Abstract
Imidacloprid is one of the frequently used insecticides. Indiscriminate use of imidacloprid makes it perilous to non-target organisms as well as the environment, including soil and water sources, thus, making its elimination from the environment an irresistible concern. Bioremediation is a technique that uses the degrading capabilities of bacteria to create an economical and reliable method of pesticide abatement. In an attempt to solve the problem arising due to imidacloprid contamination, bacterial strains possessing the ability to degrade imidacloprid were isolated from contaminated agricultural soil samples in the present study. Imidacloprid-degrading isolate, identified as Tepidibacillus decaturensis strain ST1, could effectively degrade imidacloprid in liquid media, slurry, and soil microcosms. The microcosm studies using the isolate resulted in the degradation of around 77.5% and 85% of imidacloprid (200 ppm) in sterile and unsterile soils within 45 days. In addition to biodegradation, sorption of insecticide by the plants and natural reduction of insecticide over time has also been reported. The degradation in soil follows first-order kinetics. Hydrazinecarboxamide and hydroxyurea were identified as metabolites on conducting GC–MS analysis of the degraded samples.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
As the global population is increasing continuously, pesticides have become indispensable components of modern agriculture for cost-effective pest management to improve crop yield. Only 0.1–2% of all pesticides applied reach the target organisms (Negi et al. 2014). It is expected that pesticides and other environmental contaminants will become more prevalent in the future due to their overuse. Pesticide residues have been found to persist in the environment for more than a decade in some cases, as a result of overuse and detected in water sources and soil above the permissible limit (Craddock et al. 2019; Yadav et al. 2015; Silva et al. 2019). Pesticides can be found in relatively high concentrations in potable water and soils, causing adverse impacts on human health due to their high neurotoxicity and carcinogenicity (Choi et al. 2004; Wallace and Buha Djordjevic 2020). Pesticide residues and their metabolites usually intensify in the environment and have lethal impacts on the health of humans and different ecosystems due to their recalcitrant nature (Shekher Giri et al. 2020).
Imidacloprid belongs to the neonicotinoid class of insecticides and is used in a wide range of crops. It is effective against fleas of domestic animals, sucking insects, and chewing insects (Jeschke et al. 2011). It is a potential pollutant of surface and subsurface water because of its low soil sorption and higher leaching capability (Pietrzak et al. 2020). The field dissipation rates of imidacloprid vary greatly and have been observed to break-down at a slower rate in the soil in certain cases, with half-lives in non-vegetated soil, exceeding 180 days (Anhalt et al. 2007). Imidacloprid sorption is directly proportional to the soil organic matter. It is crucial to eliminate these chemo-pollutants from the environment because of the adverse impact of imidacloprid on non-target entities such as humans, bees, and earthworms to some extent, as well as their ability to leach and contaminate groundwater (Wettstein et al. 2016).
Pesticide removal from the environment has been an area of exhaustive research in recent years. Several techniques have been adopted to eliminate or degrade pesticides from air, water, food, and soil. Several studies have focused on the development of physicochemical methods for the elimination of such contaminants from soil and water (Pang et al. 2020; Erguven and Yildirim 2019). Physico-chemical methods such as absorption, illumination, ionizing radiation, ultrasonic technology, hydrolysis, photo Fenton, photocatalysis, and oxidative breakdown have been studied for imidacloprid biodegradation (Malato et al. 2001; Pang et al. 2020; Qurie et al. 2021). Nevertheless, the conventional methods have significant drawbacks including excessive sludge generation, equipment toxicity, generation of toxic byproducts, and high operating costs (Ajiboye et al. 2020; Saleh et al. 2020). To overcome these issues, biological method have been proposed as a viable method for abatement of such recalcitrant compounds. Because of their environmentally friendly nature, biological approaches have become a method of choice for removal of a diverse range of contaminants. In the recent years, several researchers across the globe have studied the imidacloprid degrading capability of various microorganisms isolated from different sources (Anhalt et al. 2008; Pandey et al. 2009; Sharma et al. 2014; Hu et al. 2013). A large number of bacteria including Pseudomonas, Alcaligens, Sphingomonas, Stenotrophomonas, Rhodococcus, and Bacillus (Qureshi et al. 2007; Geed et al. 2018; Rank et al. 2018; Erguven and Yildirim 2019; Bhattacherjee et al. 2020; Kavitha et al. 2020; Dai et al. 2021) have been reported for bioremediation of various contaminated sites. Microbial degradation of imidacloprid in soil has been reported by various research groups using bacterial strains such as Enterobacter sp. strain ATA1 (Sharma et al. 2014), Bacillus aerophilous (Akoijam and Singh 2015), Klebsiella Pneumoniae strain BCH1 (Phugare et al. 2013), Bacillus weihenstephanensis (Shetti et al. 2014), Liefsonia strain PC21 (Anhalt et al. 2007), Hymenobacter latericoloratus CGMCC 16,346 (Guo et al. 2020), Ochrobactrum thiophenivorans, and Sphingomonas melonis (Erguven and Demirci 2021). Some researchers also observed that mixed cultures are more likely to undergo complete mineralization as compared to pure cultures (Briceño et al. 2020).
However, the success of biological methods are controlled by factors such as environmental compatibility, accessibility of the microorganisms to pesticide compounds, and identification of appropriate pesticide-degrading microorganisms (Azubuike et al. 2016; Dua et al. 2002). Pesticide biodegradation is influenced by pesticide bioavailability and microbial activity (Odukkathil and Vasudevan 2013). The low bioavailability of imidacloprid and its metabolites in soil may limit biodegradation, leading to extended half-lives. With time, imidacloprid and its metabolite become more strongly bonded to the soil (Anhalt et al. 2007).
Scaling up pesticide bioremediation technologies from the lab to the field is still a difficult challenge, despite decades of research. Artificial, simplified ecosystems called microcosms are used in controlled environments to mimic and predict the behavior of natural ecosystems. Laboratory microcosm studies have been carried out to address the issue of inconsistency and reproducibility, as well as to give a higher degree of accuracy for the prediction of environmental impacts of xenobiotics on the ecosystem.
In the present study, an attempt has been made to identify potential imidacloprid degrading bacterial species from soil, followed by batch studies in liquid, slurry, and soil. Microcosm studies have also been done to get an idea about the pattern of imidacloprid degradation in agricultural fields under natural conditions.
Materials and methods
Isolation of imidacloprid degrading bacteria
Soil samples were collected from the agricultural field of Banaras Hindu University (25.2586° N, 82.9880° E) from the subsurface layer, i.e., 5–15 cm depth. The samples were immediately taken to the laboratory for further analysis. To remove debris and plant residues, the samples were air-dried and sieved. The soil was sieved to remove particles larger than 0.5 mm and stored at 4 °C until further use. The sample was examined for physicochemical properties before being enriched with imidacloprid. Soil pH, moisture, and organic matter contents were measured. Soil sample (10 g) was suspended in a flask, containing 50 ml of modified nutritional broth enriched with imidacloprid and glucose during the broth enrichment process. Imidacloprid was filter sterilized and aseptically added to the media, and equal amount of glucose was added as a co-substrate. The amount of glucose was gradually decreased, and imidacloprid was used as the sole source of carbon after 20 days. The enrichment process was carried out for 45 days.
The bacteria obtained through the enrichment process were cultured in a specified mineral salt medium (MSM) and prepared in a liter of deionized water. The composition of MSM is as follows: KH2PO4 (1 g/L), K2HPO4. 2H2O (1 g/L), MgSO4.7H2O (0.3 g/L) NaCl (0.5 g/L), (NH4)2. SO4 (0.3 g/L), and trace elements [CaCl2 (2 g/L), MnSO4.H2O (3 g/L), FeSO4 (0.25 g/L), ZnSO4 (0.05 g/L), MgSO4 (1 g/L)]. The media pH was adjusted to 7 ± 0.2 and autoclaved at 121 °C for 15 min for sterilization. Morphologically different colonies were identified and subcultured using nutrient agar and mineral salt agar slants. The isolated bacterial colonies were further subcultured to obtain pure strains. Imidacloprid degrading capability of individual isolates was examined, and the most effective bacterial species was selected for further studies. Batch experiments were conducted to study the effect of temperature, pH, shaking speed, and inoculum dose.
Maximum tolerance level of imidacloprid in selected bacterial isolates
The isolated bacterial culture was inoculated into different sets of sterilized minimum broth (25 ml) spiked with imidacloprid varying from 50 to 250 ppm and incubated at 30 °C for 72 h at 120 rpm. The control flask was uninoculated. Growth of bacteria was assessed using absorbance at 600 nm. The imidacloprid concentration that sustained the maximum growth was recorded.
Morphological, biochemical, and molecular characterization of isolates
The pure colonies were identified morphologically for their size, and shape, Gram staining ability, and biochemical tests. The genomic DNA of isolated bacteria was extracted. The spectrophotometric quantification of the DNA sample was done at wavelengths of 260 and 280 nm, with the assumption that one absorbance unit at 260 nm wavelength is equivalent to 50 µg DNA per ml (Gallagher 2017). Agarose gel electrophoresis was used to evaluate the quality and purity of the DNA. Amplification of the collected DNA was carried out using PCR (BIO-RAD T100 Thermo Cycler) in a 50-µL reaction mixture containing 20 pmol of universal primers (8F-5′ AGAGTTTGATCCTGGCTCA3′ and 1492 R-5′GGTTACCTTGTTACGACTT3′) each, 20 ng of template DNA, and 25 µL PCR Master Mix. The thermocycling conditions were as follows: initial denaturation at 94 °C for 5 min; followed by 35 cycles of 94 °C (1 min), 50 °C (1 min), 72 °C (1 min); and final extension at 72 °C for 10 min. The amplified PCR products were sequenced (Bioraj Laboratories, Nagpur).
16S rRNA sequencing and phylogenetic analysis were done for molecular identification of the isolate. To identify the organism, the nucleotide sequences were used in a BLAST analysis against the NCBI database. Phylogenetic trees were constructed by using MEGA (version 10.2.5) software. GenBank accession number was assigned for 16S rRNA gene sequences of the isolate.
Imidacloprid extraction and quantification
After 45 days of treatment, the total imidacloprid residue in the soil, as well as uptake by the plant, was analyzed using HPLC. For determining residual imidacloprid concentration in slurry, plant parts, and soil, extraction was performed as described.
Residual imidacloprid from soil was extracted by adding 5 g of soil to 20 ml acetonitrile in a flask and shaken for 1 h. The mixture was then centrifuged and filtered. The filtrate was transferred in a round bottom flask, and it was dried in a rotary evaporator at 50 °C. The residue was dissolved in methanol and filtered by a 0.22-μ filter. Samples of imidacloprid were analyzed using HPLC. The plant parts were crushed and homogenized. A representative sample of the homogenized plant was extracted with 100 ml acetonitrile. The extract was filtered and salted out with sodium chloride, followed by drying. The dried fraction was dissolved in acetonitrile: water (4:1, v/v) and vortexed for 30 s. The supernatant was dried in a rotary evaporator. The residue was dissolved in methanol for further analysis.
The equipment used for quantification of imidacloprid was a high-performance liquid chromatograph (Shimadzu, Japan), comprising of a binary pump and photodiode array (PDA) detector. The C18 analytical column was operated isocratically at room temperature with a flow rate of 1 ml/min using 80:20 (v/v) acetonitrile and HPLC grade water. The compounds were detected using UV spectroscopy at 270 nm, by injecting 25 μL of samples. Appropriate calibration standards were used for quantitative analysis of the samples. The LC solutions software was used to collect and process data. Facilities for GC–MS were provided by the Department of Chemistry, IIT (BHU) Varanasi.
Imidacloprid degradation in liquid medium
The bacterial isolate was assessed for the capability to breakdown imidacloprid in MSM liquid medium. Flasks (250 ml) containing 100 ml of MSM were inoculated with the isolate (24-h-old bacterial culture, containing about 1 × 106 cells) and maintained for 20 days. Aseptically, 1 ml aliquots were obtained every 5 days to measure the growth of the bacteria and the level of imidacloprid in the culture media. Optical density at 600 nm (OD600) measurement was used to measure growth, and high-performance liquid chromatography (HPLC) was used to calculate the residual imidacloprid. Controls were uninoculated flasks spiked with the same concentration of imidacloprid. The experiment was carried out in triplicate.
Biodegradation of imidacloprid in soil slurry
Experiments on imidacloprid biodegradation were also conducted in soil slurry. The slurry was prepared in flasks by adding 30 g of sterile soil as well as unsterile soil separately and 70 ml of autoclaved distilled water. The resulting slurry from the sterile soil sample was autoclaved again. Slurries were spiked with imidacloprid (200 ppm) and incubated at 30 ± 2 °C in a shaking incubator (120 rpm), for 20 days. Experiments were conducted in triplicate with the corresponding biotic and abiotic controls. Residual imidacloprid concentration and microbial growth were determined at regular intervals throughout the study.
In situ biodegradation of imidacloprid in soil microcosm
A microcosm analysis was conducted to estimate the imidacloprid degrading capability of the bacterial isolate in soil under natural conditions. The collected soil sample was divided into two sets, one was sterilized and the other was unsterilized. From each set, four subsets containing 1000 g of soil sample were taken in four containers. 1000 g of dry soil was weighed and added to each container, and soil moisture was adjusted by using a mixture of imidacloprid and deionized water. The containers spiked with imidacloprid, without bacterial inoculation (sets 1 and 2) served as control. The treatments in sterile soil (A) and unsterile soil (B) were as follows: control (set 1): (soil + imidacloprid), control (set 2): (soil + imidacloprid + plant), treatment of soil with imidacloprid and bacteria but without plantation (set 3): (soil + imidacloprid + bacteria), treatment of soil using combination of bacteria and plant (set 4): (soil + imidacloprid + plant + bacteria).
A total of 24 treatments were included with 12 sets of sterilized and unsterilized soil samples each. The containers were watered regularly to maintain the moisture content of the soil. Imidacloprid was added to all of the containers in each set at a final concentration of 200 mg/kg. After mixing, a suspension of 24-h-old imidacloprid-degrading bacterial isolate (1.0 × 106 CFU/g) was added to the soil samples in sets 3 and 4. For plantation, Cicer arietinum seeds were surface sterilized with HgCl2 (0.2%) for 5 min before being thoroughly washed with sterile distilled water. The losses other than biodegradation were also noted. The experiment was conducted in triplicate in sterilized as well as unsterilized soil. Imidacloprid residue was extracted from each container separately after every 5 days and quantified using HPLC.
Kinetic analysis of imidacloprid degradation
To fit the experimental data of imidacloprid degradation kinetics, the first-order kinetic equation was used:
where Co is the initial concentration of imidacloprid in the medium, Ct is the concentration of imidacloprid at time t, k is the degradation rate constant (day−1), and t is the degradation time. The biodegradation half-life (T1/2) of imidacloprid was calculated as
The natural logarithm values of C were plotted against time t to determine k. Residual imidacloprid concentration and % degradation at different time intervals were calculated with respect to imidacloprid residue on the initial day, after the application (100%). The slope of the line was used to calculate the k of each concentration.
Results and discussion
Isolation and screening of pesticide degrading bacteria
The soil’s physicochemical properties were as follows: pH 7.2, carbon 7.5%, nitrogen content 0.5%, moisture content 20%, organic matter 8.5%, and ash content 91.5%. In the study, the optimum conditions for imidacloprid degradation using the bacterial isolate were found to be: pH 7 ± 0.2, temperature 30 °C, shaking speed 120 rpm, and inoculum dose 1 × 106 CFU/ml.
The most efficient isolated bacteria was characterized and identified as Tepidibacillus decaturensis strain ST1 (Accession number MZ208922). The phylogenetic tree obtained using MEGA (version 10.2.5) software has been represented in Fig. 1.
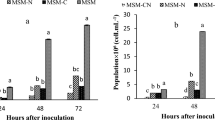
The growth pattern of isolates was studied with respect to time and imidacloprid concentration. Figure 2 shows the growth of isolates at OD600, supplemented with imidacloprid. It can be noted that there was an initial lag phase, followed by an exponential phase, where absorbance was highest at 600 nm, indicating the maximum growth of bacteria. However, after 35 h, the stationary phase was attained. The isolated bacteria used in the present study could effectively degrade imidacloprid up to 200 mg/L, beyond 200 mg/L; the isolates were not capable of metabolizing imidacloprid. Inoculum density is a critical variable influencing the effective degradation of applied pesticides. For studying imidacloprid degradation, an inoculum of 1 × 106 CFU/ml was used and found capable of removing imidacloprid proficiently.
Imidacloprid biodegradation in liquid media and slurry
The % removal of imidacloprid was found to be 18.5%, 42.5%, 75%, and 90% on the 5th, 10th, 15th, and 20th days, respectively. However, in control, 89% of the imidacloprid remained undegraded after the 20th day. On the 25th day, a negligible reduction in imidacloprid residue was observed as compared to the 20th day of degradation. Figure 3a shows the degradation of imidacloprid in a liquid medium.
The bacterial treatment was found to be considerably effective in imidacloprid remediation over control without inoculation in soil slurry. The removal of imidacloprid was least in the case of sterilized slurry (without bacteria and other microorganisms). The sterilized slurry after inoculation with the isolated bacterial species was capable of degrading more than 80% of the imidacloprid within 20 days. Figure 3b shows the biodegradation of imidacloprid in slurry.
Several studies have been conducted on soil slurries for the degradation of various organic compounds, including insecticides. The study conducted by Kumar et al. (2007) for endosulfan biodegradation resulted in about 75% removal in 10 days using mixed cultures. In another study, cypermethrin degradation was studied by Bhatt et al. (2020) and reported more than 83% degradation in 15 days.
Imidacloprid biodegradation in soil microcosm
Tepidibacillus decaturensis strain ST1, an indigenous bacteria from the collected soil sample, was identified as the predominant bacterium in the enrichment and was found to be the most effective isolate for imidacloprid biodegradation. In a similar study, conducted by Hu et al. (2013), indigenous bacteria were found to be effective for imidacloprid degradation. The selected bacterial strain was capable of degrading imidacloprid in liquid medium, slurry, and soil, in the present study. According to previous studies on pesticide degradation, microorganisms capable of degrading pesticides in culture media can also degrade pesticides in soil (Lu et al. 2013).
Pattern of imidacloprid degradation was as follows: set 4 (82%) > set 3 (77.5%) > set 2 (11%) > set 1 (5.5%) in case of sterile soil (A). The degradation of imidacloprid in case of unsterile soil sample (B) was set 4 (91%) > set 3 (85%) > set 2 (13.5%) > control (7.5%). Best % degradation of imidacloprid was observed when unsterile soil was used along with bacteria and plant (set 4 B). The difference in residual imidacloprid in sterile and unsterile soils in the case of set 3 and set 4 has been presented in Fig. 4a and b, respectively. Figure 5 represents the degradation patterns of imidacloprid in the case of sterilized and unsterilized soils in all the sets. Table 1 shows the % degradation of imidacloprid in sterile and unsterile soils in set 1, set 2, set 3, and set 4.
Results indicate that imidacloprid can be degraded to a larger extent in sterilized soil when inoculated with the isolated bacteria, indicating the efficacy of Tepidibacillus decaturensis in imidacloprid degradation. Imidacloprid degradation was observed to be higher in inoculated non-sterilized soils, with a plantation (91%) as compared to sterilized soils (82%). This study indicates that soil microflora facilitates imidacloprid degradation. The results of this research indicate that employing soil bacteria for degradation of imidacloprid is efficient. Similar results have been reported by Sabourmoghaddam et al. (2015). Imidacloprid degradation was more pronounced in inoculated non-sterile soils than in inoculated sterile soils, demonstrating the potential of indigenous microorganisms for bioremediation. Higher degradation in unsterile soil samples could be due to inoculation of degrading bacteria in the soil, which increases the catabolic potential of the soil. Furthermore, the presence of indigenous bacteria most likely performed a synergistic effect in biodegradation. This was also favored by previous studies that showed that soil bacteria play a vital role in pesticide degradation. According to previous researches, the involvement of native soil microorganisms in pesticide bioremediation processes is variable. For example, Bidlan et al. (2004) reported that the degradation of hexachlorocyclohexane (HCH) isomers by the indigenous microflora was limited during the bioremediation of soils using known bacterial consortium polluted with HCH. In contrast, Cycoń et al. (2013) investigated the biodegradation of a mixture of organophosphorus pesticide by Serratia marcescens in soils of different textures, and they reported that the indigenous microflora present in each type of soil was capable of degrading the pesticides under investigation. According to the study conducted by Fuentes et al. (2017), degradation of pesticides could be achieved by actinobacteria even in the presence of native microflora.
A lower degradation rate was observed in the case of control, i.e., set 1 and set 2, in both sterile as well as unsterile soils. The residual level of imidacloprid was 18 mg/kg in the case of unsterile soil after 45 days when treated with the isolated bacteria in combination with plantation. In control, imidacloprid residue in plant parts was 9.5 and 12.5 µg/g in sterile soil and unsterile soil respectively. Self-degradation of imidacloprid was 5.5 and 7.5% in sterile and unsterile soil respectively when spiked with 200 ppm imidacloprid. The study conducted by Sharma et al. (2014) also shows the degradation of imidacloprid in soil under autoclaved and un-autoclaved conditions.
Pesticides are degraded in the soil by physical, chemical, and biological processes. The availability of pesticides and the ability of microorganisms to use them are the major factors that influence the degradation of pesticides in soil. The efficacy of pesticide biodegradation could definitely be influenced by the physicochemical characteristics of the soil. The natural loss of imidacloprid could be due to volatilization, photocatalysis, or deep percolation. The uptake of imidacloprid by plants remains constant in inoculated as well as uninoculated soils. The degradation in the sterile soil was always found lower than in unsterile soil under same condition. This clearly indicates that the natural microflora present in the soil always favors the degradation.
Metabolites identified after GCMS analysis
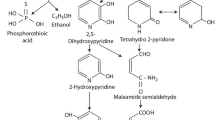
Two metabolites, hydrazinecarboxamide and hydroxyurea, were identified on conducting GCMS analysis of the degraded samples. The mass spectral profile of the identified metabolites have been presented in Fig. 6a and b.
Numerous possible imidacloprid metabolites have been identified by researchers in soil and water. Leifsonia sp. could degrade imidacloprid to (6-chloronicotinic acid) 6-CNA in three weeks (Anhalt et al. 2007). Similarly, imidacloprid can be transformed to desnitro-imidacloprid and urea metabolites by Pseudomonas sp. (Pandey et al. 2009). Imidacloprid can be hydroxylated to olefin imidacloprid by Stenotrophomonas maltophilia (Dai et al. 2006).
Kinetic analysis
Using the exponential formula (Ct = Co.ekt) as described in the previous sections, the kinetic parameters (k and T1/2) of imidacloprid degradation were calculated and are presented in Table 2. The degradation of imidacloprid followed a first-order degradation reaction at the above-described growth conditions in a soil microcosm. Figure 7a and b depict the linear plot of first and second-order kinetics respectively, indicating that the degradation in the present study follows first-order kinetics.
The initial phase of imidacloprid degradation included a lag period of 5 days during which only approximately 1% of initial imidacloprid was degraded. Bacterial strains exhibited higher imidacloprid degradation potential in non-sterilized soils as compared to sterilized soil. Tepidibacillus decaturensis strain ST1 degraded 91% of the applied imidacloprid within a time frame of 45 days with an average rate of 4.5 and 5.0 mg/kg day and rate constant of 0.039 and 0.048/day in sterile and non-sterile soils, respectively. The half-life (T1/2) of imidacloprid was found to be 12.9 days in non-sterile soil and 18.7 days in sterile soil after bacterial treatment.
Under optimum conditions, in the case of unsterile case, biodegradation using the isolates followed first-order kinetics. Imidacloprid was degraded in uninoculated soils at the rate of 0.36 mg/kg day and a rate constant of 0.0015/day, with a half-life of 291.7 days. The half-life of imidacloprid in un-inoculated planted soils was calculated to be 257.2 days, at a rate of 0.52 mg/kg day and a rate constant of 0.0027/day. In inoculated soils, 85% of applied imidacloprid was degraded within 45 days with an average rate of 4.76 mg/kg day and a rate constant of 0.042/day. Similarly, the value of k, T1/2, and reduction per day was calculated for all the samples of sets 1, 2, 3, and 4 in sterile and unsterile soils. Table 2 shows the first-order kinetic parameters calculated in the microcosm study.
The degradation kinetics were calculated using the first-order rate equation Ct = C0 × e−kt, since the removal of imidacloprid was found to be time-dependent. The rate constants for imidacloprid degradation in soil by bacterial isolates ranged from 0.015 to 0.048/day with a T1/2 decrease from 291.77 in uninoculated soil set 1 to 12.95 days in set 4 and rate constant 0.0419/d with T1/2 of 16.50 days in set 3. More than 20 times reduction in half-life period clearly indicate the potential of bacterial species for real time applications. With proper application of this technique, soil can be made imidacloprid free before start of next cultivation. The study conducted by Gupta et al. (2016) shows similar results, where degradation of imidacloprid follows first-order kinetics. In the similar study, Cycoń et al. (2013) noted that Serratia marcescens was capable of degrading imidacloprid at a rate constant ranging from 0.017 to 0.052/day with T1/2 of 13.6–37 days in different types of soils.
Conclusion
The widespread and indiscriminate use of insecticides has resulted in serious adverse impacts on the environment as well as human health. However, due to the lack of viable alternatives at present, insecticides are expected to be an indispensable part of agricultural practices. Various physicochemical approaches have been adopted for the remediation of such recalcitrant chemicals. The only environmentally acceptable way to reduce this contamination is through bioremediation. The use of indigenous bacteria from insecticide-contaminated sites can result in faster degradation in addition to protecting the soil microflora and enhancing the enzyme activity.
The overall findings reported here demonstrate the significant contribution of microorganisms in the degradation of imidacloprid. The presence and action of indigenous microorganisms are essential for the effective remediation of contaminants from the environment. The bacterial isolate, Tepidibacillus decaturensis strain ST1, used in the present study could effectively degrade imidacloprid up to 250 ppm in liquid media, slurry, and in soil microcosms. Consequently, the isolate may be effectively utilized for imidacloprid remediation in agricultural soil.
In a few previously published reports, the role of bacterial species have been studied in imidacloprid biodegradation, but, to the best of our knowledge, this is the first study in which role and contribution of factors other than bacterial degradation were identified and evaluated. It was found that volatilization, self-degradation, uptake by plant, etc., also contribute in the degradation imidacloprid along with bacterial degradation.
The presence of indigenous bacterial communities with natural remediation potential can be used to generate site-specific bioremediation of polluted soil. Laboratory microcosms are excellent alternatives to field experiments, which are typically expensive and influenced by variations in environmental conditions, and standard laboratory tests, the results of which are infrequently reflective of the real world since the experimental conditions and microorganisms used in the study may not be site-specific. Moreover, due to the relatively smaller size of microcosms, multiple replicates can be produced to study and establish a relationship between different microorganisms and the environmental conditions that suit best for the selected microbes for more efficient degradation of insecticides.
Data availability
Not applicable.
References
Ajiboye TO, Kuvarega AT, Onwudiwe DC (2020) Recent strategies for environmental remediation of organochlorine pesticides. Appl Sci 10. https://doi.org/10.3390/APP10186286
Akoijam R, Singh B (2015) Biodegradation of imidacloprid in sandy loam soil by Bacillus aerophilus. Int J Environ Anal Chem 95:730–743. https://doi.org/10.1080/03067319.2015.1055470
Anhalt JC, Moorman TB, Koskinen WC (2007) Biodegradation of imidacloprid by an isolated soil microorganism. J Environ Sci Heal - Part B Pestic Food Contam Agric Wastes 42:509–514. https://doi.org/10.1080/03601230701391401
Anhalt JC, Moorman TB, Koskinen WC (2008) Degradation and sorption of imidacloprid in dissimilar surface and subsurface soils. J Environ Sci Heal - Part B Pestic Food Contam Agric Wastes 43:207–213. https://doi.org/10.1080/03601230701771107
Azubuike CC, Chikere CB, Okpokwasili GC (2016) Bioremediation techniques–classification based on site of application: principles, advantages, limitations and prospects. World J Microbiol Biotechnol 32:1–18. https://doi.org/10.1007/s11274-016-2137-x
Bhatt P, Huang Y, Zhang W et al (2020) Enhanced cypermethrin degradation kinetics and metabolic pathway in Bacillus thuringiensis strain SG4. Microorganisms 8:1–14. https://doi.org/10.3390/microorganisms8020223
Bhattacherjee AK, Shukla PK, Dikshit A (2020) Microbial biotransformation of neonicotinoid insecticides in soil — a review. Int J Curr Microbiol Appl Sci 9:3255–3277. https://doi.org/10.20546/ijcmas.2020.907.380
Bidlan R, Afsar M, Manonmani HK (2004) Bioremediation of HCH-contaminated soil: elimination of inhibitory effects of the insecticide on radish and green gram seed germination. Chemosphere 56:803–811. https://doi.org/10.1016/j.chemosphere.2004.01.015
Briceño G, Levio M, González ME, et al (2020) Performance of a continuous stirred tank bioreactor employing an immobilized actinobacteria mixed culture for the removal of organophosphorus pesticides. 3 Biotech 10:1–11. https://doi.org/10.1007/s13205-020-02239-9
Choi SM, Yoo SD, Lee BM (2004) Toxicological characteristics of endocrine-disrupting chemicals: developmental toxicity, carcinogenicity, and mutagenicity. J Toxicol Environ Heal - Part B Crit Rev 7:1–23. https://doi.org/10.1080/10937400490253229
Craddock HA, Huang D, Turner PC et al (2019) Trends in neonicotinoid pesticide residues in food and water in the United States, 1999–2015. Environ Heal A Glob Access Sci Source 18:1–16. https://doi.org/10.1186/s12940-018-0441-7
Cycoń M, Markowicz A, Borymski S et al (2013) Imidacloprid induces changes in the structure, genetic diversity and catabolic activity of soil microbial communities. J Environ Manage 131:55–65. https://doi.org/10.1016/j.jenvman.2013.09.041
Dai YJ, Yuan S, Ge F et al (2006) Microbial hydroxylation of imidacloprid for the synthesis of highly insecticidal olefin imidacloprid. Appl Microbiol Biotechnol 71:927–934. https://doi.org/10.1007/s00253-005-0223-3
Dai ZL, Yang WL, Fan ZX et al (2021) Actinomycetes Rhodococcus ruber CGMCC 17550 degrades neonicotinoid insecticide nitenpyram via a novel hydroxylation pathway and remediates nitenpyram in surface water. Chemosphere 270:128670. https://doi.org/10.1016/j.chemosphere.2020.128670
Dua M, Singh A, Sethunathan N, Johri A (2002) Biotechnology and bioremediation: successes and limitations. Appl Microbiol Biotechnol 59:143–152. https://doi.org/10.1007/s00253-002-1024-6
Erguven GO, Demirci U (2021) Using Ochrobactrum thiophenivorans and Sphingomonas melonis for bioremediation of Imidacloprid. Environ Technol Innov 21:101236. https://doi.org/10.1016/j.eti.2020.101236
Erguven GO, Yildirim N (2019) The evaluation of imidacloprid remediation in soil media by two bacterial strains. Curr Microbiol 76:1461–1466. https://doi.org/10.1007/s00284-019-01774-w
Fuentes MS, Raimondo EE, Amoroso MJ, Benimeli CS (2017) Removal of a mixture of pesticides by a Streptomyces consortium: influence of different soil systems. Chemosphere 173:359–367. https://doi.org/10.1016/j.chemosphere.2017.01.044
Gallagher SR (2017) Quantitation of DNA and RNA with absorption and fluorescence spectroscopy. Curr Protoc Immunol 2017:A.3L.1-A.3L.14. https://doi.org/10.1002/cpim.20
Geed SR, Kureel MK, Prasad S et al (2018) Novel study on biodegradation of malathion and investigation of mass transfer correlation using alginate beads immobilized Bacillus sp. S4 in bioreactor. J Environ Chem Eng 6:3444–3450. https://doi.org/10.1016/j.jece.2018.05.025
Guo L, Dai Z, Guo J, et al (2020) Oligotrophic bacterium Hymenobacter latericoloratus CGMCC 16346 degrades the neonicotinoid imidacloprid in surface water. AMB Express 10. https://doi.org/10.1186/s13568-019-0942-y
Gupta M, Mathur S, Sharma TK, Rana M, Gairola A, Navani NK, Pathania R (2016) A study on metabolic prowess of Pseudomonas sp. RPT 52 to degrade imidacloprid, endosulfan and coragen. J Hazard Mater 301:250–258. https://doi.org/10.1016/j.jhazmat.2015.08.055
Hu G, Zhao Y, Liu B et al (2013) Isolation of an indigenous imidacloprid-degrading bacterium and imidacloprid bioremediation under simulated in situ and ex situ conditions. J Microbiol Biotechnol 23:1617–1626. https://doi.org/10.4014/jmb.1305.05048
Jeschke P, Nauen R, Schindler M, Elbert A (2011) Overview of the status and global strategy for neonicotinoids. J Agric Food Chem 59:2897–2908. https://doi.org/10.1021/jf101303g
Kavitha V, Anandhan R, Alharbi NS et al (2020) Impact of pesticide monocrotophos on microbial populations and histology of intestine in the Indian earthworm Lampito mauritii (Kinberg). Microb Pathog 139:103893. https://doi.org/10.1016/j.micpath.2019.103893
Kumar K, Devi SS, Krishnamurthi K et al (2007) Enrichment and isolation of endosulfan degrading and detoxifying bacteria. Chemosphere 68:317–322. https://doi.org/10.1016/j.chemosphere.2006.12.076
Lu P, Li Q, Liu H et al (2013) Biodegradation of chlorpyrifos and 3,5,6-trichloro-2-pyridinol by Cupriavidus sp. DT-1. Bioresour Technol 127:337–342. https://doi.org/10.1016/j.biortech.2012.09.116
Malato S, Caceres J, Agüera A et al (2001) Degradation of imidacloprid in water by photo-fenton and TiO2 photocatalysis at a solar pilot plant: a comparative study. Environ Sci Technol 35:4359–4366. https://doi.org/10.1021/es000289k
Negi G, Srivastava A, Sharma A (2014) Carbendazim using indigenous bacterial cultures of agriculture fields of Uttarakhand. India 8:973–981
Odukkathil G, Vasudevan N (2013) Toxicity and bioremediation of pesticides in agricultural soil. Rev Environ Sci Biotechnol 12:421–444. https://doi.org/10.1007/s11157-013-9320-4
Pandey G, Dorrian SJ, Russell RJ, Oakeshott JG (2009) Biotransformation of the neonicotinoid insecticides imidacloprid and thiamethoxam by Pseudomonas sp. 1G. Biochem Biophys Res Commun 380:710–714. https://doi.org/10.1016/j.bbrc.2009.01.156
Pang S, Lin Z, Zhang W et al (2020) Insights into the microbial degradation and biochemical mechanisms of neonicotinoids. Front Microbiol 11:1–20. https://doi.org/10.3389/fmicb.2020.00868
Phugare SS, Kalyani DC, Gaikwad YB, Jadhav JP (2013) Microbial degradation of imidacloprid and toxicological analysis of its biodegradation metabolites in silkworm (Bombyx mori). Chem Eng J 230:27–35. https://doi.org/10.1016/j.cej.2013.06.042
Pietrzak D, Kania J, Kmiecik E, et al (2020) Fate of selected neonicotinoid insecticides in soil–water systems: current state of the art and knowledge gaps. Chemosphere 255. https://doi.org/10.1016/j.chemosphere.2020.126981
Qureshi A, Verma V, Kapley A, Purohit HJ (2007) Degradation of 4-nitroaniline by Stenotrophomonas strain HPC 135. Int Biodeterior Biodegrad 60:215–218. https://doi.org/10.1016/j.ibiod.2007.03.004
Qurie M, Alkhatib M, Bufo SA et al (2021) Removal of imidacloprid from polluted water using adsorption and membrane separation technologies. Desalin Water Treat 242:178–186. https://doi.org/10.5004/dwt.2021.27852
Rank JK, Nathani NM, Hinsu AT et al (2018) Isolation characterization and growth response study of chlorpyrifos utilizing soil bacterium Pseudomonas putida JR16. Indian J Agric Res 52:355–361
Sabourmoghaddam N, Zakaria MP, Omar D (2015) Evidence for the microbial degradation of imidacloprid in soils of Cameron Highlands. J Saudi Soc Agric Sci 14:182–188. https://doi.org/10.1016/j.jssas.2014.03.002
Saleh IA, Zouari N, Al-Ghouti MA (2020) Removal of pesticides from water and wastewater: chemical, physical and biological treatment approaches. Environ Technol Innov 19:101026. https://doi.org/10.1016/j.eti.2020.101026
Sharma S, Singh B, Gupta VK (2014) Biodegradation of imidacloprid by consortium of two soil isolated bacillus sp. Bull Environ Contam Toxicol 93:637–642. https://doi.org/10.1007/s00128-014-1386-3
Shekher Giri B, Geed S, Vikrant K et al (2020) Progress in bioremediation of pesticide residues in the environment. Environ Eng Res 26:200446. https://doi.org/10.4491/eer.2020.446
Shetti A, Kaliwal R, Kaliwal B (2014) Imidacloprid induced intoxication and its biodegradation by soil isolate Bacillus weihenstephanensis. Br Biotechnol J 4:957–969. https://doi.org/10.9734/bbj/2014/8255
Silva V, Mol HGJ, Zomer P et al (2019) Pesticide residues in European agricultural soils — a hidden reality unfolded. Sci Total Environ 653:1532–1545. https://doi.org/10.1016/j.scitotenv.2018.10.441
Wallace DR, Buha Djordjevic A (2020) Heavy metal and pesticide exposure: a mixture of potential toxicity and carcinogenicity. Curr Opin Toxicol 19:72–79. https://doi.org/10.1016/j.cotox.2020.01.001
Wettstein FE, Kasteel R, Garcia Delgado MF et al (2016) Leaching of the neonicotinoids thiamethoxam and imidacloprid from sugar beet seed dressings to subsurface tile drains. J Agric Food Chem 64:6407–6415. https://doi.org/10.1021/acs.jafc.6b02619
Yadav IC, Devi NL, Syed JH et al (2015) Current status of persistent organic pesticides residues in air, water, and soil, and their possible effect on neighboring countries: a comprehensive review of India. Sci Total Environ 511:123–137. https://doi.org/10.1016/j.scitotenv.2014.12.041
Acknowledgements
The authors also acknowledge the Department of Chemical Engineering, Indian Institute of Technology (BHU) Varanasi, India, for providing laboratory facilities to conduct this study.
Funding
The authors received financial support from the ARFI project of the ISRO-Geosphere Biosphere Programme supported by the Indian Space Research Organization, Thiruvananthapuram.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Sonam Tiwari, Pranjal Tripathi, Ram Sharan Singh, and Devendra Mohan. The first draft of the manuscript was written by Sonam, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
All the authors declare that this study does not contain any studies with human participants or animals performed.
Consent to participate
Not applicable.
Consent for publication
All authors have approved the manuscript and agree with submission to Environmental Science and Pollution Research (ESPR).
Competing interests
The authors declare no competing interests.
Additional information
Responsible Editor: Robert Duran
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tiwari, S., Tripathi, P., Mohan, D. et al. Imidacloprid biodegradation using novel bacteria Tepidibacillus decaturensis strain ST1 in batch and in situ microcosm study. Environ Sci Pollut Res 30, 61562–61572 (2023). https://doi.org/10.1007/s11356-022-24779-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-022-24779-8