basal autophagy is unlikely to exist in a cohort where insulin resistance also likely exists.
Although the current study uses methodology that has been developed to best represent physiological basal autophagic flux, several limitations must be considered. We measured basal autophagic flux in PBMCs. It is probable that basal autophagy in different tissues responds to aging in different ways, as previously described in mice [7]. Further, basal autophagy was measured in PBMCs; it is known that subpopulations in this pool change composition with aging [46]. For example, within CD4+ T lymphocytes, naïve cells decrease with human aging, whereas central memory cells increase. This compositional change within PBMCs could result in an apparent change in basal autophagic flux that was not a change in autophagic flux per se.
For measurement of basal autophagic flux in whole blood, we used chloroquine. Chloroquine is used because it inhibits lysosomal degradation, and allows the build up of autophagic cargo for determination of flux. It may do this by preventing autophagosomes from fusing to lysosomes, and its mechanism of action is distinct from v-type ATPase inhibitor bafilomycin [47]. However, it also has side effects. For example, it may disrupt Golgi apparatus and early endosomes, and cause lipidation of LC3 onto single membrane vesicles [47, 48]. Although to our knowledge this phenomenon has yet to be explored in PBMCs, this may cause overestimation of the amount of basal autophagy occurring in this cell pool in this study. The reason for using chloroquine in whole blood is because it is soluble in water, unlike other lysosomal inhibitors such as bafilomycin. This is important as vehicles used to solubilize other lysosomal inhibitors disrupt the integrity of whole blood [26], destroying the physiological nature of the assay used in the current study.
This study was also a cross-sectional study, meaning that factors such as survivorship bias could be influencing what we observe. If survivorship bias was influencing our results, it could mean that people who had relatively less basal autophagy as a function of age were screened out of the original study design as they were already further along the path to disease [27, 28]. This would bias us to observe an increase in basal autophagy with age. Further to this, the study cohort was, on average, obese with an elevated risk of diabetes, and people with hypertension and high cholesterol were also enrolled. It is possible that basal autophagic flux would respond differently to aging in a cohort that were not overweight or at elevated risk of diabetes. This may impact the generalizability of the results and data generation from cohorts across a wide range of BMI will be important.
Basal autophagy in this study was only measured using LC3B. We did not assess MAP1LC3A/LC3A (microtubule associated protein 1 light chain 3 alpha) or MAP1LC3C/LC3C (microtubule associated protein 1 light chain 3 gamma). These three paralogs all localize to the autophagosome but are not completely functionally equivalent [49]. It is possible that use of one of one the paralogs over another could alter measurement of basal autophagy. However, the reason we used LC3B is because we trialed LC3A, LC3B, and LC3C in our original investigation into measurement of autophagy in the context of whole blood, and LC3B was the only one of the three proteins to yield a useful signal for autophagic flux using our methodology [26].
In conclusion, we have demonstrated that basal autophagic flux positively correlated with age in a cohort of people with an elevated risk of diabetes. This is the largest study (N = 114) to analyze basal autophagic flux in humans using physiologically relevant methodology. Data generated here has demonstrated that measurement of basal autophagic flux is feasible in humans and will be useful for designing future studies that look to measure basal autophagic flux and its impact on aging and disease in humans. Although this study is subject to some caveats, we show for the first time that basal autophagic flux does not decrease with aging across human healthspan in people living with being overweight or obese. This goes against the current dogma that basal autophagy decreases with age. In fact, we observed that basal autophagy increases with age in blood and we speculate that this could be in response to age-related damage and a mechanism that prevents development of age-related disease.
Data availability
The data associated with this study available from the corresponding author upon reasonable request.
Abbreviations
- AUSDRISK:
-
Australian type 2 diabetes risk assessment tool
- βHex:
-
β-Hexosaminidase
- BMI:
-
Body mass index
- ELISA:
-
Enzyme-linked immunosorbent assay
- HbA1c:
-
Glycated hemoglobin
- hs-CRP:
-
High-sensitivity C-reactive protein
- MAP1LC3B/LC3B:
-
Microtubule associated protein 1 light chain 3 beta
- PBMCs:
-
Peripheral blood mononuclear cells
References
Carosi JM, Fourrier C, Bensalem J, Sargeant TJ. The mTOR-lysosome axis at the centre of ageing. FEBS Open Bio. 2021;12:739–57.
Schmauck-Medina T, Molière A, Lautrup S, Zhang J, Chlopicki S, Madsen HB, et al. New hallmarks of ageing: a 2022 Copenhagen ageing meeting summary. Aging (Albany NY). 2022;14:6829–39.
Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20.
Cassidy LD, Young ARJ, Young CNJ, Soilleux EJ, Fielder E, Weigand BM, et al. Temporal inhibition of autophagy reveals segmental reversal of ageing with increased cancer risk. Nat Commun. 2020;11:307.
Fernández ÁF, Sebti S, Wei Y, Zou Z, Shi M, McMillan KL, et al. Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature. 2018;558:136–40.
Lim YM, Lim H, Hur KY, Quan W, Lee HY, Cheon H, et al. Systemic autophagy insufficiency compromises adaptation to metabolic stress and facilitates progression from obesity to diabetes. Nat Commun. 2014;5:4934.
Yamamuro T, Kawabata T, Fukuhara A, Saita S, Nakamura S, Takeshita H, et al. Age-dependent loss of adipose Rubicon promotes metabolic disorders via excess autophagy. Nat Commun. 2020;11:4150.
Barbosa MC, Grosso RA, Fader CM. Hallmarks of aging: an autophagic perspective. Front Endocrinol (Lausanne). 2018;9:790.
Escobar KA, Cole NH, Mermier CM, VanDusseldorp TA. Autophagy and aging: Maintaining the proteome through exercise and caloric restriction. Aging Cell. 2019;18:e12876.
Leidal AM, Levine B, Debnath J. Autophagy and the cell biology of age-related disease. Nat Cell Biol. 2018;20:1338–48.
Sarkis GJ, Ashcom JD, Hawdon JM, Jacobson LA. Decline in protease activities with age in the nematode Caenorhabditis elegans. Mech Ageing Dev. 1988;45:191–201.
Chapin HC, Okada M, Merz AJ, Miller DL. Tissue-specific autophagy responses to aging and stress in C. elegans. Aging (Albany NY). 2015;7:419–34.
Chang JT, Kumsta C, Hellman AB, Adams LM, Hansen M. Spatiotemporal regulation of autophagy during Caenorhabditis elegans aging. Elife. 2017;6:e18459.
Vittorini S, Paradiso C, Donati A, Cavallini G, Masini M, Gori Z, et al. The age-related accumulation of protein carbonyl in rat liver correlates with the age-related decline in liver proteolytic activities. J Gerontol A Biol Sci Med Sci. 1999;54:B318–23.
Donati A, Cavallini G, Paradiso C, Vittorini S, Pollera M, Gori Z, et al. Age-related changes in the regulation of autophagic proteolysis in rat isolated hepatocytes. J Gerontol A Biol Sci Med Sci. 2001;56:B288–93.
Del Roso A, Vittorini S, Cavallini G, Donati A, Gori Z, Masini M, et al. Ageing-related changes in the in vivo function of rat liver macroautophagy and proteolysis. Exp Gerontol. 2003;38:519–27.
Sebti S, Zou Z, Shiloh MU. BECN1(F121A) mutation increases autophagic flux in aged mice and improves aging phenotypes in an organ-dependent manner. Autophagy. 2023;19:957–65.
Puleston DJ, Zhang H, Powell TJ, Lipina E, Sims S, Panse I, et al. Autophagy is a critical regulator of memory CD8(+) T cell formation. Elife. 2014;3:e03706.
Nakamura S, Oba M, Suzuki M, Takahashi A, Yamamuro T, Fujiwara M, et al. Suppression of autophagic activity by Rubicon is a signature of aging. Nat Commun. 2019;10:847.
Walter S, Jung T, Herpich C, Norman K, Pivovarova-Ramich O, Ott C. Determination of the autophagic flux in murine and human peripheral blood mononuclear cells. Front Cell Dev Biol. 2023;11:1122998.
Bektas A, Schurman SH, Gonzalez-Freire M, Dunn CA, Singh AK, Macian F, et al. Age-associated changes in human CD4(+) T cells point to mitochondrial dysfunction consequent to impaired autophagy. Aging (Albany NY). 2019;11:9234–63.
Phadwal K, Alegre-Abarrategui J, Watson AS, Pike L, Anbalagan S, Hammond EM, et al. A novel method for autophagy detection in primary cells: impaired levels of macroautophagy in immunosenescent T cells. Autophagy. 2012;8:677–89.
McCormick JJ, VanDusseldorp TA, Ulrich CG, Lanphere RL, Dokladny K, Mosely PL, et al. The effect of aging on the autophagic and heat shock response in human peripheral blood mononuclear cells. Physiol Int. 2018;105:247–56.
Raz Y, Guerrero-Ros I, Maier A, Slagboom PE, Atzmon G, Barzilai N, et al. Activation-induced autophagy is preserved in CD4+ T-cells in familial longevity. J Gerontol A Biol Sci Med Sci. 2017;72:1201–6.
Garmany A, Yamada S, Terzic A. Longevity leap: mind the healthspan gap. NPJ Regen Med. 2021;6:57.
Bensalem J, Hattersley KJ, Hein LK, Teong XT, Carosi JM, Hassiotis S, et al. Measurement of autophagic flux in humans: an optimized method for blood samples. Autophagy. 2021;17:3238–55.
Teong XT, Liu K, Hutchison AT, Liu B, Feinle-Bisset C, Wittert GA, et al. Rationale and protocol for a randomized controlled trial comparing daily calorie restriction versus intermittent fasting to improve glycaemia in individuals at increased risk of developing type 2 diabetes. Obes Res Clin Pract. 2020;14:176–83.
Teong XT, Liu K, Hutchison AT, Liu B, Feinle-Bisset C, Wittert GA, et al. An update to the study protocol for a randomized controlled trial comparing daily calorie restriction versus intermittent fasting to improve glycaemia in individuals at increased risk of developing type 2 diabetes. Obes Res Clin Pract. 2021;15:306.
Chen L, Magliano DJ, Balkau B, Colagiuri S, Zimmet PZ, Tonkin AM, et al. AUSDRISK: an Australian type 2 diabetes risk assessment tool based on demographic, lifestyle and simple anthropometric measures. Med J Aust. 2010;192:197–202.
Gottlieb RA, Andres AM, Sin J, Taylor DP. Untangling autophagy measurements: all fluxed up. Circ Res. 2015;116:504–14.
Whyte LS, Fourrier C, Hassiotis S, Lau AA, Trim PJ, Hein LK, et al. Lysosomal gene Hexb displays haploinsufficiency in a knock-in mouse model of Alzheimer’s disease. IBRO Neuroscience Reports. 2022;12:131–41.
Leaback DH, Walker PG. Studies on glucosaminidase. 4. The fluorimetric assay of N-acetyl-beta-glucosaminidase. Biochem J. 1961;78:151–6.
Fourrier C, Bryksin V, Hattersley K, Hein LK, Bensalem J, Sargeant TJ. Comparison of chloroquine-like molecules for lysosomal inhibition and measurement of autophagic flux in the brain. Biochem Biophys Res Commun. 2021;534:107–13.
Demarest TG, Waite EL, Kristian T, Puche AC, Waddell J, McKenna MC, et al. Sex-dependent mitophagy and neuronal death following rat neonatal hypoxia–ischemia. Neuroscience. 2016;335:103–13.
Wang Z, Wang Y, Liu H, Che Y, Xu Y, E L. Age-related variations of protein carbonyls in human saliva and plasma: is saliva protein carbonyls an alternative biomarker of aging? Age (Dordr). 2015;37:9781.
Jana CK, Das N, Sohal RS. Specificity of age-related carbonylation of plasma proteins in the mouse and rat. Arch Biochem Biophys. 2002;397:433–9.
Gonos ES, Kapetanou M, Sereikaite J, Bartosz G, Naparło K, Grzesik M, et al. Origin and pathophysiology of protein carbonylation, nitration and chlorination in age-related brain diseases and aging. Aging (Albany NY). 2018;10:868–901.
Syslová K, Böhmová A, Mikoška M, Kuzma M, Pelclová D, Kačer P. Multimarker screening of oxidative stress in aging. Oxid Med Cell Longev. 2014;2014:562860.
Whyte LS, Hassiotis S, Hattersley KJ, Hemsley KM, Hopwood JJ, Lau AA, et al. Lysosomal dysregulation in the murine app(NL-G-F/NL-G-F) model of Alzheimer’s disease. Neuroscience. 2020;429:143–55.
Lee JH, Yang DS, Goulbourne CN, Im E, Stavrides P, Pensalfini A, et al. Faulty autolysosome acidification in Alzheimer’s disease mouse models induces autophagic build-up of Aβ in neurons, yielding senile plaques. Nat Neurosci. 2022;25:688–701.
Hassiotis S, Manavis J, Blumbergs PC, Hattersley KJ, Carosi JM, Kamei M, et al. Lysosomal LAMP1 immunoreactivity exists in both diffuse and neuritic amyloid plaques in the human hippocampus. Eur J Neurosci. 2018;47:1043–53.
James HA, O’Neill BT, Nair KS. Insulin regulation of proteostasis and clinical implications. Cell Metab. 2017;26:310–23.
Petersen KF, Morino K, Alves TC, Kibbey RG, Dufour S, Sono S, et al. Effect of aging on muscle mitochondrial substrate utilization in humans. Proc Natl Acad Sci U S A. 2015;112:11330–4.
Gastaldelli A, Gaggini M, DeFronzo RA. Role of adipose tissue insulin resistance in the natural history of type 2 diabetes: results from the San Antonio metabolism study. Diabetes. 2017;66:815–22.
Tonks KT, Ng Y, Miller S, Coster AC, Samocha-Bonet D, Iseli TJ, et al. Impaired Akt phosphorylation in insulin-resistant human muscle is accompanied by selective and heterogeneous downstream defects. Diabetologia. 2013;56:875–85.
Kverneland AH, Streitz M, Geissler E, Hutchinson J, Vogt K, Boës D, et al. Age and gender leucocytes variances and references values generated using the standardized ONE-Study protocol. Cytometry A. 2016;89:543–64.
Mauthe M, Orhon I, Rocchi C, Zhou X, Luhr M, Hijlkema KJ, et al. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14:1435–55.
Florey O, Gammoh N, Kim SE, Jiang X, Overholtzer M. V-ATPase and osmotic imbalances activate endolysosomal LC3 lipidation. Autophagy. 2015;11:88–99.
von Muhlinen N, Akutsu M, Ravenhill BJ, Foeglein Á, Bloor S, Rutherford TJ, et al. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol Cell. 2012;48:329–42.
Acknowledgements
Project conceptualization: JB, LKHeilbronn, & TJS; Experiments & data analysis: JB, XTT, KJH, LKHein, CF, KL, ATH, LKHeilbronn, & TJS. Preparation of Figures: JB. Preparation of manuscript: JB & TJS. All authors revised and approved the manuscript. This work was supported by a Diabetes Australia Research Program Grant (Y21G-SART) and an NHMRC Ideas Grant (2002608).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
TJS and JB are listed as inventors on a related patent—Australia (Provisional) 2019903187; 2019904822; PCT/AU/2020/050908; United Kingdom GB2204321.0; USA 17/637,494.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
About this article
Cite this article
Bensalem, J., Teong, X.T., Hattersley, K.J. et al. Basal autophagic flux measured in blood correlates positively with age in adults at increased risk of type 2 diabetes. GeroScience 45, 3549–3560 (2023). https://doi.org/10.1007/s11357-023-00884-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11357-023-00884-5