Abstract
A heterotrophic nitrifying aerobic denitrifying (HN-AD) strain ZQ-A1 with excellent denitrification performance, identified as Acinetobacter, was isolated from simultaneous nitrification and denitrification (SND) craft. ZQ-A1 was capable of removing NH4+, NO2–, and NO3–; the 21-hour removal rates were 84.84%, 87.13%, and 92.63%. ZQ-A1 has the ability to treat mixed nitrogen sources. In addition, ZQ-A1 can be well applied to actual sewage. According to the analysis of microbial community characteristics, the relative abundance of Acinetobacter in the experimental group increased from 0.06% to 2.38%, which is an important reason for the removal rate of NH4+ exceeding 99% within 30 days. The results of KEGG function prediction showed that with the addition of ZQ-A1, the relative abundance of pathways related to bacterial metabolism, such as tricarboxylic acid cycle metabolism, was higher. The research expanded the thinking of HN-AD bacteria in actual production and laid a foundation for its application in sewage treatment.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
A large amount of nitrogen and phosphorus are discharged, which is an important reason for water eutrophication. According to Tao’s research in 2021, more than 90% of lakes in China are in the middle trophic and eutrophic state. As an important part of agricultural sewage discharge, pig farm sewage is particularly important to reduce the environmental problems caused by its high ammonia nitrogen content, such as water eutrophication. In recent years, the process of biological denitrification process in sewage treatment technology has been paid more and more attention because of its high efficiency, no secondary pollution, low cost, and stable operation. Generally speaking, traditional processes of biological nitrogen removal is mainly divided into two parts: nitrification and denitrification (Zhao et al. 2017). The nitrification process is an aerobic oxidation process, so autotrophic microorganisms are required to participate in the process and complete the oxidation process of NH4+ to NO2− or NO3−. Denitrification is an anaerobic reduction process, which requires heterotrophic microorganisms to complete the process of reducing NO2− or NO3− to N2 (Zhao et al. 2012). However, the traditional biological denitrification needs aerobic nitrification and anaerobic denitrification, and the growth cycle of autotrophic nitrifying bacteria is long, which leads to the problems of large occupied area, high operating cost, and long start-up time of the traditional sewage treatment process (Zheng et al. 2012).
In 1980s, researchers have found that the phenomenon of nitrogen non-isomerized loss was found in the non-obvious anoxic or anaerobic section and called them simultaneous nitrification and denitrification (SND) (Robertson and Kuenen 1990). The mechanism of SND mainly includes macro-environmental theory, micro-environmental theory, and microbial theory. Macro-environmental theory means that in the process, due to the uneven aeration rate in the process design, the place with high dissolved oxygen content will undergo nitrification and the place with low dissolved oxygen will undergo denitrification. Micro-environmental theory holds that due to the existence of activated sludge or granular sludge in water, dissolved oxygen diffuses inside flocs, forming a dissolved oxygen gradient from the outside to the inside, where the dissolved oxygen concentration is high, autotrophic microorganisms will take the lead in nitrification, and where the dissolved oxygen concentration is low, anaerobic denitrification will take place.
At present, the research in the laboratory shows that the successful operation of SND process reduces the sewage treatment time, improves the system treatment efficiency, and reduces the operation cost compared with the traditional biological denitrification technology (Huan et al. 2022). Therefore, if SND process can be successfully applied to actual production, it will reduce the oxygen demand, shorten the hydraulic retention time, and reduce the energy consumption and operation cost; and a single reaction vessel can reduce the occupied area, thus saving the construction cost (Lang et al. 2019).
The discovery and successful culture of heterotrophic nitrification and aerobic denitrification (HN-AD) bacteria had broken the traditional theoretical understanding and enriched the microbial theory. HN-AD bacteria have both nitrification and denitrification functions. Therefore, HN-AD bacteria have become a research hotspot (Xi et al. 2022). HN-AD bacteria widely exist in the environment. Previous studies have successfully isolated HN-AD bacteria from pig farm sewage (Zhang et al. 2012), seabed sludge (Zhang et al. 2015), paddy soil (He et al. 2016), and activated sludge (Huang et al. 2022).
With the further research on HN-AD bacteria, more and more HN-AD bacteria have been successfully isolated and applied more and more widely, but there are few examples of HN-AD bacteria applied to pig farm sewage at present. The purpose of this study is to isolate a HN-AD strain which can be used for nitrogen removal of pig farm sewage and provide reference and experimental support for the subsequent application of HN-AD in livestock and poultry sewage.
Material and methods
Main medium
LB broth, heterotrophic nitrification medium, and aerobic denitrification medium are slightly adjusted according to the previous research (Huan et al. 2022) and preparation and the actual situation.
Evaluation of nitrogen removal characteristics with different nitrogen sources
In order to determine the nitrogen removal characteristics of the strain under different nitrogen sources, two-stage experiments were carried out.
The first part is a single nitrogen source experiment to explore the nitrogen metabolic function of the strain. The prepared ZQ-A1 bacteria were added to the culture medium with NH4Cl, NaNO2, and KNO3 as the only source of nitrogen (concentration 200 mg/L), C/N ratio of 10 and pH = 7, and cultured at 30 °C and 200 rpm for 21 h. Samples were collected every three hours for 21 hours to measure optical density (OD), ammonium nitrogen, nitrite nitrogen, and nitrate nitrogen; and three parallel experiments were conducted. Determination of NH4+, NO2-, NO3-, TN, and COD by standard method (APHA 2005).
The second stage is the mixed source of nitrogen experiment. In order to explore the simultaneous nitrification and denitrification characteristics of strain ZQ-A1 under mixed nitrogen sources, four kinds of mixed nitrogen sources were added to the culture medium (concentration of each nitrogen source was 200 mg/L), namely, (1) NH4Cl and NaNO2, (2) NH4Cl and KNO3, (3) NaNO2 and KNO3, and (4) NH4Cl, NaNO2 and KNO3, C/N ratio of 10 and pH = 7, and cultured at 30 °C and 200 rpm for 21 h, samples were collected every three hours.
Strain isolation and identification
Pig farm wastewater was inoculated into LB culture medium according to the proportion of 10%. The sample was cultured at 37 °C and 200 revolutions per minute (rpm)de for 24 hours (h); the above bacterial solution was taken out and inoculated into HNM at the ratio of 1% and domesticated at 30 °C and 150 rpm. The 1% of the inoculum was transferred to the new HNM for 4 days and acclimated for 3 rounds. The mixture was inoculated with ADM at 30 °C, 160 rpm, and 1 % inoculation rate, every 4 days as a cycle. After 3 times of acclimation, the new ADM was inoculated with 1% inoculum. After 24 h, 0.1 mL of mixed bacterial solution was collected, diluted from 10−1 to 10−10, and then coated on LB agar medium plate and cultured for 12~16 h in 37 °C incubator.
Bacterial DNA was extracted by using DNA genome extraction kit (Tiangen Biotech, China). 27F and 1492R were selected as universal primers for amplification (Table 1) and sent to Sangon Bioengineering (Shanghai) Co., Ltd. for sequencing. By using BLAST in NCBI database for homology comparison, MEGA7.0 is used to build phylogenetic tree. In addition, the nitrogen removal functional genes of the strain were tested by PCR according to the primer sequence (Table 1).
Effect of the factors on nitrogen removal efficiency
Single factor experiments were carried out to explore the culture conditions of different pH values, rotational speeds, C/N ratios, carbon sources, and temperatures (Table 4). Strain ZQ-A1 was cultivated in LB broth for 12 h and then transferred to a sterile conical flask containing 200 mL culture medium by sucking 2 mL of the bacterial solution. Experiments were carried out under different parameters, including pH values (5, 6, 7, 8, and 9), rotational speed (0 rpm, 50 rpm, 100 rpm, 150 rpm, and 200 rpm), C/N ratio (1, 5, 10, 15, and 20), different carbon sources (sodium succinate, potassium tartrate, sodium citrate, sodium acetate, and glucose), and temperature (15 °C, 20 °C, 25 °C, 30 °C, and 35 °C); and the growth and denitrification characteristics of ZQ-A1 were observed. The initial concentration of NH4Cl in the medium was 200 mg/L and pH value was 7 (except for pH experiment experiment). The NH4+-N and OD600 were measured at 15 h and 18 h, respectively. The uninoculated sterile medium was used as the control, and all experiments were repeated three times.
Preliminary application of strain ZQ-A1 in sewage treatment
The raw sewage was filtered using 0.22 μM filter. The DNA was extracted by water sample DNA extraction kit and stored at −80 °C. The pre-shaken strain ZQ-A1 was inoculated at 1% into 5 L of sewage. The experiment was carried out at room temperature and 50 rpm and chemical oxygen demand (COD), total nitrogen (TN), NH4+, NO2-, and NO3- were measured on days 0, 3, 6, 9, 12, 15, 18, 21, 24, 27, and 30. On the 30th day, water DNA was extracted and sent to Guangdong Meige Technology Gene Co., Ltd. for high-throughput sequencing together with the original sewage DNA sample. MagicHand online platform (https://www.magichand.onlineom) is used to analyze data and draw graphs. Total RNA extraction kit (Changzhou EMI Biotechnology Co., Ltd.) was used to extract RNA. RNA concentration was measured by Biotek Epoch ultraspectrophotometer. Reverse transcription amplification was performed with Nanjing Vazyme Biotechnology Co., Ltd. reverse transcription kit. The 1 μg cDNA was used to make qPCR and amplified by TSE202 kit (2× T5 rapid qPCR mixture (SYBRGreenI)). AmoA, napA, narG, nirS, norB, and nosZ gene expressions were detected, and 16SrRNA gene house-keeping gene. Primers used for quantitative PCR were listed in Table 2. The 2-△△Ct method was used to calculate the relative expression of different genes (Livak and Schmittgen 2001).
Statistical analysis
In order to analyze whether there is any difference between the experimental group and the control group, one-way ANOVA was conducted by using SPSS26.0 software. Whether the difference between the averages is significant or not is determined by using Tukey’s test. Among them, when P < 0.05, the difference is significant. The images in this study are generated by GraphPad Prism 8.0 and Origin 2021. PICRUSt software was used to standardize the OTU grace table and compare it to KEGG database through the corresponding greengene ID of each OTU and make functional prediction analysis (https://www.magichand.onlineom).
Results and discussion
Isolation, identification, and functional gene amplification
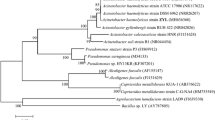
A strain with nitrification and denitrification functions was successfully isolated and cultured. It was named as Acinetobacter ZQ-A1 (hereinafter referred to as ZQ-A1), given that this strain could be classified as Acinetobacter (Fig. 1).
Analysis of nitrification and denitrification capacity
Single nitrogen source test
The nitrogen removal ability of strain ZQ-A1 was investigated by adding a single nitrogen source. When NH4Cl was used as the only source of nitrogen, ammonium nitrogen in the culture decreased from 200 mg/L to 30.33 mg/L with the enrichment and growth of ZQ-A1, and the average removal rate was 8.08 mg/(L·h) (Fig. 2a). Moreover, the accumulation of NO2- and NO3− was not detected in the whole process, which indicated that ZQ-A1 could carry out a complete nitrification and denitrification process. In addition, it can be observed that the pH gradually increases from 7 to 9, which indicates that the subsequent denitrification leads to the increase of alkalinity, which is similar to previous studies (Zhang et al. 2022); it indicated that ZQ-A1 played a denitrification role, produced more OH-, and led to the increase of pH. When NaNO2 is used as the sole source of nitrogen, because high concentration of free nitrous acid (FNA) will produce toxicity to bacteria (Kouba et al. 2019) (Fig. 2b), the growth of ZQ-A1 was inhibited 8 hours ago; in spite of this, the denitrification performance of this strain was not affected. In the first 9 hours, the average denitrification efficiency of ZQ-A1 was 11.88 mg/L·h; it can be inferred that the high-efficiency removal of NO2- in the first eight hours is to reduce the toxicity of FNA to ZQ-A1. When KNO3 is used as the sole nitrogen source, similar to NaNO2, ZQ-A1 shows a strong denitrification ability, with a denitrification efficiency of 92.62% in 21 h (Fig. 2c). ZQ-A1 can remove 8.82 mg/L NO3- per hour, which is higher than that of WT-14 in previous research (Chen et al. 2021a, 2021b). The results showed that no matter which nitrogen source was used as the only nitrogen source, ZQ-A1 showed good nitrogen removal performance (Table 3).
Compound nitrogen source test
In order to explore the nitrogen removal characteristics of ZQ-A1 under mixed nitrogen sources, four kinds of mixed nitrogen sources were added to the culture medium. When the mixed nitrogen source is NH4Cl and NaNO2, compared with a single source of nitrogen, the nitrogen removal efficiency is obviously improved, and the removal of NH4Cl and NaNO2 is more thorough which can reach 100% (Fig. 3a). To combine with the growth of ZQ-A1, it was found that its growth rate increased. Previous studies showed that although high concentration of FNA could inhibit the activity of microorganisms, when the concentration of FNA decreased, it would promote the activity of microorganisms (Chen et al. 2021a, 2021b), which may be the reason for the faster removal rate of ZQ-A1. At 2~4 h, NO2- decreased obviously, and the average removal rate reached 39.76 mg/(L·h). The reason for this result may be the strain preferentially uses NO2- for short-cut nitrification and denitrification (SCND), which has similar results with previous studies (Duan et al. 2020). On the other hand, it is concluded that the high oxygen content and acidic environment in the medium accelerated the oxidation process of NO2-. When NH4Cl and KNO3 are used as nitrogen sources in the culture concentration, it can be seen that the removal rate of NH4+ is obviously higher than that of NO2- in the first 12 hours, and NH4+ is preferentially utilized by ZQ-A1 at this time. However, the overall nitrogen removal efficiency is obviously higher than that of the former group, and the removal rates of NH4+ and NO3- can reach 100% in 12 h. This is similar to previous studies (He et al. 2022) (Fig. 3b). Interestingly, when nitrogen sources NaNO2 and KNO3 were used in the culture medium, the removal efficiency of NO3- reached 87.07% at 12 h, which was higher than that of strain HY-1 used in a recent study (Huan et al. 2022) (Fig. 3c). In 9 h, the content of NH4+ increased from 0 mg in 0 hours to 26.34 mg. At this stage, NO3- can be removed by 14.51 mg per hour. This phenomenon may be due to the phenomenon of dissimilatory nitrate reduction to ammonium (DNRA) (Lin and Stewart 1998), and some NO3- is converted into NH4+-N which can be used by organisms themselves. Because the mixing of three nitrogen sources is common in actual production, the nitrogen removal ability of ZQ-A1 is evaluated by the mixing of three nitrogen sources. The results show that NH4+, NO2-, and NO3- could be completely removed in 21 h, and the degradation rate was higher than that of the three single source of nitrogen groups. It is speculated that NH4+ in ZQ-A1 nitrogen removal process can provide electrons and improve nitrogen removal efficiency, which is similar to previous research results (Fig. 3d) (Gu et al. 2022).
Nitrogen removal functional gene amplification
Generally speaking, there are two main ways for HN-AD biological denitrification. One is the complete process of nitrogen removal, NH4+ → NH2OH →NO2−→NO3−→NO2–→ NO → N2O → N2 (Richardson et al. 1998), the other is NH4+ → NH2OH→ N2O → N2 (Zhao et al. 2010). In the above experiment, it was found that when NH4Cl was added as the only nitrogen source, no accumulation of NO2- and NO3- was found. As we all know, the enzymes related to nitrification and denitrification play an important role in the whole process of nitrogen removal. In our research, the genes encoding the enzymes related to nitrogen removal were amplified by PCR to determine the main denitrification pathway. We detected the genes related to nitrogen removal. The results showed amoA, hao, napA, nirK, nirS, norB, and nosZ genes were successfully amplified by bacterial liquid-phase PCR and gel electrophoresis C, and about 1200 bp, 850 bp, 300 bp, 480 bp, 420 bp, 600 bp, and 250 bp products were produced, respectively (Fig. S1). The napA gene encoding periplasmic nitrate reductase (NAP) was successfully amplified and can be used as a marker gene of HN-AD because it can be expressed under both anoxic and aerobic conditions (Holmes et al. 2019).
Factors influencing performance of HN-AD process
The influence of pH value on the nitrogen removal efficiency of ZQ-A1 is shown in Fig. 4a. The results show that ZQ-A1 grows faster in weak alkaline environment and can remove NH4+ more efficiently. When pH value is 5, the removal efficiency of NH4+ in 15 hours is 52.79%, and when pH value is 9, the removal rate of NH4+ can reach 83.58%. According to the growth and denitrification effect, our research found that pH mainly affects the nitrogen removal efficiency of ZQ-A1 strain by affecting its growth. The rotation speed determines the oxygen content. The faster rotation speed increased the concentration of dissolved oxygen, and a certain concentration of dissolved oxygen promoted the activity of bacterial nitrogen removal enzyme (Ren et al. 2021). In 15 h, when the rotating speed is 0 rpm, the NH4+-N removal rate of ZQ-A1 is 31.29%, and when the rotating speed rises to 200 rpm, the removal rate rises to 82.11% (Fig. 4b), which indicates that the rotating speed has a great influence on the NH4+ removal rate (Huan et al. 2022). At 18 h, the removal rate of 0 rpm group increased to 50.39%, and the OD600 also increased, which indicated that the effect of rotating speed on nitrogen removal performance of ZQ-A1 was realized by affecting its growth. The results showed that ZQ-A1 could grow under low dissolved oxygen, but its growth rate and denitrification rate were slow.
The growth rate of ZQ-A1 increases with the increase of C/N ratio, and the NH4+ removal rate also increases from 28.39% to 94.70% (Fig. 4c). At 18 h, when the C/N ratio was 1, the degradation rate of NH4+ was 3.45 mg/L·h, and when the C/N ratio was 20, the degradation rate increased to 10.57 mg/(L·h), which is similar to the previous research (Liu et al. 2022). Probably, the conversion of NH4+ into NO2- and NO3- requires more carbon sources; it may be because more carbon sources are needed as electron acceptors during nitrification. However, ZQ-A1 can remove nitrogen at a low nutritional level, which is similar to previous studies, but the strain grows relatively slowly because of relatively few nitrogen sources.
Carbon source is the growth energy source of bacteria, and it is also the electron donor in the nitrogen removal process (Hou et al. 2021). The effects of carbon sources on nitrification and denitrification of ZQ-A1 strain were studied with five different carbohydrates. The present study found that glucose could not be effectively utilized by ZQ-A1, which was consistent with the results of SNDPR-01 (Huang et al. 2022), but inconsistent with DM02 strain (Deng et al. 2021). In contrast, ZQ-A1 can make better use of sodium succinate, sodium citrate, and sodium acetate (Fig. 4d). Specifically, when the carbon source is sodium citrate, the removal efficiency is the highest, and the removal rate of NH4+ can reach 79.83% in 18 h.
In the experiment of the effect of temperature on nitrogen removal efficiency of ZQ-A1, this study found that temperature had little effect on the growth performance of ZQ-A1 (Fig. 4e). At 15 h, the NH4+ removal rate of ZQ-A1 strain at 15 °C was 80.01%, which was little different from 88.11% at 35 °C. This indicated that ZQ-A1 showed strong adaptability at low temperature, which was consistent with Acinetobacter sp. Y7 (Liu et al. 2013). It was found that at low temperature, the growth performance of ZQ-A1 was affected, but the nitrogen removal performance was not affected. As in previous studies, the key enzyme activities related to strain growth were inhibited at low temperatures (Wu et al. 2022). Overall, we conclude that temperature has little effect on the nitrogen removal of ZQ-A1. To sum up, ZQ-A1 has strong adaptability to environmental changes. Especially when the temperature and dissolved oxygen are low, ZQ-A1 can continue to grow and play its nitrogen removal role. These characteristics can reduce operating costs in wastewater treatment processes, which lays a foundation for ZQ-A1 subsequent application in pig farm sewage.
Preliminary application of strain ZQ-A1 in sewage treatment
Nitrogen removal performance of ZQ-A1 in pig farm wastewater
In our above research, we found that strain ZQ-A1 can carry out biological nitrogen removal in the environment with low rotating speed (means low dissolved oxygen), and ZQ-A1 also showed good adaptability to temperature change. In the actual pig farm sewage treatment, low oxygen demand means that the aeration rate can be reduced, so as to reduce the input of production cost. Moreover, it has a good adaptability to temperature changes, so it can better adapt to the changeable external environment.
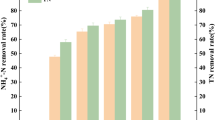
During the experiment, the rotating speed of the shaker was strictly controlled at 50 rpm, and the dissolved oxygen in sewage was controlled at 0.5~1 mg/L. As shown in Fig. 5a, after the 9th day, the NH4+ efficiency of the experimental group was significantly higher than that of the control group in a low dissolved oxygen environment. At the 27th day, the NH4+ content of the experimental group was only 11.02 mg/L, which was much lower than that of the control group (264.34 mg/L) (P < 0.0001). Also, the NH4+ removal efficiency reaches 99.58%, while control group is 88.89%. In addition, the study found that at the end of the experiment, the COD removal rate of the experimental group was 80.88% (Fig. 5b) and the TN removal rate was 98.26% (Fig. 5c), both of which were significantly higher than that of the control group (P < 0.001).
Preliminary application of strain ZQ-A1 in sewage treatment. a Within 30 days, the chemical oxygen demand (COD), total nitrogen (TN), NH4+-N, NO2—N, and NO3--N change curve (K is the control group and A is the experimental group); b–c the difference analysis of COD and TN removal efficiency on the 30th day, respectively; d relative expression levels of nitrification and denitrification functional genes (****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05)
According to the results of the relative expression of genes related to nitrogen removal on the 30th day, this study found that the relative gene expression of amoA, hao, nirS, and norB in the experimental group was higher than that in the control group that was one of the reasons why the experimental group had faster ammonia removal efficiency than the blank group (Fig. 5d). Specifically, the amoA gene was significantly higher than that in the control group (P < 0.001); it means that the flora in the experimental group can reduce the NH4+ content more efficiently. During the experiment, NO3- was found to accumulate, but no NO2- was found during the whole experiment. Combined with the expression of functional genes, the expression of nitrite reductase was higher than that of nitrate reductase, so NO2- could be better removed. Our research found that adding ZQ-A1 strain in the experimental group in low dissolved oxygen environment was helpful to the removal of COD, TN, and NH4+ and obviously shortened the time.
Analysis of community characteristics
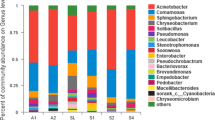
Through the analysis of community characteristics, the influence of ZQ-A1 on the flora structure in sewage was studied, and whether it was successfully colonized as a dominant flora was analyzed. At the genus level, according to PCA analysis, there are significant differences among the initial sewage, the blank group, and the experimental group. Further analysis revealed that the abundance of Desulfomonile, Nitrosomonas, Anaerovorax, and Turicibacter bacteria in the two groups has increased significantly compared with the initial sewage (Fig. 6a). Desulfomonile bacteria are one of the sulfate-reducing bacteria, which use 3-chlorobenzoic acid/fumaric acid/SO32-/S2O32-/S0/NO3- as electron acceptors under anaerobic conditions (Sun et al. 2001). Anaerovorax bacteria mainly exist in the anaerobic digestion system. Studies have shown that the abundance of Anaerovorax bacteria in anaerobic membrane bioreactors used to treat urban sewage has increased, and it has gradually become the dominant bacterial group (Kong et al. 2022). Nitrosomonas mainly include nitrite bacteria and nitrate bacteria. After 6–18 days, NO3--N increases and then gradually declines, which may be caused by the increase in the abundance of Nitrosomonas bacteria (Li et al. 2022), it may be the reason for the accumulation of nitrate nitrogen during the experiment (Li et al. 2022). In addition, the relative abundance of Nitrosomonas in the control group was 9.27%, which was significantly higher than that in the experimental group. However, due to the addition of ZQ-A1 bacteria to the experimental group, the abundance of Acinetobacter bacteria in the experimental group was 2.38%, which was significantly higher than that in the control group (0.06%). It also shows that exogenous ZQ-A1 bacteria gradually become the dominant flora and play a role in the test. It is one of the reasons for the big difference in the removal efficiency of total nitrogen and ammonia nitrogen between the two groups. It can also be found from ternary diagram (Fig. 6c) and species clustering diagram (Fig. 5d) that Acinetobacter is an endemic genus of the experimental group. This study found that that the abundance of bacteria belonging to the genus Comamonas in the experimental group increased from 0.09 to 2.61%, and the analysis of genus-level evolution showed that Acinetobacter and Comamonas were the closest in the experimental group (Fig. S3). Previous studies showed that Comamonas had denitrification function, and it was speculated that Comamonas and Acinetobacter played a role in denitrification together. It is worth noting that the concentration of denitrifying bacteria, Hydrogenophaga, enriched in low dissolved oxygen increased from 0.02% to 1.13% in this experimental group (Feng et al. 2018); this also indicates that the concentration of dissolved oxygen in this experiment has been at a relatively low level.
The application results of strain ZQ-A1 in pig farm wastewater show that ZQ-A1 not only has a good removal effect on total nitrogen and ammonia nitrogen but also can effectively reduce the COD concentration in aquaculture concentration. At the same time, it also shows that ZQ-A1 can adapt to the complex sewage environment and has a competitive advantage, which lays a theoretical foundation for its subsequent application.
In order to further analyze the correlation between the reasons for the decline of COD, TN, NH4+, and microorganisms, we made redundancy analysis (RDA) on the removal efficiency of COD, TN, NH4+, and microorganisms (Fig. 7a). The results showed that there was a strong correlation between the microbial community composition of the experimental group and the removal efficiency of NH4+. To combine with the relative abundance of microorganisms, it can be inferred that the relative abundance of Acinetobacter bacteria in the experimental group increased significantly, resulting in higher NH4+ removal efficiency. In addition, our research explained the correlation analysis between nitrogen removal functional genes and the content of TOP30 bacteria by Spearman correlation analysis (Fig. 7b). The results showed that napA and nirS genes were positively correlated with Acinetobacter (P < 0.05), and ZQ-A1 bacteria in the experimental group could express these two genes. Interestingly, the Comamonas has strong correlation with other genes except napA and nirS, so it can be further speculated that Acinetobacter and Comamonas in the experimental group have synergistic effect. Interestingly, there is a significant correlation between bacteria Hydrogenophaga and nosZ, norB, and nirK related to denitrification (P < 0.05). In this study, it was found that the concentration of NO3-increased during the experiment, presumably because the abundance of Hydrogenophaga was low in the middle of the experiment (day 6 to 18), and the concentration of NO3- began to decrease in the late stage of the experiment (day 18 to 30) due to the increase of the relative abundance of Hydrogenophaga.
Correlation analysis and KEGG function prediction. a RDA analysis, b PCA analysis of KEEG functional prediction, c correlation between nitrogen removal gene and bacterial community (red represents positive correlation, blue represents negative correlation, and the darker the color, the higher the correlation), d multi-group difference analysis of KEEG functional prediction
The PCA results show that the flora function of the experimental group and the control group is quite different from that of the initial sewage, and the functions of the experimental group and the control group are similar to each other (Fig. 7c). We further found through cluster analysis that the abundance of xenobiotics biodegradation and metabolism, metabolism of other amino acids, and lipid metabolism in the treated sewage, whether in the experimental group or the control group, is higher than before treatment. Therefore, the microbial community in the treated sewage has stronger metabolic function, so as to reduce the concentration of COD and NH4+ in the sewage. Further analysis of the functional differences between the experimental group and the control group showed that the abundance of fatty acid biosynthesis, thiamine metabolism, nicotinate and nicotinate metabolism, and fluorobenzoate degradation in the experimental group was significantly higher than that in the control group (P < 0.01) (Fig. 7d). Nicotinate forms coenzyme I and coenzyme II with ribose, phosphoric acid, and adenophorin in vivo (Romani et al. 2019). They are coenzymes of many dehydrogenases, which play a role in dehydrogenation and hydrogenation in vivo biological oxidation. In the study of carbon source utilization, we found that ZQ-A1 can make better use of carbon sources related to tricarboxylic acid cycle (e.g., sodium succinate, sodium citrate, and sodium acetate), and the abundance of fatty acid biosynthesis and nicotinate and nicotinate metabolism function is high, which indicates that the addition of ZQ-A1 in the experimental group leads to the enhancement of grape glycolysis, fat metabolism, and pyruvate metabolism.
Conclusions
In this study, a HN-AD bacterium, Acinetobacter ZQ-A1, was isolated. This bacterium has strong simultaneous nitrification and denitrification ability. Through the identification of genes related to nitrification and denitrification, it is speculated that the nitrogen removal pathway is NH4+ → NH2OH→NO2−→NO3−→NO2–→NO→N2O→N2. Through the experiment of nitrogen removal characteristics, it was found that ZQ-A1 showed high nitrogen removal ability whether facing single nitrogen source or mixed nitrogen source. ZQ-A1 can also adapt to complex environments, especially low temperature and low dissolved oxygen, which provides a theoretical basis for reducing production costs. In addition, strain ZQ-A1 has a good application in pig farm sewage treatment. On the 30th day, the concentration of ammonia nitrogen in the experimental group decreased from 1459.39 mg/L to 11.02 mg/L, and the removal rate reached 99.36%. The results of microbial community characteristics and KEGG function prediction show that strain ZQ-A1 can colonize sewage and accelerate the removal of NH4+ in sewage, which shows that ZQ-A1 has great potential in pig farm sewage treatment process.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. The data presented in this study are available upon request from the corresponding author.
References
APHA (2005) Standard methods for the examination of water and wastewater. American Public Health Association (APHA), Washington, DC
Bru D, Sarr A, Philippot L (2007) Relative abundances of proteobacterial membrane-bound and periplasmic nitrate reductases in selected environments. Appl Environ Microbiol 73(18):5971–5974. https://doi.org/10.1128/AEM.00643-07
Chen J, Xu J, Zhang S et al (2021a) Nitrogen removal characteristics of a novel heterotrophic nitrification and aerobic denitrification bacteria, Alcaligenes faecalis strain WT14. J Environ Manage 282:111961. https://doi.org/10.1016/j.jenvman.2021.111961
Chen P, Zhang F, Zhang L et al (2022) Characterization of a novel salt-tolerant strain Sphingopyxis sp. CY-10 capable of heterotrophic nitrification and aerobic denitrification. Bioresour Technol 358:127353. https://doi.org/10.1016/j.biortech.2022.127353
Chen R, Deng M, He X et al (2017) Enhancing nitrate removal from freshwater pond by regulating carbon/nitrogen ratio. Front Microbiol 8:1712. https://doi.org/10.3389/fmicb.2017.01712
Chen X, Zhang Q, Zhu Y et al (2021b) Response of wastewater treatment performance, microbial composition and functional genes to different C/N ratios and carrier types in MBBR inoculated with heterotrophic nitrification-aerobic denitrification bacteria. Bioresour Technol 336:125339. https://doi.org/10.1016/j.biortech.2021.125339
Cui Y, Cui YW, Huang JL (2021) A novel halophilic Exiguobacterium mexicanum strain removes nitrogen from saline wastewater via heterotrophic nitrification and aerobic denitrification. Bioresour Technol 333:125189. https://doi.org/10.1016/j.biortech.2021.125189
Deng M, Zhao X, Senbati Y et al (2021) Nitrogen removal by heterotrophic nitrifying and aerobic denitrifying bacterium Pseudomonas sp. DM02: removal performance, mechanism and immobilized application for real aquaculture wastewater treatment. Bioresour Technol 322:124555. https://doi.org/10.1016/j.biortech.2020.124555
Duan H, Gao S, Li X et al (2020) Improving wastewater management using free nitrous acid (FNA). Water Res 171:115382. https://doi.org/10.1016/j.watres.2019.115382
Feng L, Jia R, Zeng Z et al (2018) Simultaneous nitrification-denitrification and microbial community profile in an oxygen-limiting intermittent aeration SBBR with biodegradable carriers. Biodegradation 29(5):473–486. https://doi.org/10.1007/s10532-018-9845-x
Gu X, Leng J, Zhu J et al (2022) Influence mechanism of C/N ratio on heterotrophic nitrification- aerobic denitrification process. Bioresour Technol 343:126116. https://doi.org/10.1016/j.biortech.2021.126116
He T, Li Z, Sun Q et al (2016) Heterotrophic nitrification and aerobic denitrification by Pseudomonas tolaasii Y-11 without nitrite accumulation during nitrogen conversion. Bioresour Technol 200:493–499. https://doi.org/10.1016/j.biortech.2015.10.064
He T, Zhang M, Ding C et al (2022) New insight into the nitrogen removal capacity and mechanism of Streptomyces mediolani EM-B2. Bioresour Technol 348:126819. https://doi.org/10.1016/j.biortech.2022.126819
Henry S, Bru D, Stres B et al (2006) Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl Environ Microbiol 72(8):5181–5189. https://doi.org/10.1128/AEM.00231-06
Holmes DE, Dang Y, Smith JA (2019) Nitrogen cycling during wastewater treatment. Adv Appl Microbiol 106:113–192. https://doi.org/10.1016/bs.aambs.2018.10.003
Hou P, Sun X, Fang Z et al (2021) Simultaneous removal of phosphorous and nitrogen by ammonium assimilation and aerobic denitrification of novel phosphate-accumulating organism Pseudomonas chloritidismutans K14. Bioresour Technol 340:125621. https://doi.org/10.1016/j.biortech.2021.125621
Huan C, Yan Z, Sun J et al (2022) Nitrogen removal characteristics of efficient heterotrophic nitrification-aerobic denitrification bacterium and application in biological deodorization. Bioresour Technol 363:128007. https://doi.org/10.1016/j.biortech.2022.128007
Huang MQ, Cui YW, Huang JL et al (2022) A novel Pseudomonas aeruginosa strain performs simultaneous heterotrophic nitrification-aerobic denitrification and aerobic phosphate removal. Water Res 221:118823. https://doi.org/10.1016/j.watres.2022.118823
Kong QX, Wang XW, Jin M et al (2006) Development and application of a novel and effective screening method for aerobic denitrifying bacteria. FEMS Microbiol Lett 260(2):150–155. https://doi.org/10.1111/j.1574-6968.2006.00306.x
Kong Z, Li L, Wu J et al (2022) Unveiling the characterization and development of prokaryotic community during the start-up and long-term operation of a pilot-scale anaerobic membrane bioreactor for the treatment of real municipal wastewater. Sci Total Environ 813:152643. https://doi.org/10.1016/j.scitotenv.2021.152643
Kouba V, Svehla P, Catrysse M et al (2019) How biomass growth mode affects ammonium oxidation start-up and NOB inhibition in the partial nitritation of cold and diluted reject water. Environ Technol 40(6):673–682. https://doi.org/10.1080/09593330.2017.1403491
Lang X, Li Q, Xu Y et al (2019) Aerobic denitrifiers with petroleum metabolizing ability isolated from caprolactam sewage treatment pool. Bioresour Technol 290:121719. https://doi.org/10.1016/j.biortech.2019.121719
Li Y, Liang H, Yang W et al (2022) Enhanced nitrogen removal in mainstream deammonification systems at ambient temperature by novel modified carriers and differentiation of microbial community transformation. Bioresour Technol 366:128158. https://doi.org/10.1016/j.biortech.2022.128158
Liang Y, Wu C, Wei X et al (2021) Characterization of nirS- and nirK-containing communities and potential denitrification activity in paddy soil from eastern China. Agric Ecosyst Environ 107561. https://doi.org/10.1016/j.agee.2021.107561
Lin JT, Stewart V (1998) Nitrate assimilation by bacteria. Adv Microb Physiol 39:1–379. https://doi.org/10.1016/s0065-2911(08)60014-4
Liu B, Mao Y, Bergaust L et al (2013) Strains in the genus Thauera exhibit remarkably different denitrification regulatory phenotypes[J]. Environ Microbiol 15(10):2816–2828
Liu S, Liu Q, Wu H et al (2022) Integrative chemical and omics analysis of the ammonia nitrogen removal characteristics and mechanism of a novel oligotrophic heterotrophic nitrification-aerobic denitrification bacterium. Sci Total Environ 852:158519. https://doi.org/10.1016/j.scitotenv.2022.158519
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Ren J, Bai X, Liu Y et al (2021) Simultaneous nitrification and aerobic denitrification by a novel isolated Ochrobactrum anthropi HND19. Bioresour Technol 340:125582. https://doi.org/10.1016/j.biortech.2021.125582
Richardson DJ, Wehrfritz JM, Keech A et al (1998) The diversity of redox proteins involved in bacterial heterotrophic nitrification and aerobic denitrification. Biochem Soc Trans 26(3):401–408. https://doi.org/10.1042/bst0260401
Robertson LA, Kuenen JG (1990) Combined heterotrophic nitrification and aerobic denitrification in Thiosphaera pantotropha and other bacteria. Antonie Van Leeuwenhoek 57(3):139–152. https://doi.org/10.1007/BF00403948
Romani M, Hofer DC, Katsyuba E et al (2019) Niacin: an old lipid drug in a new NAD+ dress. J Lipid Res 60(4):741–746. https://doi.org/10.1194/jlr.S092007
Sun B, Cole JR, Tiedje JM (2001) Desulfomonile limimaris sp. nov., an anaerobic dehalogenating bacterium from marine sediments. Int J Syst Evol Microbiol 51(Pt 2):365–371. https://doi.org/10.1099/00207713-51-2-365
Wu L, Ding X, Lin Y et al (2022) Nitrogen removal by a novel heterotrophic nitrification and aerobic denitrification bacterium Acinetobacter calcoaceticus TY1 under low temperatures. Bioresour Technol 353:127148. https://doi.org/10.1016/j.biortech.2022.127148
Xi H, Zhou X, Arslan M et al (2022) Heterotrophic nitrification and aerobic denitrification process: promising but a long way to go in the wastewater treatment. Sci Total Environ 805:150212. https://doi.org/10.1016/j.scitotenv.2021.150212
Zhang QL, Liu Y, Ai GM, Miao LL, Zheng HY, Liu ZP (2012) The characteristics of a novel heterotrophic nitrification-aerobic denitrification bacterium, Bacillus methylotrophicus strain L7. Bioresour Technol 108:35–44
Zhang M, Pan L, Su C et al (2021) Simultaneous aerobic removal of phosphorus and nitrogen by a novel salt-tolerant phosphate-accumulating organism and the application potential in treatment of domestic sewage and aquaculture sewage. Sci Total Environ 758:143580. https://doi.org/10.1016/j.scitotenv.2020.143580
Zhang M, He T, Chen M et al (2022) Ammonium and hydroxylamine can be preferentially removed during simultaneous nitrification and denitrification by Pseudomonas taiwanensis EN-F2. Bioresour Technol 350:126912. https://doi.org/10.1016/j.biortech.2022.126912
Zhang Y, Shi Z, Chen M et al (2015) Evaluation of simultaneous nitrification and denitrification under controlled conditions by an aerobic denitrifier culture. Bioresour Technol 175:602–605. https://doi.org/10.1016/j.biortech.2014.10.016
Zhao B, He YL, Hughes J et al (2010) Heterotrophic nitrogen removal by a newly isolated Acinetobacter calcoaceticus HNR. Bioresour Technol 101(14):5194–5200. https://doi.org/10.1016/j.biortech.2010.02.043
Zhao B, An Q, He YL et al (2012) N2O and N2 production during heterotrophic nitrification by Alcaligenes faecalis strain NR. Bioresour Technol 116:379–385. https://doi.org/10.1016/j.biortech.2012.03.113
Zhao B, Tian M, An Q, Ye J, Guo JS (2017) Characteristics of a heterotrophic nitrogen removal bacterium and its potential application on treatment of ammonium-rich wastewater. Bioresour Technol 226:46–54. https://doi.org/10.1016/j.biortech.2016.11.120
Zheng HY, Liu Y, Gao XY et al (2012) Characterization of a marine origin aerobic nitrifying-denitrifying bacterium. J Biosci Bioeng 114(1):33–37. https://doi.org/10.1016/j.jbiosc.2012.02.025
Funding
This research was funded by the Guangdong Major Project of Basic and Applied Basic Research (No. 2020B0301030007) and the Natural Science Foundation of Guangdong Province (2022A1515012473; 2020A1515010295).
Author information
Authors and Affiliations
Contributions
Conceptualization, Z.C.; methodology, Z.C. and F.H.; formal analysis, Z.C. and F.H.; writing—original draft preparation, Z.C., X.Y.Z., and R.Y.Z.; writing—review and editing, Y.W., X.D.L., and Y.B.W.; supervision, M.J.Y., Y.Y.F., and L.T. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethical approval
The experimental project was approved by the Ethics Committee.
Informed consent
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Responsible Editor: Gerald Thouand
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cao, Z., Huang, F., Zhang, R. et al. Nitrogen removal characteristics of heterotrophic nitrification-aerobic denitrification bacterium Acinetobacter ZQ-A1 and community characteristics analysis of its application in pig farm wastewater. Environ Sci Pollut Res 30, 104029–104042 (2023). https://doi.org/10.1007/s11356-023-29556-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-023-29556-9