Abstract
Emerging evidence suggested that long non-coding RNAs (lncRNAs) play pivotal roles in tumorigenesis. LINC01133 is a newly identified lncRNA first discovered as an oncogene in lung squamous cell carcinoma. Subsequent studies further demonstrated this lncRNA was deregulated in a wide spectrum of tumors, including colorectal, gastric, lung, and pancreatic ductal adenocarcinoma as well as osteosarcoma and hepatocellular carcinoma. Intriguingly, this lncRNA exerted oncogenic or tumor-suppressive action in a tissue-dependent manner. This review sought to summarize our current understanding concerning the deregulation of LINC01133 in human tumors in relation to its molecular mechanisms and cellular functions. The clinical utilization of LINC01133 as a potential prognostic biomarker and a treatment target is also discussed.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Data from the Human Genome Project and the Encyclopaedia of DNA Elements (ENCODE) Project revealed that only about 2% of the human genome codes for protein-coding genes with the remaining transcripts from the genome as mainly non-coding RNAs with little or no protein-coding potential(Johnsson et al. 2014, Lin & Yang 2018, Zhang & Jeang 2013). Long (> 200 nucleotides in the length) and small (< 200 nucleotides in length) non-coding RNAs represent two crucial groups of regulatory RNAs that were transcribed from this so called genomic dark matter (Li et al. 2018, Li et al. 2019, Yang et al. 2018a, Zhao et al. 2018, Zhou et al. 2017). An increasing number of studies suggested that long non-coding RNAs (lncRNAs) play critical roles in diverse cancer-related cellular processes (e.g., apoptosis, cell proliferation, differentiation, migration, invasion, drug resistance) through multiple mechanisms of action, such as transcriptional regulation via recruitment of chromatin modifiers, modulation of RNA splicing, sponging of microRNAs (miRNAs), regulating mRNA translation and degradation as well as direct interactions with target proteins(Wei et al. 2017, Wu et al. 2017, Ye et al. 2017, Zhang et al. 2017b, Zhang et al. 2017c, Zheng et al. 2017). In this connection, lncRNAs were found to be differentially expressed in most, if not all, types of cancer, including colorectal cancer, gastric cancer, hepatocellular carcinoma, breast cancer, gallbladder cancer, endometrial carcinoma, and osteosarcoma (Chen et al. 2017, Guo et al. 2016, Li et al. 2016a, Liu & Lin 2016, Liu et al. 2017, Ma et al. 2015, Zhang et al. 2016). Pertinent to clinical utilization, lncRNA has emerged as new biomarkers for tumor diagnosis and prognostication as well as therapeutic aims for the development of novel anti-cancer agents (Chak et al. 2016, Li et al. 2016c, Yang et al. 2018a, Zhu et al. 2015).
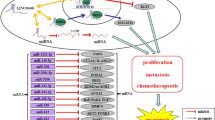
LINC01133, located in chromosome 1q23.2, is a newly discovered lncRNA originally identified as a potential oncogene in lung squamous cell carcinoma (LSCC)(Zhang et al. 2015). A growing number of studies subsequently revealed its deregulation in multitude of human cancers (Foroughi et al. 2018, Huang et al. 2018, Kong et al. 2018, Zang et al. 2016, Zhang et al. 2017a). Intriguingly, the function of LINC01133 seems to be tissue-specific, in which LINC01133 acts as an oncogene or tumor-suppressing gene in a context-dependent manner (Kong et al. 2016, Weng et al. 2019, Zeng et al. 2018, Zhang et al. 2019). This review sought to summarize our current knowledge about the aberrant expression of LINC01133 in human cancers (Table 1) with respect to its molecular mechanisms and cellular functions (Fig. 1). We also discuss the potential clinical utility of LINC01133 as a prognostic biomarker and a therapeutic target.
LINC01133 as an oncogenic lncRNA
Non-small cell lung cancer
The cancer-related function of LINC01133 was first reported by Zhang and colleagues in 2015 (Zhang et al. 2015). The investigators performed data mining of published microarray datasets to identify lncRNAs that could differentiate LSCC from lung adenocarcinoma (LAD), both of which are the two major histological types of non-small cell lung cancer (NSCLC). A total of 1646 lncRNAs were found be differentially abundant, among which LINC01133 showed the largest fold-change between the two cancer types. The authors further confirmed their results using reverse transcription-quantitative PCR (RT-qPCR) to demonstrate that the expression of LINC01133 was highly upregulated in LSCC (~ 6 fold) but not in LAD tissues. In LSCC patients, higher expression of LINC01133 was associated with shorter survival time (Zhang et al. 2015). Functionally, small interfering RNA (siRNA)-mediated repression of LINC01133 impaired the migration and invasion abilities of LSCC cell line, suggesting the oncogenic function of this lncRNA. These data provided the first glimpse into the potential function of LINC01133 in human cancers.
In an independent study, another group of investigators found that LINC01133 was significantly overexpressed in NSCLC samples and was positively correlated with tumor size, lymph node metastasis, and more advanced tumor-node-metastasis (TNM) staging as well as shorter survival time(Zang et al. 2016). Functional characterization revealed that knockdown of INC01133 suppressed the proliferation, migration, and invasion together with the induction of apoptosis and cell cycle arrest at G0/G1 phase in NSCLC cells in vitro and impaired the growth of NSCLC xenografts in vivo. Mechanistically, LINC01133 was found to interact with LSD1 (lysine specific demethylase 1, an important H3K4 demethylase of the CoREST repressor complex) and EZH2 (enhancer of zeste homolog 2, an essential H3K27 methyltransferase of the polycomb repressive complex 2) and recruit them to the promoters of CDH1 (encoding E-cadherin), CDKN1A (encoding the cyclin-dependent kinase inhibitor p21), and KLF2 (a zinc finger transcription factor) to silence their expression. RT-qPCR analysis confirmed the negative correlation of CDH1, CDKN1A, and KLF2 expression with LINC01133 levels in NSCLC tissues. In this regard, knockdown of Klf2 partially weakened the inhibitory effect of LINC01133 siRNA on NSCLC cell viability, suggesting that repression of Klf2 was functionally involved in the oncogenic action of this lncRNA (Zang et al. 2016). These findings collectively depicted the molecular pathway by which LINC01133 promotes NSCLC development.
Osteosarcoma
Osteosarcoma is the most common type of primary malignant tumor of the bone in children and young adults (Li et al. 2017, Li et al. 2016b). Zeng and colleagues investigated the role of LINC01133 in the development of osteosarcoma, in which LINC01133 expression was shown to be upregulated in the osteosarcoma cell lines and tissues as compared with a normal osteoplastic cell line and non-cancerous tissues (Zeng et al. 2018). Knockdown of LINC01133 significantly inhibited the proliferation, migration, and invasion of cultured osteosarcoma cells. Using bioinformatic prediction and dual-luciferase reporter assay, the investigators further demonstrated that miR-442a is the direct target of LINC01133 in osteosarcoma, where miR-442a functioned as a tumor-suppressing miRNA and reversed the oncogenic action of LINC01133(Zeng et al. 2018). These findings suggested that LINC01133 promoted the malignant phenotypes of osteosarcoma cells through targeting miR-442a.
Pancreatic ductal adenocarcinoma
Pancreatic ductal adenocarcinoma (PDAC) is a leading cause of cancer-related mortality, with a 5-year survival of less than 8% (Frampton et al. 2012, Szafranska-Schwarzbach et al. 2011). Huang and colleagues reported that LINC01133 was overexpressed in PDAC samples as compared with non-tumor samples and such upregulation was positively correlated with poorer prognosis (Huang et al. 2018). The oncogenic function of LINC01133 was demonstrated by the observation that LINC01133 knockdown suppressed the proliferation of cultured PDAC cells and the growth of PDAC xenografts. The transcription factor C/EBPβ was found to upregulate LINC01133 via binding to its response element within LINC01133 promoter. Consistently, C/EBPβ expression was upregulated in PDAC and its higher expression was correlated with poorer survival. Through analysis of The Cancer Genome Atlas (TCGA) dataset, the authors demonstrated a positive relationship between LINC01133 and CCNG1 (encoding cyclin G1) expression in PDAC. Concordantly, enforced expression of CCNG1 attenuated the impairment of cell proliferation induced by LINC01133 silencing (Huang et al. 2018). These findings indicated that the C/EBPβ-LINC01133-cyclin G1 axis plays a clinically relevant, oncogenic role in PDAC.
The upregulation of LINC01133 in PDAC was confirmed by another study in which the authors applied a systems biology approach known as the weighted gene co-expression network analysis (WGCNA) on published lncRNA expression data (Weng et al. 2019). Interestingly, the authors found that the gene encoding LINC01133 was frequently amplified and LINC01133 expression was significantly correlated with its copy number in PDAC. This study suggested that, in addition to the upstream regulator C/EBPβ, gene amplification is another mechanism that drives LINC01133 overexpression (Weng et al. 2019). Concerning the downstream effector pathway, Weng and colleagues reported that LINC01133 overexpression was associated with methylation and silencing of DKK1 (encoding the Wnt signaling inhibitor Dkk1), resulting in higher expression of genes in the Wnt signaling pathway in PDAC (Weng et al. 2019). Gene reporter assays revealed that LINC01133 bound to DKK1 promoter, resulting in H3K27 trimethylation and DKK1 gene silencing and increased expression of β-catenin, MMP-7, and Wnt-5a. Functionally, LINC01133 enhanced the proliferation, migration, and invasion of PDAC cells.
Hepatocellular carcinoma
Zheng and colleagues investigated the role of LINC01133 in the progression of hepatocellular carcinoma (HCC)(Zheng et al. 2019). They observed that LINC01133 expression was higher in a panel of HCC cell lines (HepG2, MHCC-97 L, Hep3B, MHCC-97H and SK-Hep-1) as compared with the normal human liver cell line HL-7702. Knockdown of LINC01133 inhibited HCC cell proliferation and colony formation together with the induction of apoptosis and cell cycle arrest at G0/G1 phase in vitro and delayed xenografts growth in vivo. Anti-tumorigenic effects of LINC01133 knockdown were paralleled by the inhibition of the phosphoinositide 3-kinase (PI3K)/AKT pathway (Zheng et al. 2019). These results showed that LINC01133 plays a pro-tumorigenic role in HCC through promoting the oncogenic PI3K/AKT signaling.
Cervical squamous cell carcinoma
Mao and colleagues developed and validated a prognostic lncRNA signature for cervical squamous cell carcinoma (CSCC), in which 15 lncRNAs of prognostic significance were identified from TCGA dataset (Mao et al. 2018). Among these lncRNAs, higher expression of LINC01133 was found to be significantly associated with shorter survival time of CSCC patients (Mao et al. 2018). Nevertheless, no experiment has been carried out so far to characterize the cellular function and molecular mechanism of LINC01133 in CSCC. Recently, Feng et al. (2019) illustrated that LINC01133 was overexpressed in the cervical tumor samples and LINC01133 induced cell proliferation, migration and EMT partly via regulating miR-4784/AHDC1.
LINC01133 as a tumor-suppressing lncRNA
Oral squamous cell carcinoma
Unlike its reported oncogenic function in other cancer types, LINC01133 functions as a tumor suppressor in oral squamous cell carcinoma (OSCC) (Kong et al. 2018). LINC01133 expression was reduced in OSCC samples as compared with paired normal tissues. OSCC patients with low tumoral expression of LINC01133 were also associated with metastasis and poorer survival. Concordantly, enforced expression of LINC01133 inhibited OSCC cell migration and invasion whereas LINC01133 knockdown produced the opposite effects. Transcriptome analysis revealed that GDF15 (a secreted ligand of the transforming growth factor (TGF)-β superfamily) was inhibited by LINC01133 in OSCC, in which GDF15 was upregulated. In this connection, enforced expression of GDF15 rescued LINC01133-induced inhibition of OSCC cell invasion and migration. Interestingly GDF15 was found to repress the expression of LINC01133, indicating a reciprocal regulation between LINC01133 and GDF15. In paired tissue analysis, there existed a significant negative correlation between these two factors (Kong et al. 2018). These findings suggested that LINC01133 functions as a tumor-suppressive lncRNA via targeting GDF15.
Esophageal squamous cell carcinoma
LINC01133 expression was lower in esophageal squamous cell carcinoma (ESCC) tissues and cell lines and such downregulation was positively correlated with ESCC progression (i.e., larger tumor size, greater depth of tumor invasion, lymph node metastasis, and more advanced TNM stage) and exposure to risk factors (i.e., smoking and alcohol drinking). LINC01133 downregulation was also found to be an independent prognostic factor predicting poorer overall survival and progression-free survival of ESCC patients (Yang et al. 2018c).
Gastric cancer
Yang and colleagues demonstrated that LINC01133 expression was decreased in gastric cancer (GC) cell lines and clinical samples and such downregulation was associated with metastasis and GC progression (Yang et al. 2018b). Functionally, knockdown of LINC01133 induced cell proliferation and epithelial-mesenchymal transition (EMT) whereas overexpression exerted opposite effects. LINC01133 directly targeted miR-106a-3p to derepress the expression of APC for inhibition of the oncogenic Wnt signaling. In this regard, LINC01133 inhibited EMT and metastasis by suppressing the Wnt/β-catenin signaling in an APC-dependent manner (Yang et al. 2018b). The downregulation of LINC01133 in GC was confirmed by an independent study which identified LINC01133 as a GC-related lncRNA in Gene Expression Omnibus (GEO) datasets followed by validation with both internal samples and external datasets. Pathway analyses indicated that genes co-expressed with LINC01133 were mostly involved in metabolic pathways as well as gastrointestinal disease and function (Yang et al. 2018b).
Colorectal cancer
LINC01133 expression was decreased in CRC samples as compared with non-tumor tissues. Importantly, downregulation of LINC0113 was associated with lymph node metastasis, distant metastasis, and advanced TNM stage as well as poorer overall survival. Further multivariate analysis indicated that LINC01133 was associated with CRC patients’ survival independent of other clinicopathological parameters (Zhang et al. 2017a). Consistently, another study found that LINC01133 was downregulated by TGF-β whereas enforced expression of LINC01133 suppressed EMT and/or metastasis of CRC cells in vitro and in vivo. LINC01133 was found to physically interact with SRSF6, of which its overexpression promoted EMT and metastasis, indicating that LINC01133-regulated EMT was mediated, at least in part, via inhibiting SRSF6. Consistent with its anti-EMT function, clinical sample analysis revealed that LINC01133 expression was negatively associated with vimentin (a mesenchymal marker) and positively associated with E-cadherin (an epithelial marker). Low tumor expression of LINC01133 was correlated with poor survival of CRC patients (Zhang et al. 2017a). These two studies indicated that LINC01133 downregulation contributes functionally to CRC progression where this lncRNA could serve as a prognostic factor.
Breast cancer
Song et al. (2019) illustrated that LINC01133 was decreased in breast tumor specimens and was correlated with poor prognosis and progression of breast tumor. Ectopic expression of LINC01133 suppressed metastasis and invasion in breast tumor both in vivo and vitro. Mechanistic studies noted that LINC01133 suppressed SOX4 expression through recruiting the EZH2 to promoter of SOX4. This study noted that LINC01133 acted as one new therapeutic target and prognostic biomarker for breast tumor.
Ovarian tumor
Liu et al. (2019) clarified that LINC01133 was downregulated in ovarian tumor cells and specimens and overexpression of LINC01133 decreased cell invasion, growth, and migtation both in vivo and in vitro. LINC01133 was noted to act as one miR-205 sponge to suppress cell invasion, growth, and migtation via enhancing LRRK2.
Future perspectives
LINC01133 is a newly identified cancer-related lncRNA with oncogenic functions in NSCLC, osteosarcoma, PDAC, HCC, and CSCC but tumor-suppressive actions in luminal cancers of the gastrointestinal tract, namely, OSCC, ESCC, GC, and ESCC (Table 1). As an oncogenic lncRNA, the high expression of LINC01133 is driven by the transcription factor C/EBPβ and gene amplification in tissues where LINC01133 promotes tumorigenesis through targeting tumor-suppressive miRNA (i.e., miR-442a) or proteins (i.e., Klf2, E-cadherin, p21, Dkk1) or upregulation of oncogenic mediators (Cyclin G1 and phosphorylated PI3K/AKT). As a tumor-suppressor, there is a reciprocal inhibition between LINC01133 and the pro-EMT TGF-β signaling. LINC01133 also inhibited the oncogenic Wnt/β-catenin signaling via targeting miR-106a-3p (Fig. 1).
Despite its paradoxical role in tumorigenesis, deregulation of LINC01133 has been shown to be associated with clinicopathological parameters and patients’ survival in multiple cancer types, suggesting the prognostic value of this lncRNA. However, next study of its performance in cohorts of more specimen sizes is still needed to promote the clinical use of LINC01133. Moreover, the tissue-specific function of LINC01133 might preclude its systemic inhibition or reactivation for cancer therapy. Development of methods for tissue-specific modulation of LINC01133 is therefore warranted.
Taken together, LINC01133 is a crucial tumor-related lncRNA with the clinical potentials. Indeed, more research is needed to accelerate the translation of LINC01133 from basic research into clinical use.
Data availability statement
Research data are not shared.
References
Chak WP, Lung RW, Tong JH, Chan SY, Lun SW, Tsao SW, Lo KW, To KF (2016): Downregulation of long non-coding RNA MEG3 in nasopharyngeal carcinoma. Mol Carcinog
Chen LS, Yao HB, Wang K, Liu XF (2017) Long non-coding RNA MALAT1 regulates ZEB1 expression by sponging miR-143-3p and promotes hepatocellular carcinoma progression. J Cell Biochem 118:4836–4843
Feng Y, Qu L, Wang X, Liu C (2019) LINC01133 promotes the progression of cervical cancer by sponging miR-4784 to up-regulate AHDC1. Cancer Biol Ther 20:1453–1461
Foroughi K, Amini M, Atashi A, Mahmoodzadeh H, Hamann U, Manoochehri M (2018): Tissue-specific down-regulation of the long non-coding RNAs PCAT18 and LINC01133 in gastric cancer development. Int J Mol Sci 19
Frampton AE, Krell J, Zhang Y, Stebbing J, Castellano L, Jiao LR (2012) The role of miR-10b in metastatic pancreatic ductal adenocarcinoma. Surgery 152:936–938
Guo Q, Qian Z, Yan D, Li L, Huang L (2016) LncRNA-MEG3 inhibits cell proliferation of endometrial carcinoma by repressing notch signaling. Biomed Pharmacoth Biomed Pharmacoth 82:589–594
Huang CS, Chu JJ, Zhu XX, Li JH, Huang XT, Cai JP, Zhao W, Yin XY (2018) The C/EBP beta-LINC01133 axis promotes cell proliferation in pancreatic ductal adenocarcinoma through upregulation of CCNG1. Cancer Lett 421:63–72
Johnsson P, Lipovich L, Grander D, Morris KV (2014) Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta 1840:1063–1071
Kong JL, Zhang HH, Xu EP, Huang Q, Chen J, Lai MD (2016): Long non-coding RNA LINC01133 inhibits epithelial-mesenchymal transition and metastasis in colorectal cancer by interacting with SRSF6. Cancer Res 76
Kong JL, Sun WJ, Zhu WY, Liu CX, Zhang HH, Wang HM (2018) Long noncoding RNA LINC01133 inhibits oral squamous cell carcinoma metastasis through a feedback regulation loop with GDF15. J Surg Oncol 118:1326–1334
Li P, Xue WJ, Feng Y, Mao QS (2016a) Long non-coding RNA CASC2 suppresses the proliferation of gastric cancer cells by regulating the MAPK signaling pathway. Am J Transl Res 8:3522–3529
Li Z, Yu X, Shen JX (2016b) Long non-coding RNAs: emerging players in osteosarcoma. Tumor Biol 37:2811–2816
Li ZW, Zhao L, Wang QG (2016c) Overexpression of long non-coding RNA HOTTIP increases chemoresistance of osteosarcoma cell by activating the Wnt/beta-catenin pathway. Am J Transl Res 8:2385–2393
Li Z, Shen JX, Chan MTV, Wu WKK (2017) MicroRNA-379 suppresses osteosarcoma progression by targeting PDK1. J Cell Mol Med 21:315–323
Li Z, Li XY, Chen C, Li SG, Shen JX, Tse G, Chan MTV, Wu WKK (2018) Long non-coding RNAs in nucleus pulposus cell function and intervertebral disc degeneration. Cell Prolif 51:e12483
Li Z, Li XY, Chen X, Li SG, Ho IHT, Liu XD, Chan MTV, Wu WKK (2019) Emerging roles of long non-coding RNAs in neuropathic pain. Cell Prolif 52:e12528
Lin C, Yang L (2018) Long noncoding RNA in cancer: wiring signaling circuitry. Trends Cell Biol 28:287–301
Liu CB, Lin JJ (2016) Long noncoding RNA ZEB1-AS1 acts as an oncogene in osteosarcoma by epigenetically activating ZEB1. Am J Transl Res 8:4095–4105
Liu J, Song ZW, Feng C, Lu YL, Zhou Y, Lin Y, Dong CY (2017) The long non-coding RNA SUMO1P3 facilitates breast cancer progression by negatively regulating miR-320a. Am J Transl Res 9:5594–5602
Liu M, Shen C, Wang C (2019) Long noncoding RNA LINC01133 confers tumor-suppressive functions in ovarian cancer by regulating leucine-rich repeat kinase 2 as an miR-205 sponge. Am J Pathol 189:2323–2339
Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY, Gong W, Quan ZW (2015) Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis 6:e1583
Mao XG, Qin XM, Li L, Zhou JT, Zhou M, Li XX, Xu Y, Yuan LY, Liu QN, Xing H (2018) A 15-long non-coding RNA signature to improve prognosis prediction of cervical squamous cell carcinoma. Gynecol Oncol 149:181–187
Song Z, Zhang X, Lin Y, Wei Y, Liang S, Dong C (2019) LINC01133 inhibits breast cancer invasion and metastasis by negatively regulating SOX4 expression through EZH2. J Cell Mol Med 23:7554–7565
Szafranska-Schwarzbach AE, Adai AT, Lee LS, Conwell DL, Andruss BF (2011) Development of a miRNA-based diagnostic assay for pancreatic ductal adenocarcinoma. Expert Rev Mol Diagn 11:249–257
Wei W, Liu Y, Lu YB, Yang B, Tang L (2017) LncRNA XIST promotes pancreatic Cancer proliferation through miR-133a/EGFR. J Cell Biochem 118:3349–3358
Weng YC, Ma J, Zhang J, Wang JC (2019) Long non-coding RNA LINC01133 silencing exerts antioncogenic effect in pancreatic cancer through the methylation of DKK1 promoter and the activation of Wnt signaling pathway. Cancer Biol Ther 20:368–380
Wu ZH, He YY, Li DL, Fang X, Shang T, Zhang HK, Zheng XT (2017) Long noncoding RNA MEG3 suppressed endothelial cell proliferation and migration through regulating miR-21. Am J Transl Res 9:3326–3335
Yang C, Wu K, Wang S, Wei G (2018a) Long non-coding RNA XIST promotes osteosarcoma progression by targeting YAP via miR-195-5p. J Cell Biochem 119:5646–5656
Yang XZ, Cheng TT, He QJ, Lei ZY, Chi J, Tang Z, Liao QX, Zhang H, Zeng LS, Cui SZ (2018b) LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/beta-catenin pathway. Mol Cancer 17:126
Yang XZ, He QJ, Cheng TT, Chi J, Lei ZY, Tang Z, Liao QX, Zhang H, Zeng LS, Cui SZ (2018c) Predictive value of LINC01133 for unfavorable prognosis was impacted by alcohol in esophageal squamous cell carcinoma. Cell Physiol Biochem 48:251–262
Ye KS, Wang SK, Zhang HH, Han H, Ma B, Nan W (2017) Long noncoding RNA GAS5 suppresses cell growth and epithelial-mesenchymal transition in osteosarcoma by regulating the miR-221/ARHI pathway. J Cell Biochem 118:4772–4781
Zang CS, Nie FQ, Wang Q, Sun M, Li W, He J, Zhang ML, Lu KH (2016) Long non-coding RNA LINC01133 represses KLF2, P21 and E-cadherin transcription through binding with EZH2, LSD1 in non small cell lung cancer. Oncotarget 7:11696–11707
Zeng HF, Qiu HY, Feng FB (2018) Long noncoding RNA LINC01133 functions as an miR-422a sponge to aggravate the tumorigenesis of human osteosarcoma. Oncol Res 26:335–343
Zhang Q, Jeang KT (2013) Long non-coding RNAs (lncRNAs) and viral infections. Biomed Pharmacoth Biomed Pharmacoth 3:34–42
Zhang J, Zhu N, Chen XD (2015) A novel long noncoding RNA LINC01133 is upregulated in lung squamous cell cancer and predicts survival. Tumor Biol 36:7465–7471
Zhang YH, Fu J, Zhang ZJ, Ge CC, Yi Y (2016) LncRNA-LINC00152 down-regulated by miR-376c-3p restricts viability and promotes apoptosis of colorectal cancer cells. Am J Transl Res 8:5286–5297
Zhang JH, Li AY, Wei N (2017a) Downregulation of long non-coding RNA LINC01133 is predictive of poor prognosis in colorectal cancer patients. Eur Rev Med Pharmacol Sci 21:2103–2107
Zhang LM, Wang P, Liu XM, Zhang YJ (2017b) LncRNA SUMO1P3 drives colon cancer growth, metastasis and angiogenesis. Am J Transl Res 9:5461
Zhang Y, Dang YW, Wang X, Yang X, Zhang R, Lv ZL, Chen G (2017c) Comprehensive analysis of long non-coding RNA PVT1 gene interaction regulatory network in hepatocellular carcinoma using gene microarray and bioinformatics. Am J Transl Res 9:3904–3917
Zhang WJ, Du MY, Wang TT, Chen W, Wu J, Li Q, Tian XK, Qian LX, Wang Y, Peng FY, Fei Q, Chen J, He X, Yin L (2019) Long non-coding RNA LINC01133 mediates nasopharyngeal carcinoma tumorigenesis by binding to YBX1. Am J Cancer Res 9:779
Zhao J, Gao Z, Zhang C, Wu H, Gu R, Jiang R (2018): Long non-coding RNA ASBEL promotes osteosarcoma cell proliferation, migration and invasion by regulating microRNA-21. J Cell Biochem
Zheng JL, Yi D, Liu Y, Wang MQ, Zhu YL, Shi HZ (2017) Long nonding RNA UCA1 regulates neural stem cell differentiation by controlling miR-1/Hes1 expression. Am J Transl Res 9:3696–3704
Zheng YF, Zhang XY, Bu YZ (2019) LINC01133 aggravates the progression of hepatocellular carcinoma by activating the PI3K/AKT pathway. J Cell Biochem 120:4172–4179
Zhou DD, Liu XF, Lu CW, Pant OP, Liu XD (2017) Long non-coding RNA PVT1: emerging biomarker in digestive system cancer. Cell Prolif 50:e12398
Zhu J, Liu S, Ye F, Shen Y, Tie Y, Wei L, Jin Y, Fu H, Wu Y, Zheng X (2015) Long noncoding RNA MEG3 interacts with p53 protein and regulates partial p53 target genes in hepatoma cells. PLoS One 10:e0139790
Author information
Authors and Affiliations
Contributions
Zheng Li, Derong Xu, Shugang Li, M T.V. Chan, and Xin Chen have all contributed to design and write manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible editor: Lotfi Aleya
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zheng Li, Derong Xu, and Xin Chen are co-first authors.
Rights and permissions
About this article
Cite this article
Li, Z., Xu, D., Chen, X. et al. LINC01133: an emerging tumor-associated long non-coding RNA in tumor and osteosarcoma. Environ Sci Pollut Res 27, 32467–32473 (2020). https://doi.org/10.1007/s11356-020-09631-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-020-09631-1