Abstract
In China, harmful algal blooms (HABs) are one of the most prominent ecological disasters in the coastal areas. Among the harmful algae species that cause HABs, Karen mikimotoi is a kind of algae that appear frequently. It can secrete hemolytic toxins and fish toxins such as glycolipids and unsaturated fatty, posing a significant threat to marine life. In order to establish a fast and effective detection technology for Karen mikimotoi that can be promoted and applied on site, we have developed a test strip which is based on monoclonal antibody technology and the colloidal gold immune-chromatography assay (GICA). The experimental results show that this test strip can detect different growth stages including growth, and stable and recession period of Karen mikimotoi. The detection results can be displayed within 3–15 min. It had high sensitivity and specificity, with a detection limit of 754 cells/mL. A colorimetric card was made to further determine the concentration of algae detected. What is more, we had developed a method that can be used for on-site enrichment of algae cells using a syringe to detect lower concentrations of Karen mikimotoi, with a minimum detection concentration of 100 cells/mL. Also the test strip was used for on-site testing along the coast of China. This test strip not only serves as a warning for the outbreak of red tide, but also provides a new approach for the development of rapid detection technology for red tide algae.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Harmful algal blooms (HABs) mainly refer to an environmental abnormal phenomenon caused by the proliferation of microalgae and other organisms, which leads to the death of fish, shellfish, and other aquatic products (Furuya et al. 2018; Glibert et al. 2018). HABs may change the structure and function of the ecosystem and endanger human health and safety (Yu et al. 2018).
In recent decades, with the development of human science and technology and the improvement of productivity, some human activities have led to frequent outbreaks of harmful algal blooms in the world (Anderson et al. 2002). It has become a common ecological phenomenon that seriously endangers the ecological environment and human economy, and has aroused widespread concern (Reise and van Beusekom 2008).
Karen mikimotoi, belonging to Gymnodiniales and Karen, was a harmful algal that cause the harmful algal bloom worldwide (Hansen 2003). The outbreak area was all over the world’s offshore waters, usually occurring in spring and summer, when the sea water was obviously stratified (Brand et al. 2012). It has been proved that Karen mikimotoi is a typical ichthyotoxic species. It can produce hemolytic toxins such as glycolipids and unsaturated fatty (Gentien and Arzul 1990; Mooney et al. 2007; Parrish et al. 1998). It is lethal to both wild and cultured fish and also associated with mortality of shellfish and other invertebrates (Botes et al. 2003; Mitchell and Rodger 2007; Zou et al. 2010). Karen mikimotoi has been found in multiple parts of the world (Davidson et al. 2009).
In China, the largest scale of HABs of Karenia mikimotoi broke out in 2012, resulting in the almost death of abalone in the disaster-stricken aquaculture waters in Quanzhou, Putian, Fuzhou, and other places in southeast China (Ministry of Natural Resources of the People’s Republic of China 2009–2020). This HAB had caused more than 2 billion yuan of direct economic losses (Deng et al. 2016). In the coastal waters of China, Karenia mikimotoi has become the second largest cause of toxic and harmful algal blooms (Lu et al. 2014).
So far the most commonly used method for the detection of microalgae is based on morphological identification, using common optical microscope for microscopic inspection, but this method is time consuming, labor intensive, and difficult to achieve large-scale detection (Xiao-Kun et al. 2022). In recent years, the molecular biological technology of detecting marine microalgae from the molecular perspective has become a research hotspot such as rolling ring amplification technology (Chen et al. 2015), fluorescent quantitative PCR, in situ hybridization (Chen et al. 2011), dot acrobatics, etc. (Al-Kandari et al. 2011; Antonella and Luca 2013; Chen et al. 2013; Dyhrman et al. 2006; Nezan et al. 2014). These technologies have their own advantages in the detection of red HABs and play an important role in their respective fields (Anderson et al. 2012). But they have a common deficiency that they need special instruments to assist in the detection and require professional personnel to operate. These technologies are not suitable for daily large-scale monitoring, especially for the high-frequency detection during the peak period of HABs in China from April to June every year (Guo et al. 2015).
Golloidal gold immune-chromatography assay (GICA) is a rapid immunoassay technology developed after three major labeling technologies (fluorescein, radioisotope, and enzyme) in the 1980s (Glad and Grubb 1981; Zuk et al. 1985). It is a solid phase immunolabeling detection technology based on immunochromatography, monoclonal antibody technology, and colloidal gold labeling technology (Wan et al. 2022). Colloidal gold is a kind of gold particles with specific size polymerized by HAuCl4 under the action of reducing agents such as trisodium citrate, ascorbic acid, tannic acid, etc., and becomes a stable colloidal state due to electrostatic action (Gao et al. 2005). The principle of GICA is that colloidal gold first binds specifically to the substance to be tested, then binds to the corresponding antigen or antibody, and finally enriches in the detection band to display color. Therefore, the test results can be determined by visual observation of the presence of color (Putalun et al. 2004; Yuhi et al. 2006). Compared with other detection methods, GICA has the advantages of fast, agile, sensitive, and accurate, no need for any combination, and low cost. The technology has been rapidly promoted in many fields for the detection of immunogenic macromolecular substances such as hormones, antibodies in serum, and viruses (Sun et al. 2018). However, the application of this technology to the detection of red tide algae is relatively rare.
In this study, to develop a technology that can quickly detect harmful red tide algae and promote its application in the field, we have developed a test strip for rapid detection of Karenia mikimotoi based on GICA. The test strip only takes 3–15 min to complete the results of ELISA that take several hours to develop color. The tested samples do not require complex processing. It also has good stability, specificity, and high sensitivity, and can realize the on-site rapid detection of red tide.
Materials and methods
Cultivation and collection of Karenia mikimotoi
Karenia mikimotoi (CCMA-028) used in this study was provided by Center for Collections of Marine Algae, Xiamen University. The algae was cultured with seawater and f/2 medium (without silicon) in a 2 L conical flask at a temperature of 20 ± 1 ℃ with 55 µmol photons m−2 s−1 of a light/dark regime of 12 h/12 h in a light incubator (Laifu: PGX-330A-3H, Ningbo, China) (Guillard and Ryther 1962). The natural seawater used for algae culture was filtered through a 0.45-µm Millipore membrane and autoclaved at 121 ℃ for 30 min by High-Pressure Steam Sterilization Pot (Huxi: BXM-30R, Shanghai, China).
During cultivation, shake once a day during the culture period, and take 1 mL of algal at 16:00 every day for counting three times, and take the average (Huang et al. 2021). Samples for algal cell count were fixed with Lugol reagent, and microscopic cell count was performed under a × 10 magnification microscope (Nikon: E200MV, Nanjing, China). The number of Karenia mikimotoi was counted by a counting chamber (capacity 0.5 mL; made in Henan Taiheng Plastic Industry Co., Ltd., China) (Lu et al. 2014).
Preparation of polyclonal antibody and monoclonal antibody
Karenia mikimotoi was collected at 1700 × g for 10 min by the centrifuge (Cence: H2050R, Hunan, China) when it grew to a stable stage. After collection, the algae was washed with PBS twice and collected after centrifugation at 1700 × g for 10 min after each washing. After washing, it was resuspended with PBS to prepare immune adjuvant. Two milliliter Karenia mikimotoi (about 1 × 106 cells/mL) was mixed with the same amount of Freund’s complete adjuvant and Freund’s incomplete adjuvant (F5506—10 × 10 mL, USA), and then fully mixed with a 1-mL sterile syringe to prepare complete adjuvant immunogen and incomplete adjuvant immunogen (Li et al. 2022).
Then the prepared adjuvant was used as immunogen to immunize the Balb/c mice. The mice were immunized with complete adjuvant immunogen by multi-point injection on the back. After 10 days, the mice were immunized with incomplete adjuvant immunogen four times with an interval of 1 week. One week after the end of the last immunization with incomplete adjuvant immunogen, blood was taken from mice. After the collected blood was left at 4 ℃ overnight, the serum was obtained after centrifugation at 1120 × g rpm for 10 min. The titer of serum (polyclonal antibody) was determined by indirect ELISA.
Karenia mikimotoi was diluted to 1 × 105 cells/mL with carbonate buffer (pH 9.6) and then the algae was coated on an ELISA plate for overnight. Wash with PBST 3 times for 5 min each time. Next, seal the ELISA plate with 5% skimmed milk for 2 h at 37 ℃. After sealing, wash the plate with PBST for 3 times, 5 min each time. Then add the first antibody. The serum was diluted to different concentration gradients with 5% skim milk and added in ELISA plate. Incubate it for 2 h at 37 ℃. After incubation, wash the plate with PBST 6 times, 5 min each time. After that, dilute the HRP enzyme-labeled antibody IgG with 5% skim milk for 2000 times and add it into the ELISA plate. Incubate at 37 ℃ for 2 h. And then, wash the plate with PBST 6 times, 5 min/time. After washing, add TMB for light-proof reaction for 10–15 min, and then 2 mol L−1 H2SO4 was added to terminate the reaction. Finally, the OD450nm per well was determined by an enzyme-linked immunosorbent assay (PERLONG: DNM-9602, Beijing, China). When the titer of the serum met the requirements, the spleen of the mice was used to prepare monoclonal antibodies through the in vitro hybridoma technique (Hnasko and Stanker 2015; Zhang 2012).
Preparation of colloidal gold test strip
Colloidal gold was prepared by trisodium citrate reduction method and labeled with monoclonal antibodies. Prepare 100 mL of 0.01% HAuCl4 aqueous solution, heat, and boil it; then add 1 mL of 1% trisodium citrate aqueous solution, boil it for 15 min, cool it to room temperature after the color was stable to wine red. After that, add distilled water to 100 mL, and store it at 2–8 ℃ (Li et al. 2009; Shi et al. 2008). Then, the monoclonal antibody was labeled with colloidal gold and purified according to a previously reported method (Xu et al. 2006).
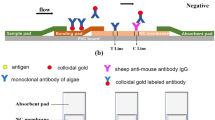
The strip consisted of four parts, including a glass fiber membrane sample pad, a glass fiber membrane antibody labeled bonding pad, the nitrocellulose membrane detection layer (NC membrane), and a glass fiber membrane water absorbent pad (Hu and Zhang 2017). The monoclonal antibody of Karenia mikimotoi labeled with colloidal gold was subscribed on the binding pad and dried. The antibody against Karenia mikimotoi and the sheep anti-mouse antibody IgG were sprayed on the NC membrane as the test line and the control line, respectively. The four pads were successively pasted onto a plastic carrier plate. The assembly of the test strip was carried out as previously reported (Lin et al. 2020).
Materials for preparing colloidal gold test strips, such as glass cellulose pads, nitrocellulose membranes, PVC boards, and absorbent pads, were purchased from Shanghai Jieyi Biotechnology Co, Ltd. The drawing software and data analysis software used in the article are Image J and Origin2021.
Verification of test strip
The effectiveness test of the test strip
The effectiveness of the test strip was verified by taking the Karenia mikimotoi in the growth period, the stable period, and the recession period. Take sterilized seawater as control. In order to further test whether the test strip can detect the different cell states of Karenia mikimotoi (about 1 × 105 cell/mL), the supernatants of removing algal cells, algal cell breaking fluid, and breaking supernatant obtained from the treatment of algal cells were used to test. Centrifuge 1700 × g for 15 min, collect Karenia mikimotoi at the stable period, and collect the supernatant after centrifugation (Fraga et al. 2012). Algae cells were resuspended with PBS and then broken with ultrasonic wave breaker (Shanghai Hushi Industrial Co., Ltd: JY92-IIN, Shanghai, China) at 4 ℃, 200 w, for 10 min. After crushing, centrifuge at 7000 × g for 10 min to separate the broken cells and the broken supernatant, and verify the effectiveness of the test strip. PBS was used as control (Fang et al. 2023).
The sensitivity test of the test strip
Take Karenia mikimotoi in the stable stage and dilute it by 2, 5, 10, 50, and 100 times with PBS, respectively, to verify the sensitivity of the test strip. Each gradient was verified three times. After the color test strip was developed, use Image J to scan the T line and C line of gray scale and analyze the color development results (Wang et al. 2015).
The specificity test of the test strip
Select several common red tide algae in the coastal areas of southeast China and red tide algae of similar species and genera to Karenia mikimotoi for specific verification (Jing et al. 2021). PBS was used as control. All kinds of algae cells were collected by centrifugation and then diluted and resuspended with PBS. Adjust the concentration of each algae to 5000–7000 cells/mL. In order to further verify the specificity of the test strip, we mixed the seven kinds of common red tide algae, including Karenia mikimotoi, Gymnodinium catenatum, Alexandrium catenella, Skeletonema costatum, Prorocentrum lima, Prorocentrum donghaiense, and Alexandrium tamarense (Fraga et al. 2012). The mixed algal liquid was divided into two types, one was the mixture of seven kinds of algae containing Karenia mikimotoi, and the other was the mixture of six kinds of algae without Karenia mikimotoi.
Production of color comparison card
Concentrations of 3 × 104 cells/mL, 2 × 104 cells /mL, 1 × 104 cells /mL, 5000 cells /mL, and 2500 cells/mL of Karenia mikimotoi algae solution were used for the detection of test strips. Each concentration was tested using 10 test strips. Color rendering and results were used in the production of color comparison card. During the production process of color comparison cards, we used Photoshop6.0 software developed by Adobe Company for image processing. White light in nature was composed of three different intensities of light: red, green, and blue. The color mode (RGB mode) used by Photoshop software was also a color mode with red, green, and blue as the primary colors. Each color had 0–255 brightness variations. After the color of the C-line on the test strip was stable, take photos in the same light. Then, in Photoshop6.0 software, use a pipette tool to take the color of the C-line. At this time, the number displayed in the data box was the current color value (Du et al. 2017; Fan et al. 2020). Each color had information on three colors: red, green, and blue. Take the color of the C-line of the 10 test strips with concentration gradients for each group. Then take the average value of each group’s RGB, and draw a color card.
A rapid enrichment method suitable of Karenia mikimotoi
Dilute the concentration of 400 cell/mL of Karenia mikimotoi with PBS to 300 cells/mL, 200 cells/mL, 100 cells/mL, 50 cells/mL, and 25 cells/mL. A nylon filter membrane (Xingya Purification Materials Factory, Shanghai) with a pore size of 10 μm was loaded into a detachable needle filter (Ruihua Scientific Instrument Co., Ltd, Jinan). Then, 50 mL of the above algae solution was aspirated using a 50-mL syringe (Shanghai Zhiyu Medical Equipment Co., Ltd.). The needle was discarded, and a detachable needle filter with a filter membrane was installed. Filter the algae solution through a detachable needle filter, collect the algae cells on the filter membrane, and then place the filter membrane in a 1.5-mL centrifuge tube. Add 1 mL of PBS, and shake to wash off the collected algae cells on the membrane. Finally, 60 μL of the algal solution was taken to be tested by the test strip.
On site application of the test strip
Conduct on-site monitoring at two locations in Pingtan County (119.74342705E, 25.63092909N) and Lianjiang County (119.81082399E, 26.32169884N) (Fig. 1), Fujian Province, on May 6, 2023 and May 17–22, 2023. Take 500 mL of surface water at the testing site and filter seawater using a vacuum filtration device (Tianjin Jinteng Experimental Equipment Co., Ltd., GM-0.33A) and a 3-μm-diameter filter membrane (Merk Millipore Ltd.). After filtration was completed, collect the filter membrane and place it in a centrifuge tube. Add 5 mL of PBS and shake. Take 60 μL for testing.
Results and discussion
Growth of Karenia mikimotoi
The growth of Karenia mikimotoi entered a rapid growth stage on the fourth day after inoculation and reached a stable growth stage on the 16th day (Fig. 2). And then the algae entered a decline period around the 24th day. The trend of growth changes was mainly related to the content of nutrients.
The titer of polyclonal and monoclonal antibody of Karenia mikimotoi
After five immunizations, the serum of the immunized mice was positive, and the serum of the non-immunized mice was negative. Titer of the serum was determined by indirect ELISA. The titer of polyclonal antibody was the dilution ratio of serum with P/N < = 2.1 and OD450 nm > 0.1. As shown in Table 1, when the dilution ratio of serum was 50,000 times, P/N = 5.88 > 2.1. When the dilution ratio of serum was 250,000 times, P/N = 2.1, but the value of positive OD450 nm was 0.0489 < 0.1. This indicated that the dilution ratio of serum was too large during the experiment. Therefore, the titer of Karen mikimotoi polyclonal antibody was between 50,000 and 250,000. Since P/N = 2.1 when the serum was diluted 250,000 times, it can be inferred from experience that the titer of Karenia mikimotoi polyclonal antibody was about 200,000.
Through the in vitro hybridoma technique, the ascites of mice injected with hybridoma cells were positive and the ascites of mice not injected with hybridoma cells were negative. Then the titer of monoclonal antibodies was determined by indirect ELISA. The titer of monoclonal antibody was determined by the dilution ratio of ascites with P/N < = 2.1. As can be seen from Table 2, when the dilution ratio of ascites was 100,000 times, P/N = 13.91 > 2.1; when the dilution ratio of ascites was 1,000,000, P/N = 12.78 > 2.1, but the OD450 nm value of positive ascites was very small. It was 0.115 and close to 0.1. It was indicated that the dilution ratio of ascites was too large. Even though the P/N value was large, the dilution ratio of ascites was meaningless. Therefore, it can be inferred that the titer of Karenia mikimotoi monoclonal antibody was about 100,000.
The preparation of monoclonal antibodies is a key step in the preparation of test strips. The higher the titer of monoclonal antibodies, the higher the specificity and sensitivity of antibodies. At present, many scholars have applied immunologic techniques to study red tide, but most of them use polyclonal antibodies to detect toxins such as saxitoxin (Dietrich et al. 1996; Micheli et al. 2002; Usleber et al. 1994). Because of its high specificity, monoclonal antibodies have been widely used in red tide research. For example, the preparation of monoclonal antibodies against DSP (diarrhetic shellfish poisoning) has been widely used (Liu et al. 2009; Usagawa et al. 1989). However, at present, it is relatively rare to use monoclonal antibody of red tide algae and colloidal gold technology to detect red tide.
Assembly and detection principle of test strip
To assemble the test strip, first fix the NC membrane on the PVC board. One side of the NC membrane was connected with a binding pad containing colloidal gold labeled antibody, and the other side was connected with an absorbent pad. As shown in the Fig. 3, the bonding pad contains the monoclonal antibody of Karenia mikimotoi labeled with colloidal gold; the T line contains the monoclonal antibody of Karenia mikimotoi, and the C line contains the sheep anti-mouse antibody IgG.
When the sample to be tested contains Karenia mikimotoi, the solution diffuses from the sample pad to the bonding pad. Karenia mikimotoi first combines with the antibody labeled with colloidal gold on the bonding pad, and then combines with the monoclonal antibody on the T line to form a double antibody sandwich structure of “gold-labeled antibody–antigen–antibody,” which makes the T line color. The gold-labeled antibody without binding antigen continues to spread forward and combine with the sheep anti-mouse antibody IgG on the C line, making the C line color (Fig. 3a). Therefore, when the sample to be tested contains Karenia mikimotoi, the T line and C line of the test strip will be colored, and the result is determined as positive. When the sample to be tested does not contain Karenia mikimotoi or contains other algae, the colloidal gold-labeled antibody will only bind with the sheep anti-mouse antibody IgG on the C line during the diffusion process, making the C line color, but the T line does not color, and the result is negative (Fig. 3b). If the T line and C line of the test strip do not show color or only T line shows color after dropping the sample, the test strip was invalid.
Applying similar principles, colloidal gold strips were also widely used in many fields, such as the detection of residues in food or water (Zhu et al. 2008), pesticide residues (Deng et al. 2022; Shi et al. 2008), viruses (Jing et al. 2021), pathogenic bacteria (Wu et al. 2017), etc.
Test strip effectiveness verification result
Karenia mikimotoi in the growth period (the 9th day, the 13th day), the stable period (the 17th day, the 21st day), and the recession period (the 24th day, the 28th day) were tested. It could be seen from Fig. 4 that the C line and T line of the test strip could be colored by the Karenia mikimotoi at different growth stages, that was, they could be detected by the test strip. Sterilized seawater in the control group could only make the C line color. Therefore, Karenia mikimotoi test strip was effective.
The results of detection of Karenia mikimotoi at different growth stages: a growth period: 17,200 cells/mL; b growth period: 22,700 cells/mL; c stable period: 37,000 cells/mL; d stable period: 31,000 cells/mL; e recession period: 10,000 cells/mL; f recession period: 4250 cells/mL; g sterilized seawater
From the results of Fig. 5, it showed that algal cells after centrifugation, the supernatant of removing algal cells, algal cell breaking fluid, and breaking supernatant could make the T line of the test strip color. It showed that the test strip could detect different cell states of Karenia mikimotoi.
Algal cells after centrifugation contained the surface antigen of intact algal cells, the supernatant of removing algal cells contained the metabolites produced during the growth of algal cells, and the broken algal cell solution contained the surface antigen and internal antigen of algal cells. The broken supernatant contains some substances secreted by algae cells and was also the internal antigen of algae. Whether it was the internal antigen or the surface antigen of algae cells, the test strip can detect. It showed that the test strip was effective. In addition, the supernatant of removing algal cells may contain toxins secreted by Karenia mikimotoi such as hemolytic toxins (Gentien et al. 2007; Yang et al. 2011). Some studies have shown that Karenia mikimotoi can produce at least three hemolytic active substances (Gentien and Arzul 1990). So we speculate that the test strip may be able to detect the toxins secreted by Karenia mikimotoi, but whether it was toxin needed further verification, such as through cell hemolysis test.
Sensitivity test results of the test strip
It can be seen from Fig. 6 that the color of the C line of the test strip was basically the same, while the color of the T line was different. The detection limit of the test strip of Karenia mikimotoi was 754 cells/mL. The research showed that in the sea area where the red tide zone of Karenia mikimotoi passed, most of the fish in the culture area died rapidly (concentration ≥ 104 cell/mL). Compared with the basic concentration of 1 × 104 cell/mL of the outbreak of red tide of Karenia mikimotoi, the test strip had higher sensitivity.
In addition, the gray scale of the T line and the C line of the test strip was scanned by Image J, and the ratio of the gray scale value of the T line to C line was used to compare the test strip with different concentrations of Karenia mikimotoi. It can be seen from Fig. 7 that with the decrease of the concentration of Karenia mikimotoi added, the ratio of the gray scale of the T line to the C line of the test strip showed a downward trend. The ratio of the gray value of the T line to C line at the concentration of 37,700 cells/mL was slightly lower than that at the concentration of 18,850 cells/mL (Fig. 7). This was because there were some fluctuations in the gray scan value of the C line.
In general, the experimental results showed that the test strip had good sensitivity.
Specificity test results of the test strip
The experimental results show that only Karenia mikimotoi could color the T line of the test strip. When other algae were used for detection alone, they could not make the T line color (Fig. 8b–i). When mixed with other algae except Karenia mikimotoi for detection, the T line of the test strip also did not show color (Fig. 8k). However, when the mixed solution to be tested includes Karenia mikimotoi, the T line of the test strip also did not show color (Fig. 8l). The possible reason was that the concentration of Karenia mikimotoi was reduced when mixed with various algal liquids, which may be lower than the detection limit of the test strip (Fig. 6).
The result of the specificity test: (a) Karenia mikimotoi; (b) Gymnodinium catenatum; (c) Alexandrium tamarense; (d) Prorocentrum donghaiense; (e) Alexandrium catenella; (f) Thalassiosira weissflogii; (g) Skeletonema costatum; (h) Phaeodactylum tricornutum Bohlin; (i) Nitzschia closterium; (j) PBS only; (k) mixing of the above algae (except Karenia mikimotoi); (l) mixing of the above algae
The results of algae mixing experiment showed that the mixed solution of algae cells with approximate concentration of 1 × 105 cell/mL, 1 × 104 cell/mL, and 1 × 103 cell/mL containing Karenia mikimotoi can make the T line of the test strip color, while the mixed solution of algae cells without Karenia mikimotoi cannot make the test strip color (Fig. 9). In addition, the higher the concentration of Karenia mikimotoi in the mixture, the darker the color of the T line (Fig. 9a, c, e). The test solution with the mixed solution concentration of 1 × 102 cell/mL, whether containing Karenia mikimotoi or not, cannot make the T line of the test strip color (Fig. 9g). This was mainly because the concentration of Karenia mikimotoi was lower than the detection limit of the test strip.
Specific test results of mixed algae: (a) mixture of seven algae (concentration of each kind of algae was about 8 × 105 cell/mL); (b) mixture of six algae except Karenia mikimotoi (concentration of each kind of algae was about 8 × 105 cell/mL); (c) mixture of seven algae (concentration of each kind of algae was about 8 × 104 cell/mL); (d) mixture of six algae except Karenia mikimotoi (concentration of each kind of algae was about 8 × 104 cell/mL); (e) mixture of seven algae (concentration of each kind of algae was about 8 × 103 cell/mL); (f) mixture of six algae except Karenia mikimotoi (concentration of each kind of algae was about 8 × 103 cell/mL); (g) mixture of seven algae (concentration of each kind of algae was about 8 × 102 cell/mL); (h) mixture of six algae except Karenia mikimotoi (concentration of each kind of algae was about 8 × 102 cell/mL); (i) PBS only
The experimental results showed that within the concentration range that can be detected by the test strip, as long as the sample contained Karenia mikimotoi, the color of T line can be developed. It showed that the test strip has high specificity.
Results of color comparison card production
The drawn color card was shown in the following Fig. 10, and as the detection concentration decreases, the color of the C-line gradually became lighter from red. At a concentration of 2500 cells/mL, the color of the C-line gradually approached gray.
The test strip developed in this study served as a rapid qualitative and semi-quantitative detection tool for Karenia mikimotoi, which can roughly determine the concentration of the algae based on the color depth of the C-line. Therefore, drawing this test strip can serve as a rapid concentration determination for on-site monitoring of Karenia mikimotoi, facilitating monitoring and early warning of the bloom of Karenia mikimotoi. However, to determine a more accurate concentration of algae cells in water samples, quantitative testing such as QPCR was necessary.
Results of needle syringe enrichment
The experimental results showed that the Karenia mikimotoi with a concentration of 400 cells/mL, 300 cells/mL, 200 cells/mL, and 100 cells/mL enriched by a syringe and resuspended on 1 mL PBS could cause the T-line of the test strip to develop color, and the color of the T-line became lighter as the detection concentration decreased (Fig. 11). After enrichment, Karenia mikimotoi with a concentration of 50 cell/mL cannot cause the T line to show color (Fig. 11e). The experimental results indicated that the minimum concentration of Karenia mikimotoi that can be detected by the test strip using a syringe enrichment device was 100 cell/mL.
Enrichment of 50 mL of algae solution from the syringe onto the membrane was followed by resuspension with 1 mL of PBS. In theory, during this process, the concentration of algae cells increased by 50 times. Therefore, by combining the use of test strips and using a syringe enrichment method, the test strips can detect lower concentrations of Karenia mikimotoi. Moreover, this enrichment method was easy to operate and has the potential to be promoted and applied in monitoring sites.
On site monitoring results of the test strip
The experimental results showed that the water samples collected from the Pingtan area on May 20, 21, and 22 after enrichment treatment with a suction pump can cause the C-line of the test strip to develop color (Fig. 12). In the Lianjiang area, test strips also detected Karenia mikimotoi on May 20 and 22. However, due to the relatively low concentration, the color development of the C-line was relatively light. Therefore, through on-site testing applications, it had been proven that the test strip developed in this study was effective.
Karenia mikimotoi blooms peaked generally in May (late spring) and June (early summer), with multiyear average water temperatures of 19–24 °C in spring and 25–28 °C in summer (Baohong et al. 2021). It was reported that the main biological groups that trigger red tides along the coast of Fuzhou include dinoflagellates and diatoms, totaling 10 species such as Prorocentrum donghaiense, Skeletonema costatum, and Karenia mikimotoi. The frequency and area of red tides caused by Karenia mikimotoi account for 15.2% and 6.8% of the total red tides in the area (Li 2012).
Conclusions
Based on the preparation technology of monoclonal antibody and colloidal gold immunochromatography technology, a colloidal gold strip for rapid detection of Karenia mikimotoi was developed in this research. The validation results of the test strip in the laboratory indicated that the test results can be directly observed within 3–5 min. The different growth stages and different cell states of Karenia mikimotoi can be detected by the strip. This test strip also had high specificity and sensitivity, which the minimum concentration of Karenia mikimotoi can be detected was 754 cells/mL. In addition, based on the color development of C-line test strips with different algal concentrations detected, a color comparison card had been made to facilitate rough judgment of the detected algal concentrations in the future. And we also applied the test strip to two coastal areas in Fujian Province, and the results showed that the test strip was effective.
Data availability
The data generated during and/or analyzed during the current study is available from the corresponding author on reasonable request.
References
Al-Kandari MA, Highfield AC, Hall MJ et al (2011) Molecular tools separate harmful algal bloom species, Karenia mikimotoi, from different geographical regions into distinct sub-groups. Harmful Algae 10(6):636–643
Anderson DM, Glibert PM, Burkholder JM (2002) Harmful algal blooms and eutrophication: nutrient sources, composition, and consequences. Estuaries 25(4B):704–726
Anderson DM, Cembella AD, Hallegraeff GM (2012) Progress in understanding harmful algal blooms: paradigm shifts and new technologies for research, monitoring, and management. In Carlson CA, SJ Giovannoni (Eds) Annual review of marine science, Vol 4 (Vol. 4, pp. 143–176). https://doi.org/10.1146/annurev-marine-120308-081121
Antonella P, Luca G (2013) The quantitative real-time PCR applications in the monitoring of marine harmful algal bloom (HAB) species. Environ Sci Pollut Res 20(10):6851–6862
Baohong C, Kang W, Huige G et al (2021) Karenia mikimotoi blooms in coastal waters of China from 1998 to 2017. Estuar Coast Shelf Sci 249:107034
Botes L, Smit A, Cook PA (2003) The potential threat of algal blooms to the abalone (Haliotis midae) mariculture industry situated around the South African coast. Harmful Algae 2(4):247–259
Brand LE, Campbell L, Bresnan E (2012) Karenia: the biology and ecology of a toxic genus. Harmful Algae 14:156–178
Chen G, Zhang C, Zhang B et al (2011) Development of a PNA probe for fluorescence in situ hybridization detection of Prorocentrum donghaiense. Plos One 6(10):e25527
Chen GF, Liu Y, Zhang CY et al (2013) Development of rRNA-targeted probes for detection of Prorocentrum micans (Dinophyceae) using whole cell in situ hybridization. J Appl Phycol 25(4):1077–1089
Chen G, Cai P, Zhang C et al (2015) Hyperbranched rolling circle amplification as a novel method for rapid and sensitive detection of Amphidinium carterae. Harmful Algae 47:66–74
Davidson K, Miller P, Wilding TA et al (2009) A large and prolonged bloom of Karenia mikimotoi in Scottish waters in 2006. Harmful Algae 8(2):349–361
Deng H, Guan W, Cao Z et al (2016) Analysis of hydrological and meteorological factors causing Karenia miki motoi bloom in 2012 along Fujian coast. J Mar Sci 34:28–38
Deng H, Chen D, Li X et al (2022) Development of a colloidal gold immunochromatographic test strip for the rapid detection of iprodione. Anal Methods 14(43):4370–4376
Dietrich R, Usleber E, Burk C et al (1996) Immunochemical approaches to the analysis of paralytic shellfish poisoning toxins. In: Beier RC, Stanker LH (Eds) Immunoassays for residue analysis: food safety (Vol. 621, pp. 395–403). <Go to ISI>://WOS:A1996BF24E00031
Du J, Ge H, Gu Q et al (2017) Gold nanoparticle-based nano-probe for the colorimetric sensing of Cr3+ and Cr2O72- by the coordination strategy. Nanoscale 9(48):19139–19144
Dyhrman ST, Erdner D, La Du J et al (2006) Molecular quantification of toxic Alexandrium fundyense in the Gulf of Maine using real-time PCR. Harmful Algae 5(3):242–250
Fan K, Wang X, Yang H et al (2020) Semiquantitative naked-eye detection of Cu(ii) with a standard colorimetric card via a hydrogel-coated paper sensor. Anal Methods 12(12):1561–1566
Fang J, Li S, Liu X et al (2023) Development of a test strip for rapid detection of Gymnodinium catenatum. Environ Monit Assess 195(1):83
Fraga S, Rodriguez F, Bravo I et al (2012) Review of the main ecological features affecting benthic dinoflagellate blooms. Cryptogam Algol 33(2):171–179
Furuya K, Iwataki M, Lim PT et al (2018) Overview of harmful algal blooms in Asia. In: Glibert PM, Berdalet E, Burford MA, Pitcher GC, Zhou M (Eds) Global ecology and oceanography of harmful algal blooms (Vol. 232, pp. 289–308). <Go to ISI>://BIOSIS:PREV201900059750
Gao DJ, Tian Y, Bi SY et al (2005) Studies on the interaction of colloidal gold and serum albumins by spectral methods. Spectrochim Acta A-Mol Biomol Spectrosc 62(4–5):1203–1208
Gentien P, Arzul G (1990) Exotoxin production by gyrodinium cf aureolum (dinophyceae). J Mar Biol Assoc UK 70(3):571–581
Gentien P, Lunven M, Lazure P et al (2007) Motility and autotoxicity in Karenia mikimotoi (Dinophyceae). Phil Trans R Soc B-Biol Sci 362(1487):1937–1946
Glad C, Grubb AO (1981) Immuno-capillary-migration with enzyme-labeled antibodies - rapid quantification of c-reactive protein in human-plasma. Anal Biochem 116(2):335–340
Glibert PM, Berdalet E, Burford MA et al (2018) Harmful algal blooms and the importance of understanding their ecology and oceanography. In: Glibert PM, Berdalet E, Burford MA, Pitcher GC, Zhou M (Eds) Global ecology and oceanography of harmful algal blooms (Vol. 232, pp. 9–25). https://doi.org/10.1007/978-3-319-70069-4_2
Guillard RR, Ryther JH (1962) Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can J Microbiol 8:229–239
Guo H, Ding D, Lin F et al (2015) Characteristics and patterns of red tide in China coastal waters during the last 20a. Adv Mar Sci 33(4):547–558 (in Chinese, abstract in English)
Hansen G (2003) Comparative study of Gymnodinium mikimotoi and Gymnodinium aureolum, comb. nov. (=Gyrodinium aureolum) based on morphology, pigment composition, and molecular data. J Phycol 36(2):394–410
Hnasko RM, Stanker LH (2015) Hybridoma technology. In: Hnasko R (Ed) ELISA: Methods and Protocols (Vol. 1318, pp. 15–28). https://doi.org/10.1007/978-1-4939-2742-5_2
Hu X, Zhang G (2017) An immunochromatographic test strip to detect ochratoxin A and zearalenone simultaneously. In: Holst O (Ed) Microbial Toxins, 2 Edition:Methods and Protocols (Vol. 1600, pp. 95–105). https://doi.org/10.1007/978-1-4939-6958-6_9
Huang K, Zhuang Y, Wang Z et al (2021) Bioavailability of organic phosphorus compounds to the harmful dinoflagellate Karenia mikimotoi. Microorganisms 9(9):961
Jing H, Li X, Xu L et al (2021) Development of a colloidal gold immunochromatographic strip for the rapid detection of channel catfish virus. J AOAC Int 104(5):1350–1354
Li X (2012) Occurrence characteristics of the red tide in Fujian coastal waters during the last two decades. Mar Environ Sci 33(07):2210–2216
Li D, Wei S, Yang H et al (2009) A sensitive immunochromatographic assay using colloidal gold-antibody probe for rapid detection of pharmaceutical indomethacin in water samples. Biosens Bioelectron 24(7):2277–2280
Li H, Meng K, Yu Q et al (2022) Rapid detection method of Skeletonema pseudocostatum and preparation of test strip. Environ Sci Pollut Res 29(46):70202–70208
Lin L, Song S, Wu X et al (2020) A colloidal gold immunochromatography test strip based on a monoclonal antibody for the rapid detection of triadimefon and triadimenol in foods. Food Hydrocolloids 31(1):475–488
Liu RY, Xu DY, Liang YB et al (2009) Preparation of monoclonal antibody against okadaic acid and development of an sensitive ELISA for detection diarrhetic shellfish poisoning in shellfish. [2009 international conference on environmental science and information application technology, vol ii, proceedings]. International Conference on Environmental Science and Information Application Technology (ESIAT 2009), Wuhan, Peoples R China
Lu D, Qi Y, Gu H et al (2014) Causative species of harmful algal blooms in Chinese coastal waters. Algol Stud 145:145–168
Micheli L, Di Stefano S, Moscone D et al (2002) Production of antibodies and development of highly sensitive formats of enzyme immunoassay for saxitoxin analysis. Anal Bioanal Chem 373(8):678–684
Ministry of Natural Resources of the People’s Republic of China (2009–2020) Bulletin of China marine disaster. http://www.mnr.gov.cn/sj/sjfw/hy/gbgg/zghyzhgb/. Accessed 18 Jul 2021
Mitchell S, Rodger H (2007) Pathology of wild and cultured fish affected by a Karenia mikimotoi bloom in Ireland, 2005. Bull Eur Assoc Fish Pathol 27(1):39–42
Mooney BD, Nichols PD, de Salas MF et al (2007) Lipid, fatty acid, and sterol composition of eight species of Kareniaceae (Dinophyta): chemotaxonomy and putative lipid phycotoxins. J Phycol 43(1):101–111
Nezan E, Siano R, Boulben S et al (2014) Genetic diversity of the harmful family Kareniaceae (Gymnodiniales, Dinophyceae) in France, with the description of Karlodinium gentienii sp nov.: a new potentially toxic dinoflagellate. Harmful Algae 40:75–91
Parrish CC, Bodennec G, Gentien P (1998) Haemolytic glycoglycerolipids from Gymnodinium species. Phytochemistry 47(5):783–787
Putalun W, Morinaga O, Tanaka H et al (2004) Development of a one-step immunochromatographic strip test for the detection of sennosides A and B. Phytochem Anal 15(2):112–116
Reise K, van Beusekom JEE (2008) Interactive effects of global and regional change on a coastal ecosystem. Helgol Mar Res 62(1):85–91
Shi C, Zhao S, Zhang K et al (2008) Preparation of colloidal gold immunochromatography strip for detection of methamidophos residue. J Environ Sci 20(11):1392–1397
Sun Z, Shi B, Meng F et al (2018) Development of a colloidal gold-based immunochromatographic strip for rapid detection of H7N9 influenza viruses. Front Microbiol 9:2069. https://doi.org/10.3389/fmicb.2018.02069
Usagawa T, Nishimura M, Itoh Y et al (1989) Preparation of monoclonal-antibodies against okadaic acid prepared from the sponge halichondria-okadai. Toxicon 27(12):1323–1330
Usleber E, Dietrich R, Martlbauer E et al (1994) Effect of heterologous paralytic shellfish poisoning toxin-enzyme conjugates on the cross-reactivity of a saxitoxin enzyme-immunoassay. Lett Appl Microbiol 18(6):337–339
Wan Y, Shi Z, Peng G et al (2022) Development and application of a colloidal-gold dual immunochromatography strip for detecting African swine fever virus antibodies. Appl Microbiol Biotechnol 106(2):799–810
Wang J-Y, Chen M-H, Sheng Z-C et al (2015) Development of colloidal gold immunochromatographic signal-amplifying system for ultrasensitive detection of Escherichia coli O157:H7 in milk. RSC Adv 5(76):62300–62305
Wu W-D, Li M, Chen M et al (2017) Development of a colloidal gold immunochromatographic strip for rapid detection of Streptococcus agalactiae in tilapia. Biosens Bioelectron 91:66–69
Xiao-Kun H, Zhuo-Ru L, Qing-Chun Z et al (2022) Detection of bloom-forming dinoflagellates Karenia mikimotoi and Prorocentrum donghaiense using qPCR assays. J Appl Phycol 34(3):1483–1496
Xu C, Wang H, Peng C et al (2006) Colloidal gold-based immumochromatographic assay for detection of diethylstilbestrol residues. Biomed Chromatogr 20(12):1390–1394
Yang W, Zhang N, Cui W et al (2011) Effects of co-existing microalgae and grazers on the production of hemolytic toxins in Karenia mikimotoi. Chin J Oceanol Limnol 29(6):1155–1163
Yu R-C, Lu S-H, Liang Y-B (2018) Harmful algal blooms in the coastal waters of China. In: Glibert PM, Berdalet E, Burford MA, Pitcher GC, Zhou M (Eds) Global ecology and oceanography of harmful algal blooms (Vol. 232, pp. 309–316). https://doi.org/10.1007/978-3-319-70069-4_15
Yuhi T, Nagatani N, Endo T et al (2006) Gold nanoparticle based immunochromatography using a resin modified micropipette tip for rapid and simple detection of human chorionic gonadotropin hormone and prostate-specific antigen. Sci Technol Adv Mater 7(3):276–281
Zhang C (2012) Hybridoma technology for the generation of monoclonal antibodies. Methods Mol Biol (Clifton, N.J.) 901:117–135
Zhu J, Chen W, Lu Y et al (2008) Development of an immunochromatographic assay for the rapid detection of bromoxynil in water. Environ Pollut 156(1):136–142
Zou Y, Yamasaki Y, Matsuyama Y et al (2010) Possible involvement of hemolytic activity in the contact-dependent lethal effects of the dinoflagellate Karenia mikimotoi on the rotifer Brachionus plicatilis. Harmful Algae 9(4):367–373
Zuk RF, Ginsberg VK, Houts T et al (1985) Enzyme immunochromatography - a quantitative immunoassay requiring no instrumentation. Clin Chem 31(7):1144–1150
Acknowledgements
The authors gratefully acknowledge comments from Kefu Zhou that greatly improved the manuscript.
Funding
This study was supported by the Fujian fishery resources monitoring centre offshore toxic (harmful) red tide algae and toxins rapid detection method development and application service procurement project [FZJZ-2020–1].
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Shuyue Li, Xiaoxiao Liu, and Weixin He did main experiments (algal culture, algal antigen treatment, and test strip verification); Lingyue Li and Jiazhao Zhang were involved in data analysis; Kefu Zhou was involved in the preparation of polyclonal antibody; Changgong Zhang was involved in the preparation of monoclonal antibody and assembly of test strip; Jiahang Mu participated in sampling at sea; the first draft of the manuscript was written by Junhua Fang. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
The ethical code of animal experiment in this paper is XMULAC20220020.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Responsible Editor: Lotfi Aleya
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fang, J., Liu, X., He, W. et al. Establishment of colloidal gold immunochromatography strip for rapid detection of Karen mikimotoi and its application. Environ Sci Pollut Res 30, 109123–109134 (2023). https://doi.org/10.1007/s11356-023-29968-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-023-29968-7