Abstract
The increasing emergence of antibiotic-resistant genes (ARGs) represents a global threat to human health. Land application of animal manure is known to contribute considerably to the propagation and dispersal of antibiotic resistance in agro-ecosystems. Yet, the primary determinants of the fate of the soil resistome remain obscure. In this study, a pot experiment was conducted to examine temporal changes in ARGs, mobile genetic elements (MGEs), and bacterial communities in a weakly developed loamy soil (an entisol known as calcareous purple soil) upon addition of pig or chicken manure. On the day of manure application, substantial increases in the diversity and relative abundance of ARGs were observed in soil amended with raw pig manure. At the same time, no obvious changes were observed for soil amended with chicken manure. Antibiotic resistance in pig manure-amended soils rapidly decreased over time to a level that was still higher than that of unamended soil at 100 days after manure application. The results of the Mantel test and Procrustes analysis indicated that ARG profiles in soil were significantly correlated with the structure of the bacterial phylogeny. Variation partitioning analysis further revealed that the bacterial community played a major role in regulating the temporal changes in ARGs in soil following manure application. Increased numbers and relative abundances of MGEs and their significant positive correlations with ARGs were observed, which suggest that a potential contribution from lateral gene transfer to the persistence and spread of ARGs should not be overlooked. Overall, our findings provide a better understanding of the mechanisms underlying the dynamics of ARGs in entisols following manure application and have practical implications for managing manure applications in entisols of the study area and other areas.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
A growing body of evidence suggests that the widespread use of antibiotics for human therapy, veterinary medicine, and growth augmentation has dramatically facilitated the occurrence and dissemination of antibiotic-resistant genes (ARGs) in various environments (Mahnert et al. 2019; Tiedje et al. 2019). More worryingly, clinically relevant pathogens have shown the ability to recruit environmental ARGs through horizontal gene transfer (HGT) mediated by mobile genetic elements (MGEs) and thus severely diminish the efficacy of antibiotics (Forsberg et al. 2012; Beka et al. 2018). Due to the alarmingly slow development of novel antibiotics, the spread of ARGs in human pathogens may eventually result in worldwide increases in morbidity and mortality rates related to infections by resistant bacteria (Mahnert et al. 2019). Therefore, antibiotic resistance is regarded as a major global public health crisis in the twenty-first century (Tiedje et al. 2019).
Soil, hosting the richest and most diverse microbes, is a natural source and the major reservoir of ARGs (D’Costa et al. 2011; Zhu et al. 2019). Apart from indigenous resistance, ARGs derived from animal manure represent a major contributor to allochthonous groups in the soil resistome (Duan et al. 2019; Pu et al. 2020). There is compelling evidence that manure application on land can markedly accelerate ARG propagation in soils (Liu et al. 2021; Duan et al. 2019). Long-term manure application significantly increases the abundance and diversity of ARGs in arable and greenhouse soils, and genes encoding resistance to nearly all major classes of antibiotics have been detected (Chen et al. 2016; Pu et al. 2020). The results of laboratory microcosm experiments lasting for several months in previous studies indicated that, relative to untreated soils, ARGs in manured soils were highly enriched at the initial phase and then declined over time to different extents at the end of the incubation depending on the soil type and manure source (Zhang et al. 2017; Gou et al. 2018; Han et al. 2018; Zhang et al. 2018; Xu et al. 2019). China is one of the largest antibiotic consumers in the world, and half of the total usage of antibiotics was for animal husbandry in 2013 (Zhang et al. 2015). Accordingly, a number of studies have been conducted with Chinese soils, and increases in the diversity and abundance of ARGs by manure application, in the short or long term, have been observed in brown soil (Han et al. 2018), red soil (Xie et al. 2018; Xu et al. 2019), fluvo-aquic soil (Chen et al. 2016; Wang et al. 2020), and black soil (Li et al. 2019). Nonetheless, the temporal ARG changes following manure application remain unknown for many other agricultural soils. This lack of knowledge has limited our ability to determine the time windows that exhibit high risks of ARG dissemination and transport and to identify the major influencing factors.
The dominant mechanism that underpins the reported increases of ARG levels after manure application is still elusive for dissimilar soils (Gao et al. 2020). It has been well documented that ARG patterns are strongly related to bacterial community compositions in manured soils (Chen et al. 2016; Han et al. 2018; Xu et al. 2019). Likewise, bacterial communities made the largest contribution to the variations of ARG abundance observed in soil, earthworm guts, and phyllosphere (Zhou et al. 2020). In contrast, some other studies have shown that the increase in ARGs in manured soils is independent of the succession of the microbial community (Liu et al. 2017) and that the bacterial community diversities and those of ARGs are regulated by distinct soil environmental factors (Xie et al. 2018). Recent comparative studies of soils from different geographical locations (Zhang et al. 2018; Pu et al. 2020; Wang et al. 2020) have indicated that bacteria contribute in distinct ways to resistance formation in different manured soils. Given the inconsistency of these findings, as reported in the literature, there is a need for more research into the responses of ARGs and microbes to manure application and the underlying mechanisms in other soils.
Purple soil, a loamy entisol, is widely distributed in the hilly areas of the Sichuan Basin, Southwest China (Li et al. 1991). Antibiotic usage in Southwest China accounts for 11.30% of total national consumption (Zhang et al. 2015). Moreover, Sichuan Province is the largest pig manure producer and is a consumption hotspot of veterinary antibiotics in China (Van Boeckel et al. 2015; Zhou et al. 2017a). Nevertheless, the impact of manure application on the dynamics and fate of ARGs in purple soils and the underlying mechanisms remain largely unknown. These factors represent essential knowledge for integrated risk assessments of ARG dissemination in manured purple soils and for the development of ARG pollution control strategies. We hypothesize that bacterial communities play a dominant role in regulating temporal changes in antibiotic resistomes following manure application. By using high-throughput quantitative polymerase chain reaction (HT-qPCR) and Illumina sequencing, the occurrence and persistence of ARGs and their links with bacterial community compositions in calcareous purple soil amended with pig or chicken manure were evaluated with a pot experiment in the present study to test this hypothesis. The objectives of this study were (1) to determine the time–course patterns of ARGs, MGEs, and bacterial structures in manured purple soils and (2) to identify the main factors that regulate the temporal succession of ARGs.
Material and methods
Soil and manure collection
The surface soil (0–15 cm) used for the pot experiment was collected in July 2018 from the cultivated layer of wheat–corn rotation farmland at the Yanting Agro-Ecological Experimental Station of Purple Soil (31n rotation f28′ E), Chinese Academy of Sciences. The farmland had been treated with inorganic chemical fertilizers over the last ten years before soil sampling. The collected soil was air-dried and passed through a 2-mm mesh prior to use. Raw pig manure (PM) and composted chicken manure (CM) were purchased from two local animal farms in July 2018 and were air-dried and milled and sieved through a 2-mm mesh before use. The basic properties of the soil and manures are provided in Table S1.
Glasshouse pot experiment
Three sets of treatments were established in plastic pots: (1) soil without manure application (CK), (2) soil amended with 2% (dw/dw) pig manure (PMS), and (3) soil amended with 2% (dw/dw) chicken manure (CMS). After being mixed by sieving through a 2-mm mesh, 2.5 kg of soil was placed into each pot. Each treatment was carried out in three replicates. All pots were placed in a glasshouse and were assigned in a randomized block design. The soils were thoroughly watered and preincubated overnight. Day 0 soil samples were collected from each pot on the next day. The experiment lasted for 100 days, and the soils in pots experienced natural evaporation. Every two or three days, deionized water was added to adjust the soil moisture content to 70% of the water-holding capacity. Soil samples were also collected from the pots on days 23, 45, 62, and 100. Approximately 5 g of soil was taken from five random locations in each pot to a depth of 10 cm by using a sterilized soil auger (5-mm inner diameter) and mixed to form a composite sample. Then, three composite samples from the triplicate pots of each treatment were mixed thoroughly to form a representative sample (Zhang et al. 2018), which was then freeze-dried and frozen at −20°C when not in use.
Soil DNA extraction and HT-qPCR of ARGs
Total DNA was extracted in duplicate from 0.25 g of soil with the DNeasy PowerSoil Kit (Qiagen, Germany) according to the manufacturer’s guidelines. The quality and quantity of the combined DNA extract were assessed using an ND-1000 spectrophotometer (NanoDrop Technology Inc., USA) and were then stored at -20°C before downstream analysis.
HT-qPCR was conducted with the SmartChip Real-time PCR system (Warfergen Inc., USA) to characterize the profiles of ARGs and MGEs in soils. The targeted genes included 16S rRNA genes, 283 ARGs, 8 transposase genes, and 4 integrase genes (Li et al. 2019). The thermal cycle procedure was 95°C for 10 min, followed by 40 cycles at 95°C for 30 s and at 60°C for 30 s (Chen et al. 2017; Han et al. 2018). All reactions for each primer set were performed in triplicate with a no-template negative control. Wells with multiple melting peaks or efficiencies outside the range of 90–110% were excluded. A threshold cycle (CT) of 31 was set as the detection limit, and the relative copy number was computed as \( {10}^{\left(31-{C}_{\mathrm{T}}\right)\left(10/3\right)} \). Only genes showing CT < 31 cycles and having amplification in all replicates were regarded as positive. The relative abundances of ARGs/MGEs were calculated by normalizing the copy numbers of each ARG/MGE to that of the 16S rRNA genes detected in the same sample.
High-throughput sequencing of 16S rRNA genes and data analysis
To evaluate the bacterial community structures, the variable regions (V4–V5) of the 16S rRNA genes were amplified with the primer set 515F/909R. Twenty-five microliter of PCR mixture containing 1 × PCR buffer, 0.5 U of TaKaRa ExTaq, 1.5 mM MgCl2, each deoxynucleoside triphosphate at 0.4 μM, each primer at 1.0 μM, and 10 ng of DNA template (Han et al. 2018). PCR was conducted in duplicate for each sample by performing an initial denaturation at 94°C for 3 min, 30 cycles at 94°C for 40 s, 56°C for 60 s, and 72°C for 60 s, and a final extension at 72°C for 10 min. After amplification, the resultant PCR products were purified, quantified, pooled at equal concentrations, and sequenced on the Illumina MiSeq platform (Illumina Inc., USA). After removing reads containing ambiguous nucleotides (three or more) or of low quality (average quality score < 30, length < 200 bp), the raw paired-end reads were assembled to generate clean reads. The high-quality sequences obtained were analyzed using QIIME with the default parameters (Caporaso et al. 2010). Operational taxonomic units (OTUs) were identified at 97% similarity using UPARSE (Edgar 2013), and singletons were culled. All samples were rarefied to 13,374 sequences for downstream analyses. Taxonomic assignment was performed using the RDP classifier with a confidence threshold of 0.80 (Wang et al. 2007). Raw sequences were uploaded to the National Center for Biotechnology Information Sequence Read Archive under the accession number SRP191351.
Statistical analysis
The alpha diversity of the bacterial community in each sample was assessed using the metrics of the observed species and Shannon index. Differences in bacterial community compositions among samples from different treatments were evaluated by the linear discriminant analysis (LDA) effect size (LEfSe: http://huttenhower.sph.harvard.edu/galaxy/) (Segata et al. 2011), and only taxa meeting the significance threshold (i.e., Kruskal−Wallis test α value < 0.05, LDA score > 3.5) were recorded in each treatment. Other statistical analyses were performed by R software (version 3.6.3) (R Core Team 2019), and a P-value < 0.05 was considered to be statistically significant. Principal coordinate analysis (PCoA) and the Adonis test, based on the Bray–Curtis distances, were conducted in R 3.6.3 with the “vegan” package (Oksanen et al. 2018). Variation partitioning analysis (VPA), the Mantel test, the Procrustes test, redundancy analysis (RDA), and Pearson correlation analysis were performed to determine the relationships among ARGs, MGEs, and bacterial communities using the “vegan” and “psych” (Revelle 2018) packages in R 3.6.3. To illustrate the co-occurrence patterns among ARGs, MGEs, and bacterial classes, network analysis was implemented and visualized using the “igraph” package (Csardi and Nepusz 2006) in R 3.6.3 with statistically robust Spearman correlations defined as r > 0.8 and FDR-corrected P-value < 0.01. SourceTracker (Knights et al. 2011) was employed to examine the relative contributions of microbial communities from the source environments (e.g., pig manure, chicken manure, and untreated purple soil on day 0 to sink environments (manured soils) with the default parameters). Other plots were generated in R 3.6.3 with the “ggplot2” package (Wickham 2016).
Results
Antibiotic resistomes in manures and untreated soil on day 0
A total of 185 ARGs and 12 MGEs were detected across all manure and soil samples during the incubation period. Most of the ARGs identified in the two animal manures conferred resistance to aminoglycosides, macrolide–lincosamide–streptogramin B (MLSB), tetracyclines, and beta-lactams, which are commonly administered during livestock breeding (Fig. S1a). These genes accounted for 73% and 71% of the total number of ARGs in pig manure and chicken manure, respectively. By comparison, genes encoding resistance to multiple drugs (36%) were the most frequently detected ARGs in untreated purple soil, followed by beta-lactams (23%). The proportions of the other classes of ARGs were all less than 10%. Pig manure harbored the most diverse ARGs (142) and MGEs (12), followed by chicken manure (76 ARGs and 6 MGEs), while the untreated soil had the fewest ARGs (21) and MGEs (1) (Fig. S1b and d). The differences in the relative abundances of these genes among different sample types exhibited similar patterns (Fig. S1c and e).
Dynamics of ARGs and MGEs in soils following manure application
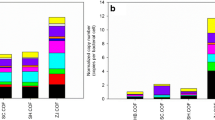
The temporal changes in ARGs and MGEs in the calcareous purple soil following amendment with different animal manures are shown in Fig. 1. The number of detected ARGs in soil amended with pig manure (106) was much higher than in untreated soil (21) at the beginning of incubation (day 0). This number showed a gradual decrease over the first 23 days and then remained unchanged afterwards. Apparently, application of pig manure introduced substantial amounts of resistance genes into the purple soil. Notably, the number and relative abundance of ARGs in pig manure-treated soil were markedly higher than those in untreated soil throughout the experimental period. In contrast, the amendment with chicken manure resulted in a marginal increase in the detected ARG number, which remained constant throughout the whole incubation period. For each treatment, similar temporal trends were observed for the diversities and relative abundances of MGEs during incubation.
The temporal changes in the composition of ARGs, as percentages of the total relative abundance of detected resistance genes, were also examined (Fig. S2). In accordance with the ARG diversities, aminoglycoside, MLSB, and tetracycline resistance genes were the most prevalent ARGs in manures, while untreated soil was dominated by multidrug resistance (93%). Intriguingly, despite the similar ARG compositions of pig manure and chicken manure, application of these two manures resulted in distinct ARG profiles in the treated soils. Specifically, the ARG compositions of PM and PMS were similar and obviously different from those of the CK samples, which remained largely stable (except for the CK sample on day 45) during the whole incubation period. In contrast, the CMS on days 0 and 62 exhibited similar ARG compositions, while on other days, the CMS generally exhibited more complex ARG profiles than the CK samples with elevated percentages of ARGs that conferred resistance to classes of antibiotics other than multidrug resistance. These results were consistent with the PCoA analysis with one-way PERMANOVA results according to the factor of treatment. As shown in Fig. 2a and Table S2, the PMS exhibited clear divergence from the CK and CMS (Adonis test, P < 0.05), while the CMS on days 0 and 62 clustered together with the CK samples on most sampling days and were separated from the CMS on other days.
Bacterial community dynamics in soils following manure application
A total of 410,135 high-quality sequences generated from 16S rRNA gene amplicons were obtained from all the manure and soil samples, with 14,536 to 35,457 sequences per sample. These sequences were clustered into 19,400 OTUs at a 97% similarity level. Good’s coverage values ranged from 0.794 to 0.854 for the soil samples and from 0.915 to 0.948 for the manure samples. Application of animal manures visibly reduced the alpha diversity of the bacterial community (as represented by the observed species and Shannon index) in the PMS, which harbored the least diverse bacteria (Figs. 3a and S3a). During incubation, the alpha diversities of the soil bacterial communities in all the treatments increased in the first 23 days and remained largely unchanged afterwards.
Impacts of manure application on soil bacterial diversity (a). Cladograms of the LEfSe analysis. For each taxon, red, green, and blue denote markedly higher relative abundances (α < 0.05, LDA score > 3.5) in the CK, CMS, and PMS treatments, respectively. Yellow denotes taxa that were not significantly different among the different treatments (b). The relationship between bacterial diversity and the relative abundance of ARGs (c)
In total, microbial communities were assembled into 43 phyla, and phyla with abundances of less than 1% were categorized as “Other” (Fig. S4). The dominant phyla in chicken manure were Firmicutes (70.90%) and Actinobacteria (20.93%), while pig manure and untreated soil (on day 0) were dominated by Proteobacteria (32.06% vs. 26.75%), Actinobacteria (29.00% vs. 23.53%), and Firmicutes (22.58% vs. 14.04%). Interestingly, the bacterial community compositions of the CK and CMS treatments (at the same sampling time) were nearly identical, although chicken manure showed taxonomic compositions that were very different from those of untreated soil. In contrast, amendments with pig manure caused an obvious effect on the soil bacterial composition, which featured a markedly increased relative abundance of Firmicutes. The time–course patterns of the soil bacterial community compositions during incubation were similar under the three treatments. From day 23 on, the soils showed significantly stable bacterial community compositions and harbored higher relative abundances of Proteobacteria but lower relative abundances of Firmicutes compared to day 0. These findings were also consolidated by the results of the PCoA analysis and the Adonis test based on the Bray–Curtis distance (Fig. 2b and Table S3). The PMS (P < 0.05) was clearly separated from the CK and CMS along PC1 (explaining 24.96% of the variance), while for all three soils, day 0 (P < 0.01) was obviously separated from the other sampling time points along PC2 (explaining 16.68% of the variance). Moreover, LEfSe analysis revealed that the phylum Firmicutes and class Gammaproteobacteria were markedly enriched in the PMS, while the class Acidimicrobiia, family Micromonosporaceae (e), and order Rhizobiales (u) were enriched in the CMS (Fig. 3b).
The results of the SourceTracker analysis revealed the fate of manure-derived bacteria that were applied in soils during incubation (Fig. S5). Chicken manure addition accounted for only 1−2% of the bacterial community in treated soil throughout the incubation period. In contrast, pig manure contributed to 35% of the bacterial community in the PMS treatment on day 0, and its proportion decreased afterwards to 8% at the end of the incubation experiment. In addition, the bacterial communities in the manured soils were mainly ascribed to “unknown source” (46−76%), which likely represented a large part of the autochthonous soil bacteria that were undetectable at the beginning of the incubation experiment (day 0) (Gou et al. 2018).
Correlations among ARGs, MGEs, and bacterial community
The co-occurrence patterns among ARGs, MGEs, and bacterial taxa obtained by network analysis are shown in Fig. 4. Interestingly, ARGs and bacteria were better clustered among themselves than with each other. At the class level, Clostridia, Bacilli, Sphingobacteriia, and Gammaproteobacteria showed close correlations with multiple ARGs and MGEs (Table S4), which suggest that these bacteria were possible hosts of relevant co-occurring genes (Han et al. 2018). This result can be corroborated with higher accuracy and greater confidence using other methods such as emulsion, paired isolation, and concatenation PCR (epicPCR); genomic cross-linking methods (e.g., Hi-C based on chromosome conformation capture technology); metagenomic sequencing and assembly; and isolation of antibiotic-resistant strains (Zhu et al. 2017; Rice et al. 2020). In addition, the network graph indicated that two integrase genes, intI-1LC and intI-1 (clinic) were significantly clustered with diverse ARG encoding resistance to different classes of antibiotics (Table S5). Pearson correlation analysis revealed that the relative abundance of ARGs was closely and positively correlated with those of both integrase and transposase genes, respectively (P < 0.0001) (Fig. S6).
Network analysis depicting co-occurrence patterns among ARGs, MGEs, and bacterial taxa (class level). Connections indicate strong (Spearman’s correlation coefficient > 0.8) and significant (P < 0.01) correlations. Edges are weighted according to the correlation coefficients. Nodes with different colors represent different ARG classes, MGEs, and bacterial taxa, and node sizes are weighted according to the number of significant connections. A more detailed correlation between ARG, MGEs, and bacterial taxa was provided in Tables S4 and S5
The results of the RDA and VPA analyses showed that the first two axes accounted for 77.01% of the total variation in ARGs. The structural variations of the ARG profiles in the PMS treatment were mainly contributed by Firmicutes and MGEs (Fig. 5a). A total of 85.30% of the observed variations in ARGs could be explained by the selected factors (Fig. 5b). The bacterial community was the most dominant contributor (explaining 66.27%), while MGEs contributed only 3.14% to the ARG variations. Meanwhile, the results of the Procrustes analysis based on the Bray–Curtis distance indicated that the ARG profiles were closely correlated with bacterial communities (sum of squares M2 = 0.5910, r = 0.6396, P = 0.0018), which was confirmed by the Mantel test (r = 0.4499, P < 0.001). In addition, the relative ARG abundances were found to be negatively correlated with the alpha diversities of the bacterial community (Figs. 3c and S3b).
Redundancy analysis revealing the relationships of soil ARGs (blue arrow) with major microbial phyla (> 1% in any sample; red arrow) and MGEs (red arrow) (a) and variation partitioning analysis (VPA) differentiating the effects of the bacterial community (BC) and MGEs on soil resistome variations (b). In VPA, a total variance of 85.30% could be explained by three selected factors: the BC alone group (with MGEs as the covariate), the MGE alone group (with BC as the covariate), and the MGE and BC group, which reflects the contribution of the interaction between the two variables to the total variance
Discussion
Significance of the present study
It is well known that animal manure application can elevate ARG abundance and diversity in soil (Zhang et al. 2017; Han et al. 2018; Pu et al. 2020). However, there have been discrepant observations regarding the temporal changes in the manuring effects on the levels of soil ARGs. As a weakly developed soil type with a low organic matter content (mostly <1.5%), low plant-available water holding capacity and low nutrient content (Wang et al. 2015), entisol is distributed in many areas/countries around the world (Tedrew and Robert 1986; Mou et al. 2009). Amendment with animal manures is a recommended agronomic practice for improving soil quality in areas of purple soil. However, the extent to which ARG levels in manured purple soil can persist has not been reported in the literature. The present study provides insight into the dominant mechanisms that underlie ARG dynamics in soils upon manure application. Our findings have practical implications for managing manure applications in the entisols of the study area and other areas. Moreover, the results are critical for evaluating the offsite environmental impacts of manure application. In particular, knowledge of ARG dynamics in soils following manure application is essential for reliable predictions of ARG transport, either in free forms or in association with sediments, from farmland to water bodies via hydrological processes upon rainfall (Cheng et al. 2020).
ARGs in the untreated soil
The detection of 43 ARG subtypes in unamended calcareous purple soils supports the long-standing view that ARGs are ubiquitous in the soil environment prior to the clinical use of antibiotics (D’Costa et al. 2011). These ARGs can be recognized as the intrinsic resistome in the tested soil and are mostly related to resistance to multiple drugs (Fig. S2). The dominance of multidrug resistance genes has also been reported in diverse types of soils (Chen et al. 2016; Xie et al. 2018; Li et al. 2019). This is probably because in addition to encoding resistance against antibiotics, these genes also play important roles in multiple bacterial functions (Piddock 2006).
Occurrence and persistence of soil ARGs following manure application
Land application of animal manures can improve crop production and alleviate soil degradation caused by chemical fertilizers (Xie et al. 2018). However, it has been recognized that manure application substantially stimulates the dissemination of antibiotic resistance in soils by introducing allochthonous ARGs and by boosting autochthonous soil ARGs because it often contains abundant ARGs, relevant selective agents, and biogenic elements (Xie et al. 2018; Liu et al. 2021). Consistently, in this study, we found that pig manure application greatly enhanced both the number and the relative abundance of soil ARGs, which were constantly higher than those in untreated soil throughout the entire experiment (Fig. 1). However, our previous field study, which was conducted on sloping farmland at the same site, showed that long-term pig manure application did not accelerate ARG dispersal in manured purple soils (Cheng et al. 2019). Several reasons could explain this disagreement. First, differences in the properties of the pig manures used in the present study (from a commercial farm) and the previous long-term field experiment (from local family pig farms) may be partially responsible for the inconsistent results. Generally, family livestock farms consume significantly fewer antibiotics than commercial livestock farms. Therefore, animal manures obtained from family farms likely harbor much less diverse and abundant ARGs (Chen et al. 2019b) and show a limited effect on ARG profiles in the applied soils (Zhou et al. 2020). Second, the elevation of ARGs in manured soils is dose dependent (Han et al. 2018), and a low manure input may be insufficient to effectively change the soil antibiotic resistome (Tang et al. 2015; Lin et al. 2016). Thus, the high application rate of pig manure employed in the present glasshouse pot experiment, which was approximately eight times higher than that used in the previous field study, may also be partly responsible for the inconsistent findings. Third, many differences in experimental setups and conditions between a microcosm study and field-scale experiment could contribute to the discrepancies in their results, such as differences in manure application methods and the frequent occurrence of storm events in the field during the time between manure application and soil sampling, which may lead to a loss of ARGs from the soil via runoff and leaching processes (Barrios et al. 2020; Cheng et al. 2020; Hall et al. 2020).
Despite the initial marked enrichment in the PMS relative to untreated soil, the abundance and diversity of ARGs dramatically decreased by day 23 (Fig. 1). Some previous studies have similarly observed that the elevated level of ARGs in soil did not remain for a long time following manure application (Gou et al. 2018; Han et al. 2018). The crucial role of manure-borne microorganisms carrying ARGs in the elevation of soil ARGs following manure application has been reported (Chen et al. 2017; Liu et al. 2021). However, manure-borne microorganisms in soil could not persist for a long time following application due to their lack of adaptability to the soil environment and the restraint by native soil microorganisms (Chen et al. 2019a). Therefore, the transient effect of manure on soil resistance could be explained by a prompt decrease of manure-derived microorganisms after application, as corroborated by the analogous temporal patterns between ARGs and manure-derived bacteria in the PMS (Figs. 1 and S5). The small proportion (8%) of pig manure-borne bacteria that survived at the end of the incubation experiment may be partially responsible for the enrichment of ARGs in pig manure-treated soil relative to untreated soil (Fig. S5). This finding implies that the risk of resistance transmission may still exist three months after pig manure application.
Unexpectedly, application of chicken manure did not cause an obvious enrichment of ARGs in soil during incubation (Fig. 1), likely due to the relatively low abundance of ARGs in chicken manure (Fig. S1). Previous studies have suggested that manures with low levels of ARGs, especially when close to the soil background level, could barely impact the abundance of ARGs in manured soils (Rutgersson et al. 2020). In addition, the lack of establishment of chicken manure-derived bacteria in the soil environment may in part lead to comparable ARG levels between manured soil and untreated soil (Fig. S5). Nonetheless, obvious temporal variations of the soil ARG composition were observed in the CMS (Figs. 2a and S2). Of particular concern was that application of chicken manure led to increases in the percentage of vancomycin-resistant genes (van) in treated soil (except for the samples on days 0 and 62), which is an ARG class that is scarcely detected in chicken manure. Vancomycin is considered to be one of the last resort defenses against “superbugs,” such as methicillin-resistant Staphylococcus aureus (Jovetic et al. 2010). This indicates that application of the chicken manure tested in the present study may pose a health risk, although no obvious effect on ARG abundances was observed.
Factors shaping ARG profiles in manured soils
Since the survival of most manure-borne bacteria is transient following manure application, HGT of ARGs from manure to resident soil bacteria may be a critical mechanism for the dispersion and persistence of ARGs in manured soils (Wang et al. 2020). In the present study, all the targeted MGEs were detected in pig manure with high abundances. The number and relative abundance of soil MGEs were highly responsive to pig manure amendment, and their temporal tendencies were similar to those of ARGs. Additionally, the relative abundance of total ARGs was tightly connected with those of total integron–integrase or transposase genes. Network analysis also demonstrated that class 1 integrons were intensively correlated with several ARG subtypes (Fig. 4 and Table S5), including some resistance determinants known to be located within integron cassettes, such as qacEdelta and the aad family of genes (Zhu et al. 2017). Altogether, these results indicated that pig manure-derived MGEs could be established in the soil and thus lead to elevated soil antibiotic resistance.
This work demonstrated that soil bacterial diversity was decreased by manure application and was negatively correlated with the relative abundance of total ARGs (Figs. 3 and S3), suggesting that the loss of microbial diversity might be partially responsible for the prevalence and spread of ARGs in manured soil (Mahnert et al. 2019). This interpretation is supported by the reported crucial role of the resident soil microbial community in controlling ARG dissemination in manured soil (Chen et al. 2019a). Interestingly, Firmicutes, which was markedly enriched in soil after application of pig manure (Fig. 3b), was the key contributor to the variations in ARGs in the PMS (Fig. 5a). Additionally, Clostridia and Bacilli of the phylum Firmicutes were possible carriers of ARGs, as was revealed by the network analysis (Fig. 4 and Table S4). These results indicated that the changes in the soil bacterial community that resulted from pig manure application could be another important reason for the shifts in ARGs, which was supported by the significant correlation between the ARG profile and bacterial community structure revealed by Procrustes analysis and the Mantel test. VPA analysis, which was conducted to decipher the key factors affecting the profile of ARGs (Fig. 5b), showed that the bacterial community explained a much larger portion of the total variation in the ARG profiles than MGEs. Therefore, it can be inferred that ARG formation in manured soils was mainly driven by changes in bacterial community composition rather than HGT through MGEs, which demonstrates our hypothesis. Similar results have been reported in some previous studies across different habitats for ARGs, such as drinking water systems (Jia et al. 2015), sewage sludge during composting (Su et al. 2015), soils amended with organic fertilizers (Chen et al. 2016; Zhou et al. 2020), and urban rivers (Zhou et al. 2017b).
Limitations and implications
The present study, however, has some limitations. First, the pig and chicken manures used here were sourced from local commercial farms with different manure management practices. This limitation restricts us from attributing the differences in the results between the two manures solely to differences in the producing animal species or management methods. In particular, comparative studies are needed in the future to reveal how manure types and composting treatments affect soil ARGs. Second, microcosm studies have their own limitations. They fail to reproduce all the factors of an actual field that could affect the fate of ARGs. In particular, previous studies have observed the offsite transport of soil ARGs during field-scale rainfall simulation tests (Barrios et al. 2020; Hall et al. 2020). Considering that purple soil is prone to erosion and that there is ample rainfall in the hilly study area (Zhang et al. 2016), rainfall-induced ARG loss via hydrological processes may contribute to the attenuation of ARGs following manure application in the field, which is also worth exploring in the future.
Despite these limitations, based on the results obtained from this study, in particular the temporary enrichment of soil ARGs and the negative correlation between the ARG content and bacterial diversity, we suggest some agricultural practices for minimizing the dispersal of resistance in soil and to human: (1) crops can be planted or harvested one month after manure application, although this safe time needs to be verified by field tests and (2) agronomic operations that could increase or maintain the diversity of native soil bacteria can be implemented to help control the prevalence of ARGs.
Conclusions
A pot experiment was conducted to explore the changes in ARGs in a calcareous purple soil over time following application of pig or chicken manure. Our results showed that the application of raw pig manure caused lasting (100 days) elevated soil ARG levels despite the rapid decreases in ARG abundance and diversity over time by day 23; however, no such changes were observed after the amendment of composted chicken manure. Microbial community composition could explain most of the variations in the soil ARGs and was the primary factor that regulated the profile of soil ARGs. These findings contribute to a comprehensive understanding of manuring effects on the composition and persistence of ARGs in soils. Future field studies of temporal changes in soil ARGs upon application of different animal manures and their various composts to different soils are required to inform best practices for minimizing the dissemination of ARGs through manure management.
References
Barrios RE, Khuntia HK, Bartelt-Hunt SL, Gilley JE, Schmidt AM, Snow DD, Li X (2020) Fate and transport of antibiotics and antibiotic resistance genes in runoff and soil as affected by the timing of swine manure slurry application. Sci Total Environ 712:136505
Beka L, Fullmer MS, Colston SM, Nelson MC, Talagrand-Reboul E, Walker P, Ford B, Whitaker IS, Lamy B, Gogarten JP, Graf J (2018) Low-level antimicrobials in the medicinal leech select for resistant pathogens that spread to patients. Mbio 9:e01328–e01318
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Chen QL, An XL, Li H, Su JQ, Ma YB, Zhu YG (2016) Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ Int 92-93:1–10
Chen QL, An XL, Li H, Zhu YG, Su JQ, Cui L (2017) Do manure-borne or indigenous soil microorganisms influence the spread of antibiotic resistance genes in manured soil? Soil Biol Biochem 114:229–237
Chen QL, An XL, Zheng BX, Gillings M, Peñuelas J, Cui L, Su JQ, Zhu YG (2019a) Loss of soil microbial diversity exacerbates spread of antibiotic resistance. Soil Ecol Lett 1:3–13
Chen CQ, Pankow CA, Oh M, Heath LS, Zhang LQ, Du P, Xia K, Pruden A (2019b) Effect of antibiotic use and composting on antibiotic resistance gene abundance and resistome risks of soils receiving manure-derived amendments. Environ Int 128:233–243
Cheng JH, Tang XY, Cui JF (2019) Effect of long-term manure slurry application on the occurrence of antibiotic resistance genes in arable purple soil (entisol). Sci Total Environ 647:853–861
Cheng JH, Tang XY, Liu C (2020) Occurrence and distribution of antibiotic resistance genes in various rural environmental media. Environ Sci Pollut Res 27:29191–29203
Csardi G, Nepusz T (2006) The igraph software package for complex network research. InterJ Complex Syst 1695:1–9
D’Costa VM, King CE, Kalan L, Morar M, Sung WWL, Schwarz C, Froese D, Zazula G, Calmels F, Debruyne R, Golding GB, Poinar HN, Wright GD (2011) Antibiotic resistance is ancient. Nature 477:457–461
Duan ML, Gu J, Wang XJ, Li Y, Zhang RR, Hu T, Zhou BB (2019) Factors that affect the occurrence and distribution of antibiotic resistance genes in soils from livestock and poultry farms. Ecotox Environ Saf 180:114–122
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998
Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MO, Dantas G (2012) The shared antibiotic resistome of soil bacteria and human pathogens. Science 337:1107–1111
Gao Q, Dong Q, Wu L, Yang Y, Hale L, Qin Z, Xie C, Zhang Q, Van Nostrand JD, Zhou J (2020) Environmental antibiotics drives the genetic functions of resistome dynamics. Environ Int 135:105398
Gou M, Hu HW, Zhang YJ, Wang JT, Hayden H, Tang YQ, He JZ (2018) Aerobic composting reduces antibiotic resistance genes in cattle manure and the resistome dissemination in agricultural soils. Sci Total Environ 612:1300–1310
Hall MC, Mware NA, Gilley JE, Bartelt-Hunt SL, Snow DD, Schmidt AM, Eskridge KM, Li X (2020) Influence of setback distance on antibiotics and antibiotic resistance genes in runoff and soil following the land application of swine manure slurry. Environ Sci Technol 54:4800–4809
Han XM, Hu HW, Chen QL, Yang LY, Ma YB (2018) Antibiotic resistance genes and associated bacterial communities in agricultural soils amended with different sources of animal manures. Soil Biol Biochem 126:91–102
Jia SY, Shi P, Hu Q, Li B, Zhang T, Zhang XX (2015) Bacterial community shift drives antibiotic resistance promotion during drinking water chlorination. Environ Sci Technol 49:12271–12279
Jovetic S, Zhu Y, Marcone GL, Marinelli F, Tramper J (2010) β-Lactam and glycopeptide antibiotics: first and last line of defense? Trends Biotechnol 28:596–604
Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, Kelley ST (2011) Bayesian community-wide culture-independent microbial source tracking. Nat Methods 8:761–763
Li ZM, Zhang XW, He YR, Tang SJ (1991) Purple soil in China (A). Science Press, Beijing
Li S, Yao Q, Wei D, Zhou BK, Zhu P, Cui XA, Jin J, Liu XB, Wang GH (2019) Profiles of antibiotic resistome with animal manure application in black soils of northeast China. J Hazard Mater 384:121216
Lin H, Sun WC, Zhang ZL, Chapman SJ, Freitag TE, Fu JR, Zhang X, Ma JW (2016) Effects of manure and mineral fertilization strategies on soil antibiotic resistance gene levels and microbial community in a paddy-upland rotation system. Environ Pollut 211:332–337
Liu P, Jia SY, He XW, Zhang XX, Ye L (2017) Different impacts of manure and chemical fertilizers on bacterial community structure and antibiotic resistance genes in arable soils. Chemosphere 188:455–464
Liu WB, Ling N, Guo JJ, Ruan Y, Wang M, Shen QR, Guo SW (2021) Dynamics of the antibiotic resistome in agricultural soils amended with different sources of animal manures over three consecutive years. J Hazard Mater 401:123399
Mahnert A, Moissl-Eichinger C, Zojer M, Bogumil D, Mizrahi I, Rattei T, Martinez JL, Berg G (2019) Man-made microbial resistances in built environments. Nat Commun 10:968
Mou SS, Qing CL, Duan WX, Wei CF, Wang DY, Li H, Zhu B, Pu FY, Xie DT (2009) More insights into purple soil II- global purple soil. J Southwest Univ 31(7):126–130
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin RP, O’Hara BR, Simpson GL, Solymos P, Stevens HHM, Szoecs E, Wagner H (2018) vegan: Community Ecology Package. R package version 2:5–3 https://cran.r-project.org/src/contrib/Archive/vegan/vegan_2.5-3.tar.gz
Piddock LJ (2006) Multidrug-resistance efflux pumps - not just for resistance. Nature Reviews Microbiology 4:629–636
Pu Q, Zhao LX, Li YT, Su JQ (2020) Manure fertilization increase antibiotic resistance in soils from typical greenhouse vegetable production bases, China. J Hazard Mater 391:122267
R Core Team (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria https://cran.r-project.org/src/base/R-3/R-3.6.3.tar.gz
Revelle M (2018) psych: procedures for psychological, psychometric, and personality research. R Package Version 1.8.10. https://cran.r-project.org/src/contrib/Archive/psych/psych_1.8.10.tar.gz
Rice EW, Wang P, Smith AL, Stadler LB (2020) Determining hosts of antibiotic resistance genes: a review of methodological advances. Environ Sci Tech Let 7(5):282–291
Rutgersson C, Ebmeyer S, Lassen SB, Karkman A, Fick J, Kristiansson E, Brandt KK, Flach CF, Larsson DGJ (2020) Long-term application of Swedish sewage sludge on farmland does not cause clear changes in the soil bacterial resistome. Environ Int 137:105339
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12:R60
Su JQ, Wei B, Ou-Yang WY, Huang FY, Zhao Y, Xu HJ, Zhu YG (2015) Antibiotic resistome and its association with bacterial communities during sewage sludge composting. Environ Sci Technol 49:7356–7363
Tang XJ, Lou CL, Wang SX, Lu YH, Liu M, Hashmi MZ, Liang XQ, Li ZP, Liao YL, Qin WJ (2015) Effects of long-term manure applications on the occurrence of antibiotics and antibiotic resistance genes (ARGs) in paddy soils: evidence from four field experiments in south of China. Soil Biol Biochem 90:179–187
Tedrew JCF, Robert E (1986) Soil of New Jersey. Krieger Publishing Company, Malabar
Tiedje JM, Wang F, Manaia CM, Virta M, Sheng HJ, Ma LP, Zhang T, Topp E (2019) Antibiotic Resistance Genes in the Human-Impacted Environment: A One Health Perspective. Pedosphere 29:273–282
Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, Teillant A, Laxminarayan R (2015) Global trends in antimicrobial use in food animals. Proc Natl Acad Sci USA. 112:5649–5694
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microb 73:5261–5267
Wang HL, Tang XY, Zhang W, Song SB, McKenzie BM (2015) Within-year changes in hydraulic properties of a shallow entisol in farmland and forestland. Vadose Zone J. 14
Wang L, Wang J, Wang J, Zhu L, Conkle JL, Yang R (2020) Soil types influence the characteristic of antibiotic resistance genes in greenhouse soil with long-term manure application. J Hazard Mater 392:122334
Wickham H (2016) ggplot2: elegant graphics for data analysis. Springer, New York
Xie WY, Yuan ST, Xu MG, Yang XP, Shen QR, Zhang WW, Su JQ, Zhao FJ (2018) Long-term effects of manure and chemical fertilizers on soil antibiotic resistome. Soil Biol Biochem 122:111–119
Xu M, Stedtfeld RD, Wang F, Hashsham SA, Song Y, Chuang Y, Fan J, Li H, Jiang X, Tiedje JM (2019) Composting increased persistence of manure-borne antibiotic resistance genes in soils with different fertilization history. Sci Total Environ 689:1172–1180
Zhang QQ, Ying GG, Pan CG, Liu YS, Zhao JL (2015) Comprehensive evaluation of antibiotics emission and fate in the river basins of China: source analysis, multimedia modeling, and linkage to bacterial resistance. Environ Sci Technol 49:6772–6782
Zhang W, Tang XY, Xian QS, Weisbrod N, Yang JE, Wang HL (2016) A field study of colloid transport in surface and subsurface flows. J Hydrol 542:101–114
Zhang YJ, Hu HW, Gou M, Wang JT, Chen D, He JZ (2017) Temporal succession of soil antibiotic resistance genes following application of swine, cattle and poultry manures spiked with or without antibiotics. Environ Pollut 231:1621–1632
Zhang JY, Sui QW, Tong J, Zhong H, Wang YW, Chen MX, Wei YS (2018) Soil types influence the fate of antibiotic-resistant bacteria and antibiotic resistance genes following the land application of sludge composts. Environ Int 118:34–43
Zhou YT, Niu LL, Zhu SY, Lu HJ, Liu WP (2017a) Occurrence, abundance, and distribution of sulfonamide and tetracycline resistance genes in agricultural soils across China. Sci Total Environ 599-600:1977–1983
Zhou ZC, Zheng J, Wei YY, Chen T, Dahlgren RA, Shang X, Chen H (2017b) Antibiotic resistance genes in an urban river as impacted by bacterial community and physicochemical parameters. Environ Sci Pollut Res 24:23753–23762
Zhou SYD, Zhu D, Giles M, Daniell T, Neilson R, Yang XR (2020) Does reduced usage of antibiotics in livestock production mitigate the spread of antibiotic resistance in soil, earthworm guts, and the phyllosphere? Environ Int 136:105359
Zhu YG, Zhao Y, Li B, Huang CL, Zhang SY, Yu S, Chen YS, Zhang T, Gillings MR, Su JQ (2017) Continental-scale pollution of estuaries with antibiotic resistance genes. Nat Microbiol 2:16270
Zhu YG, Zhao Y, Zhu D, Gillings M, Penuelas J, Ok YS, Capon A, Banwart S (2019) Soil biota, antimicrobial resistance and planetary health. Environ Int 131:105059
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Funding
This study was financially supported by the National Key Research and Development Program of China (Grant No. 2016YFD0800203), the Major Science and Technology Program of Sichuan Province (Grant No. 2018SZDZX0029), the National Natural Science Foundation of China (Grant Nos. 42007361 and 41771521), and the 135 Strategic Program of the CAS Institute of Mountain Hazards and Environment (Grant No. SDS-135-1702).
Author information
Authors and Affiliations
Contributions
Jianhua Cheng and Chen Liu did the incubation experiments and laboratory analyses. Jianhua Cheng performed the statistical analyses and wrote the first draft of this manuscript. Xiangyu Tang supervised the project and completed the final version of the manuscript. All authors improved and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Responsible Editor: Robert Duran
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 658 kb)
Rights and permissions
About this article
Cite this article
Cheng, J., Tang, X. & Liu, C. Bacterial communities regulate temporal variations of the antibiotic resistome in soil following manure amendment. Environ Sci Pollut Res 28, 29241–29252 (2021). https://doi.org/10.1007/s11356-021-12746-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-021-12746-8