Abstract
Plants adapt to metal stress by modifying their metabolism including the production of secondary metabolites in plant tissues. Such changes may impact the diversity and functions of plant associated microbial communities. Our study aimed to evaluate the influence of metals on the secondary metabolism of plants and the indirect impact on rhizosphere bacterial communities. We then compared the secondary metabolites of the hyperaccumulator Pteris vittata L. collected from a contaminated mining site to a non-contaminated site in Vietnam and identified the discriminant metabolites. Our data showed a significant increase in chlorogenic acid derivatives and A-type procyanidin in plant roots at the contaminated site. We hypothesized that the intensive production of these compounds could be part of the antioxidant defense mechanism in response to metals. In parallel, the structure and diversity of bulk soil and rhizosphere communities was studied using high-throughput sequencing. The results showed strong differences in bacterial composition, characterized by the dominance of Proteobacteria and Nitrospira in the contaminated bulk soil, and the enrichment of some potential human pathogens, i.e., Acinetobacter, Mycobacterium, and Cupriavidus in P. vittata’s rhizosphere at the mining site. Overall, metal pollution modified the production of P. vittata secondary metabolites and altered the diversity and structure of bacterial communities. Further investigations are needed to understand whether the plant recruits specific bacteria to adapt to metal stress.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The presence of heavy metals in the environment, which is largely due to anthropic activities in urban and industrial areas, represents a stress for all living organisms, impacting ecosystem structure and function, including soil microbial communities (Jaishankar et al. 2014; Singh et al. 2014). Although some metallic trace elements are essential for bacterial physiology (e.g., zinc (Zn), iron (Fe), copper (Cu)), others (e.g., cadmium (Cd), chrome (Cr), arsenic (As)) are toxic for bacterial cells and can alter diversity and function of bacterial communities depending on their concentrations in the environment (Epelde et al. 2012; Lenart-Boroń and Boroń 2014). Metal pollution then represents a potential risk for bacterial community diversity and functions and then soil functioning (Azarbad et al. 2013; Singh et al. 2014).
Plants adapt to heavy metal stress by modifying their metabolism including the production of secondary metabolites in plant tissues, although less studied than primary ones (Singh et al. 2016). On the other hand, plants are known to be major drivers of soil microbial community structure and functioning through root exudation (Hartmann et al. 2008; Michalet et al. 2013). Changes in metabolism might then impact diversity and functions of microbial communities associated to leave (i.e., phyllosphere) or root (i.e., rhizosphere) systems. A recent theory named “Plants call for support,” hypothesizes that pollution-induced changes in root exudation might select for microbial communities bearing beneficial traits for plants (e.g., degradative capacities), allowing a better adaptation under contaminant stress (Thijs et al. 2016).
In Vietnam, heavy metal pollution in soils resulting from anthropization, including mining activities, has been increasing in the last few decades. Recent study summarized that about 5000 mines of ore have been explored, and the area of abandoned mines was more than 3700 ha (Anh et al. 2011). In recent years, many researches have been focused on phytoremediation by hyperaccumulators in order to remove heavy metals from mining sites in Vietnam (Nguyen et al. 2011; Anh et al. 2011). However, it has not yet been evaluated how metal contamination alters the secondary metabolism of these plants as well as their effect on associated rhizosphere communities. Here we have used Pteris vittata L. as a plant model since this indigenous fern has been reported for its great potential to uptake As and to tolerate high Zn and Pb level in Ha Thuong mining site (Anh et al. 2011).
Pteris vittata L., commonly known as Chinese brake fern or Ladder brake fern, is a hyperaccumulator belonging to the Pteridaceae family and is found in tropical and sub-tropical regions like South East Asia, Africa, or Australia. This plant was reported as the first known As hyperaccumulator (Ma et al. 2001). Arsenic is essentially accumulated in its fronds at very high levels (up to 22,630 mg/kg), but the concentration of this metal in the roots is also non-negligible (100 mg/kg) (Ma et al. 2001; Wang et al. 2002). P. vittata has been extensively studied for the phytoremediation of As-contaminated soils, due to its strong As accumulation as well as its capacity to take up many forms of As (Ma et al. 2001; Tu and Ma 2002; Tu et al. 2002; Danh et al. 2014). P. vittata is also able to accumulate other metals such as Zn (up to 737 mg/kg in leaves) (An et al. 2006), and its tolerance to high contamination level of Pb, Zn, and Cu was also reported (Kachenko et al. 2007; Anh et al. 2011). Besides, this plant has been explored for its pharmacological properties like hypoglycemic (Paul et al. 2012), anti-inflammatory, platelet aggregation, and antitumor activities (Gong et al. 2007). Some P. vittata secondary metabolites were also examined for their antimicrobial properties (rutin) (Singh et al. 2008) and their impacts on visual process (rutin, flavones glycosides…) (Wahid et al. 2016). To our knowledge, although several studies have investigated the phytochemical composition of P. vittata extracts and the impacts of P. vittata on heavy metal content in soils (Tu et al. 2002; Nguyen et al. 2011; Gracelin et al. 2012; Jaishee and Chakraborty 2015; Wahid et al. 2016), the effect of these pollutants on its metabolism remains unknown.
Thus, our objectives were (i) to compare the secondary metabolite profiles of each part of P. vittata grown on contrasted contaminated areas, in order to evaluate the effect of metal pollution on secondary metabolism of plants colonizing metalliferous sites, and (ii) to evaluate the indirect influence of these changes in metabolism on the rhizosphere bacterial community composition. Metabolic profiling of roots, stems, and leaves of P. vittata was based on UV-absorbing compounds, thus targeting more specifically phenolics and other aromatic compounds. Changes in bacterial community structure were assessed using a metataxogenomic analysis approach.
Materials and methods
Site, plant, and soil sampling
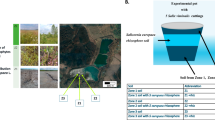
The studied site is located at Ha Thuong Commune, Dai Tu district, Thai Nguyen Province, Vietnam (Fig. 1). At that site, Sn-ore mining activities have been operated from 1988 to 2007. These operations led the contamination with heavy metals including As, Pb, Zn, Cu, and Cd in soils. In 2007, in the Vietnam National Research Projet, No. KC08.04/06-10, for phytoremediation of mining areas in Thai Nguyen, P. vittata, Pityrogramma calomelanos, and Dicranopteris linearis were cultured to remove As and Cd from soil. However, the concentration of Cu and Pb in soil still exceeds the allowable limit according to “National technical regulation on the allowable limits of heavy metals in the soils” (Table 1).
Ten mature P. vittata individuals were collected in Ha Thuong mining site and five individuals were sampled at the same growth stage in a non-contaminated site (Academy of Science and Technology, in Hoang Quoc Viet Street, Cau Giay district, Hanoi, Vietnam). All samples were identified by Prof. Tran Huy Thai, Institute of Ecology and Biological Resources, Vietnam Academy of Science and Technology. Voucher specimens were deposited in the Department of Bioactive Products, Institute of Marine Biochemistry, Vietnam Academy of Science and Technology.
From the mining site, rhizosphere soil samples were collected under P. vittata (PVP) and D. linearis (DLP) (another fern cultured in this site and used in this study as a comparative polluted rhizosphere soil). Nearby, bulk soil samples (BSP) (i.e., soil with no vegetal cover) were collected. Rhizosphere soil samples were also collected under P. vittata at the Vietnam Academy of Science and Technology and will be further considered as a control (PVC) (non-polluted soil). All soils were collected from the upper layer (0–15 cm) and stored at ambient temperature for no longer than 2 weeks before use. Soil characteristics are shown in Table 1 and were measured by the Laboratory of Soil Analysis (INRA Arras, France) using standard methods. Metal content in soil was analyzed by inductively coupled plasma mass spectrometry (ICP-MS) at the Centre des Recherches Pétrographiques et Géochimiques (CRPG) of Vandoeuvre-les-Nancy (France) and metal content in plant parts was analyzed by the Institute of Natural Products Chemistry, Vietnam Academy of Science and Technology.
Sample preparation for the analysis of plant metabolites
Each plant was carefully excavated and dried, and each part (roots, stems, and leaves) was separated before being crushed into fine powder using mortar and liquid nitrogen. Plant metabolites were extracted by immersion of 5 g of dried samples in 100 ml of absolute MeOH in an Erlenmeyer, followed by ultra-sonication for 15 min at room temperature and then filtration. Extraction process was repeated three times. The extracts were pooled and evaporated under reduced pressure, stored at −20 °C, and then dissolved in absolute MeOH to a concentration of 10 mg/mL before analysis.
For UHPLC-DAD-ESI/QTOF analysis, stem and leave extracts were prepared in the same conditions described above while root extracts were dissolved in MilliQ water and further divided into fractions by using solid phase extraction (SPE) on Bond Elut® C18 SPE cartridges (Agilent Technologies®, Santa Clara, CA, USA). Elution was performed with a gradient of H2O/MeOH (100:0; 80:20; 0:100). The fraction H2O/MeOH (80:20) was evaporated then dissolved in MeOH absolute with a final concentration of 10 mg/mL before analysis.
UHPLC-DAD-ESI/QTOF analysis
UHPLC-DAD-ESI/QTOF analysis was performed on an Agilent Infinity® 1290 system (Agilent Technologies®) coupled to a UV/vis DAD detector and equipped with a QTOF 6530 (Agilent Technologies®) detector controlled by MassHunter® software (Agilent Technologies®). Analyte separation was carried-out on a Poroshell® 120 EC-C18 column (100 mm × 3.0 mm, 2.7 μm). The following gradient was run using 0.4% aqueous formic acid (solvent A) and acetonitrile (solvent B): 0 min, 1% B; 1.5 min, 1% B; 12 min, 30% B; 18 min, 100% B; 19 min, 100% B. The flow rate and column temperature were 1.2 mL/min and 50 °C, respectively, and 2.0 μL of sample extracts was injected. The signals were detected at four wavelength λ 254, 280, 320, and 360 nm and UV spectra recorded between 190 and 600 nm. The chromatograms recorded at λ 280 nm were selected for statistical analysis.
The ESI source was optimized as follows: positive and negative ionization mode in auto-MSMS, scan spectra from m/z 100 to 3000, capillary voltage 2.5 kV, nozzle voltage 2 kV, fragmentor 120 V, and fixed collision-induced dissociation (CID) energy at 20 eV. Nitrogen was used as the nebulizing gas with a flow rate of 11 L/min and a temperature of 310 °C at 40 psi.
Metabolites annotation
The annotation of metabolites in P. vittata extracts was based on UV, HRMS, MSMS spectra, and relative RTs. Data were compared with previous literature report for compounds in Pteris spp. or in other genus.
Statistical analysis
To assess differences in metabolite composition between plants growing in non-polluted and in polluted area, RTs of peaks in chromatograms at wavelength λ 280 nm were aligned and their relative intensities integrated in a matrix for each plant part to perform the principal component analysis (PCA) using R® software, version 3.1.2 (R Core Team 2014). The compounds responsible for the discrimination between polluted and non-polluted samples were further analyzed using ANOVA followed by Tukey’s honest significant difference (HSD) tests after data were controlled for normality and homoscedasticity. Those analyses were conducted at p < 0.05.
Soil DNA extraction and metagenomic analysis
Total DNA was extracted from 0.5 g of each bulk and rhizosphere soil sample with three replicates using a FastDNA® SPIN Kit for Soil (MP Biomedicals, Solon, OH, USA) and then purified on S-400-HR mini-columns (Pharmacia, St Quentin Yvelines, France) following the manufacturer’s instructions. The DNA extracts were resolved by electrophoresis in 0.8% agarose gels, stained with ethidium bromide, and photographed using a Gel Doc 1000 camera (Bio-Rad, Ivry sur Seine, France). DNA concentration was estimated using a NanoDrop® ND-1000 spectrophotometer (Labtech International, Paris, France) at a 260 nm wavelength. Triplicate DNA extractions were performed from all samples. Replicates were then pooled for further metagenomic analysis.
16S rRNA genes of pooled DNA samples were sequenced at the DTAMB/ Biofidal platform (University of Lyon 1, France) using an Illumina’s Miseq platform and paired end reads. In total, 1,254,982*2 valid sequences for bacteria were obtained from four samples. Metagenomic analysis was carried-out following the Mothur MiSeq standard operating procedure (SOP) pipeline v1.35.1 (http://www.mothur.org/wiki/MiSeq_SOP) developed by the Schloss lab (Department of Microbiology & Immunology, University of Michigan, USA) (Schloss et al. 2009; Kozich et al. 2013). Briefly, the contigs of paired end reads 1 and 2 were constructed using the make.contigs command, and 1,245,982 sequences with an average length of 457 bp were produced. After removing the sequences with ambiguous bases, and those longer than 550 bp or shorter than 400 bp with the screen.seqs command, a total of 338,298 high-quality sequences were obtained with an average length of 448 bp per sequence. The only unique sequences were obtained using unique.seqs command and the align.seqs command were then operated to align the data with mothur’s SILVA bacterial 16S reference alignment v123 (https://www.arb-silva.de/) (Quast et al. 2013). After screening the unique sequences, the remaining reads were then pre-clustered to denoise sequences within each sample and chimeric sequences were removed using chimeric.uchime command (http://drive5.com/uchime) (Edgar et al. 2011). Finally, a range from 24,458 to 66,753 sequences with an average number of 38,566 sequences per sample were obtained and then subsampling to 24,458 sequences. Each sequence was then classified against the RDP 16S rRNA training set v9 using a naïve Bayesian classifier, at an 80% confidence level (Wang et al. 2007). The sequences were regrouped into operational taxonomic units (OTUs) with a 3% divergence threshold using cluster.split command (http://www.mothur.org/wiki/Cluster.split). The Chao1 richness estimator and the Np Shannon diversity were also calculated.
Bacterial quantification using culture enumeration and qPCR
Microorganisms were extracted by blending 5 g of soil samples with 50 mL of a 0.8% (w/v) sterile NaCl solution for 90 s in a Waring blender (Eberbach Corporation, Michigan, USA). The homogeneous soil suspension was serially diluted tenfold in sterile saline solution and 100 μL of appropriate dilutions were spread. The viable heterotrophic microflora was enumerated on tenfold diluted tryptic soy agar medium (TSA1/10) (Oxoïd, Dardilly, France) supplemented with cycloheximide (200 mg/L) to impair the growth of fungi. Three plates were inoculated per dilution and bacterial colonies were counted after 5 days of incubation at 28 °C.
Real-time PCR amplification (qPCR) was performed using a LightCycler 480 system (Roche Diagnostics, Meylan, France) with the LightCycler® 480 software, version LCS480 1.5.0.39 (Roche Diagnostics). The primers 5′-TCCATGAAGTCGGAATCGCTAG-3′ and 5′-CACTCCCATGGTGTGACGG-3′ were used (Invitrogen, Cergy Pontoise, France). The mix reactions were performed with the SsoFast™ EvaGreen® Supermix (Bio-Rad) as specified by the manufacturer. Five microliters of template DNA (corresponding to 0.2 and 1 ng of DNA) was added, and deionized water was used to reach a final volume of 25 μL. Negative controls without template DNA were run in triplicate. Each reaction was run in duplicate with both DNA template concentrations using the following cycle conditions: 1 cycle at 95 °C for 10 min followed by 40 cycles of 95 °C for 10 s and 60 °C for 10 s.
Results
Metabolic profiling of UV-absorbing compounds in P. vittata extracts
UV-based metabolite profiling was carried out on root, stem, and leave extracts of fifteen P. vittata samples, ten of which were collected from contaminated soil, in order to compare the metabolite profiles of plants from metal-polluted and non-polluted areas. Chromatograms at λ 280 nm showed strong differences between samples from these two areas as shown by the comparison of typical UHPLC/DAD chromatograms (Figs. S1, S2, S3 ). Concentrations of compounds (i.e., absolute areas) were also affected but in order to compare the variations in global extract compositions, relative area was chosen (which also enabled to attenuate the variability in total extract concentrations). This suggests that there are qualitative and quantitative changes in the secondary metabolism of plants from diversely contaminated soils.
PCA allowed to highlight differences between the metabolic profiles of plant grown on contaminated soils from those grown in non-contaminated area, and this was especially the case for stems and leaves (Fig. 2). Concerning roots, the discrimination was less pronounced (Fig. 2a), probably because of the variability observed in compound relative intensities, which might be due to the extra SPE separation procedure realized for these extracts. In leaves and stems (Fig. 2b, c), the discrimination was better and each group could be well separated along axis 1.
PCA performed on UV chromatographic profiles obtained for each extract of P. vittata: roots (a), stems (b), and leaves (c) grown on non-polluted and polluted soil. Each point on the left plots represents one extract and the circles indicate the type of soil on which plants grew. Each point on the right plots represents one peak and the circles indicates the discriminant compounds
Secondary metabolites affected by metal contamination
Based on PCA, the compounds involved in the different group segregation were highlighted (Fig. 2). In order to identify the discriminating peaks in different conditions, ANOVA followed by Tukey’s HSD test was carried-out for these compounds and only those that showed a statistically significant difference and representing at least 1% of the total area in one condition are highlighted. In root extracts, six discriminant compounds were characterized, two of which were found significantly in higher proportions in the polluted condition compared to the non-polluted one (8a, 32a), while four were found in lower proportions (26a, 28a, 34a, 38a) (Fig. 3a, b). Regarding stems, five compounds were identified with significantly increased proportions in the case of polluted soil (13b, 20b, 30b, 32b, 33b), while three were found with decreased proportions (24b, 38b, 39b) (Fig. 3c, d). Concerning leaves, six peaks were found as discriminating, of which four were in higher proportions with contamination (28c, 36c, 45c, 49c) and two presenting lower proportions (33c, 37c) (Fig. 3e, f).
P. vittata root, stem, and leave secondary metabolites affected by the type of soil. Relative intensities of discriminant chromatographic peaks: increasing compounds (a, c, e) and decreasing compounds (b, d, f) in P. vittata roots, stems, and leaves, respectively. Green bars indicate extract obtained from non-polluted soils and red bars from polluted ones. Statistical differences between type of soil are indicated using characters *, **, *** (Tukey’s HSD test, significativity codes ‘***’ p < 0.001, ‘**’ p < 0.01, ‘*’ p < 0.05). Compound numbers are indicated within brackets below Rts.
Annotation of discriminant compounds
The discriminant compounds are listed in Tables 2 and 3 with their identification based on relative RTs, UV, and mass spectra. In roots, most of the peaks involved in the segregation of different groups were identified as hydroxycinnamic acid derivatives. Considering stems and leaves, flavonol glycoside derivatives seemed to be the major discriminant compounds.
Annotation of discriminant compounds in root extracts
Discriminant compounds annotated in root extracts were distributed in two major classes: hydroxycinnamic derivatives (8a, 26a, 28a, 34a, 38a) and procyanidin (32a), according to their respective specific UV spectra (i.e., a strong absorption band at 310–320 nm with a shoulder at 300 nm and a weak absorption band at 276–278 nm, respectively). The most abundant compound, 32a, was detected at RT 8.328 min and found in higher proportion in roots of plants growing in metal-polluted area. Its mass spectra showed a pseudo-molecular ion at m/z 863.1826 ([M−H]−) and m/z 865.2013 ([M+H]+) corresponding to the molecular mass of 864 Da with the formula C45H36O18 suggesting a procyanidin trimer. Moreover, the ESI−/MS2 spectra of this compound produced the fragmentation ions at m/z 773 and m/z 289, indicative of the presence of an A-type inter-flavonoid linkage. Based on the literature data (Gu et al. 2003; Qiang et al. 2015), this compound was tentatively identified as A-type procyanidin trimer, with the (epi)catechin-A-(epi)catechin-(epi)catechin connection.
Among the hydroxycinnamic derivatives, one compound (8a) was found to be increased and four compounds (26a, 28a, 34a, 38a) were found to be decreased in metal-polluted area. For compound 8a (RT 3.623 min), the ([M−H]−) ion at m/z 353.0878 and the ([M+H]+) ion at m/z 355.1041 suggested the molecular weight of 354 Da corresponding to the formula C16H18O9. ESI+/MS2 spectra of this compound produced a base peak at m/z 163.0394 ([M+H-C7H11O6]+), corresponding to the loss of a quinic subunit. Moreover, ESI−/MS2 data generated a base peak at m/z 191.0533 accounting for the loss of a caffeic acid subunit ([M-H-C9H6O3]−). Considering the relative intensity of two secondary ions (m/z 179 and m/z 135) in ESI−/MS2 spectra, compound 8a was identified as 3-O-caffeoylquinic acid (chlorogenic acid) according to published data (Clifford et al. 2003). Using the same reasoning, compound 28a was tentatively identified as 4-O-p-coumaroylquinic acid.
Among the other hydroxycinnamic acids, compound 26a and 38a, detected at RT 7.285 and 11.828 min respectively, showed the same UV characteristic and ESI+/MS data as compound 8a (Table 2), but with a molecular formula of C18H20O10 (M = 396), indicative of an acetylation. ESI−/MS2 allowed to identify both a major product ion at m/z 233, corresponding to the acetylated quinic acid moiety (C9H14O7 = 234), and a major ESI+/MS2 product ion at m/z 163, accounting for the caffeic acid moiety. To our knowledge, the 3-O-acetyl-4-O-caffeoylquinic acid and the 3-O-caffeoyl-5-O-acetylquinic acid were identified using 1H-NMR and ESI/MS data (Whitaker and Stommel 2003), but no literature allow to differentiate these isomers based on ESI/MSn spectra. Thus, these two compounds could be annotated as acetylated caffeoylquinic acid isomers.
Using the same reasoning, compound 34a was annotated as acetylated coumaroylquinic acid.
Annotation of discriminant compounds in stem and leave extracts
The most discriminant and abundant compounds detected in P. vittata stem and leave extracts were identified as flavonol glycosides derivatives, a major class of plant secondary metabolites which have already been well described in P. vittata (cf cited literature in Table 3). Four quercetin derivatives (peak 30b in stems and 28c, 33c, 45c in leaves) as well as five kaempferol derivatives (peak 32b, 33b in stems and 36c, 37c, 49c in leaves) were identified according to their typical MSn fragmentations (providing fragmentation ions at m/z 303 and m/z 287 in positive mode corresponding to respective aglycones) together with their specific UV spectra (i.e., absorption band II lower than 260 nm for quercetin derivatives and higher than this value for kaempferol derivatives; (Mabry et al. 1970)). Among these compounds, two were found in higher proportion in both stems and leaves: 30b and 28c (RT 7.150 min); 33b and 36c (RT 7.980 min).
Compound 30b/28c possess UV characteristic of 3-O-substituted quercetin (Mabry et al. 1970) and showed pseudomolecular ions at m/z 609.1416 ([M−H]−) and at m/z 611.1636 ([M+H]+) corresponding to the molecular formula C27H30O16. The ESI/MS2 spectra show major ion at m/z 303 in positive mode and m/z 301 in negative mode, accounting for a loss of 308 Da corresponding to the loss of a deoxyhexose and a hexose unit. The comparison with literature suggested that the disaccharide moiety is a rutinoside type (Vukics and Guttman 2010), thus this compound was putatively identified as quercetin-3-O-rutinoside (= rutin) (Crupi et al. 2014).
Compound 45c (RT 9.518) showed pseudomolecular ions at m/z 773.1944 ([M+H]+) and at m/z 771.1770 ([M-H]−) allowing to establish the molecular formula of C36H36O19. Its ESI−/MS2 spectra showed product ions at m/z 609 (indicative of the loss of a caffeoyl unit), m/z 609 at very low intensity and m/z 301 (indicating the loss of 308 Da corresponding to a disaccharide of type rutinoside). The ESI+/MS2 product ion at m/z 303 and ESI−/MS2 product ion at m/z 301 reveal the presence of a quercetin aglycone. In addition, the fragment ions at m/z 325 and m/z 163 correspond to a caffeoyl-hexose unit and a caffeoyl unit, respectively. Compound 45c was thus annotated as quercetin-3-O-(caffeoyl)rutinoside. Using the same reasoning and by comparison with literature, compound 49c was tentatively identified as kaempferol-3-O-(caffeoyl)rutinoside (Martucci et al. 2014).
Compound 33c (RT 7.555), the only quercetin derivatives identified in lower proportions in leaves with the influence of metal, showed pseudomolecular ions at m/z 935.2477 [M+H]+ and at m/z 933.2289 [M−H]− with a molecular formula of C42H46O24. The ESI−/MS2 spectra showed product ions at m/z 771 (corresponding to the loss of a hexose unit), m/z 609 (loss of a caffeoyl unit), m/z 463 (indicative of the loss of a rhamnosyl unit), m/z 303 (loss of a hexosyl unit). This led to a putative identification of this compound as quercetin-3-O-(caffeoyl) rutinoside-7-O-hexoside. Using the same reasoning and by comparison with literature concerning its characteristic UV spectrum (Lin and Harnly 2010), compound 37c was tentatively identified as kaepferol-3-O-(caffeoyl) rutinoside-7-O-hexoside.
Compound 33b/36c detected at RT 7.98 min showed pseudomolecular ions at m/z 461.0718 ([M−H]−) and at m/z 463.0893 ([M+H]+), corresponding to the molecular formula of C21H18O12. The ESI−/MS2 spectra showed a base peak at m/z 285 corresponding to the kaempferol aglycone and to the loss of a glucuronide moiety (176 Da). Therefore, following literature, compound 33b/36c was tentatively identified as kaempferol-3-O-glucuronide (Kajdžanoska et al. 2010).
Although the majority of abundant and discriminant compounds in P. vittata stems and leaves were putatively annotated, minor UV-absorbing compounds still remain unidentified, in part due to their difficulty to be ionized.
Bacterial abundance and community structure
Total heterotrophic microflora varied between 7.01 (±5.06) × 103 colonies forming unit (CFU)/gram in the bulk soil of the mining site (sample BSP) and 46.6 (±13.9) × 106 CFU/g in the rhizosphere of P. vittata at the mining site (sample PVP) (Table 4). Based on qPCR quantification, the number of 16S rDNA copies varied from 0.557 (±0.0680) × 109 to 26.8 (±1.3) × 109.
Data from the metataxogenomic analysis (Table 5) clearly showed the very low diversity of the bacterial community from the bulk soil of the mining site (sample BSP). A total of 24,458 sequences were obtained and 11 phyla were detected in that soil with Proteobacteria and Nitrospira being very dominant (68 and 32%, respectively) (Fig. 4). The bacterial communities of the rhizospheres of D. linearis (DLP) and P. vittata (samples PVP and PVC) were more diverse whatever the taxonomic level considered, i.e., phylum, class, genus, or out, highlighting an important plant effect. On the other hand, there were only weak differences in terms of diversity between the non-polluted (PVC) and polluted (PVP) rhizosphere communities of P. vittata. As for the bulk soil, rhizosphere communities were consistently dominated by Proteobacteria. On the opposite, Nitrospira were detected at a much lower level. The other most abundant phyla within the rhizosphere community of P. vittata at the mining site were Acidobacteria (19%), Bacteroidetes (9%), Actinobacteria (6%), and Chloroflexi (5%). At the control site, the dominant phyla were Acidobacteria (14%), Bacteroidetes (12%), Chloroflexi (7%), and Planctomycetes (45%). In the rhizosphere communities of D. linearis (DLP), the five dominant phyla were Proteobacteria (34%), Bacteroidetes (17%), Actinobacteria (10%), Firmicutes (9%), and Chloroflexi (9%). Finally, based on diversity indexes (Table 5) as well as on prevalence of phylum/class/genus/OTU, there were more differences between the rhizosphere communities of the two plants collected at the mining site than between the rhizosphere of P. vittata at the mining site and that at the control site. Indeed, he H′ index of PVP and PVC sample were at similar level (8.24 and 8.58, respectively), whereas that of DLP sample was lower (5.98). The same trend was observed in the numbers of phylum/class/genus/OTUs.
The total sequence reads obtained for these four soils clustered into 10,796 OTUs and 11,911 OTUs, for PVP and PVC, respectively, and 140 OTUs and 4955 OTUs for BSP and DLP, respectively. Among the OTUs identified at a 3% divergence threshold, we looked then for the 50 dominant ones within each soil community. The Acidithiobacillus and Leptospirillum genera were highly dominant in the community of the mine BSP representing 67 and 32% of the dominant taxons, respectively. The comparison between the two rhizosphere communities of P. vittata showed the dominant genera to be Gp2, Gp3, and Gp4 from the Acidobacteria division, TM7 and Rhizomicrobium at the polluted site whereas the dominant ones were Gp4, Gp6, Thiobacter, and Nitrosospira at the control one. Some genera were only detected in the rhizosphere communities of P. vittata such as Bacillus (0.01% in PVP, 0.02% in PVC), Cupriavidus (0.12% in PVP, 0.01% in PVC), and Pseudomonas (0.05% in PVP, 0.34% in PVC). In addition, it has to be noted that several genera known to harbor opportunistic pathogen species were more abundant under metal pollution. For instance, Burkholderia was more prevalent in all polluted soils, PVP, BSP, and DLP than in the control one PVC. Similarly, Ralstonia, Cupriavidus, Acinetobacter, and Mycobacterium (Fig. 4) were more abundant in the rhizosphere community of P. vittata at the polluted site (PVP) than that of the control site.
Discussion
Impact of metal pollution on P. vittata secondary metabolites
Our data demonstrated that metal pollution and more precisely Cu pollution resulted in modifications of secondary metabolite production in P. vittata. While chlorogenic acid derivatives were found to be significantly increased or decreased (depending on acetylation) in roots under heavy metal pollution, procyanidin was found in higher proportions when metals were present. This could be correlated with metal chelating ability of procyanidins which was reported previously (Karamać 2009; Mendoza-Wilson et al. 2016). Catechic tannins may be able to bind metal ions available in polluted soil such as Cu(II) or Pb(II), forming complexes thus allowing metal accumulation in roots. The concentration of Cu in some P. vittata roots from polluted areas are twofold higher than in the corresponding soils, whereas the Cu level in stems and leaves are much lower (data shown in Table S1) suggesting the accumulation of this metal in roots, but not in stems or leaves. These secondary metabolites were also shown to possess radical scavenging and antioxidant activities (Maldonado et al. 2005; Qiang et al. 2015) that could be useful to prevent the oxidative stress induced by metals (Sharma and Dietz 2009). Therefore, the overproduction of procyanidins in P. vittata roots could play a role in the adaptation process of this plant to metal pollution. The exudation of catechic tanin derivatives was also shown to affect bacterial denitrification processes in Fallopia japonica rhizosphere soil (Bardon et al. 2014). On the other hand, antibacterial activities were described for chlorogenic acid derivatives (Marques and Farah 2009; Lou et al. 2011). As only chlorogenic acid was increased in polluted sites and its acetylated derivatives decreased, all this suggests that the metal-induced changes observed in root metabolic composition could have an impact on the structure and functioning of bacterial communities from P. vittata rhizosphere. Whether these changes allow plants to recruit beneficial microbes that may help to adapt to this stress as hypothesized by Thijs et al. (2016) remains to be further investigated, but the fact that Cupriavidus, a bacterial genera known to be able to tolerate very high concentrations of Cu, was found in higher relative proportions in polluted soil could be a part of adaptation process of P. vittata under metal pollution.
In the aerial parts of P. vittata, while di-C-glycosylated flavones and 3-O-glycosylated flavonols (whether caffeoylated or not) were found in higher proportion under metal stress, only caffeoyl flavonol 3,7 diglycosides (quercetin- and kampferol-3-O-(caffeoyl)rutinoside-7-O-hexoside) were found to be decreased. This diminution may be due to the increase use of these compounds by the plant, including roots were some chlorogenic acid derivatives were found in higher proportions. Some of these metabolites (kaempferol and quercetin-3-O-rutinoside (rutin), kaempferol-3-O-glucuronide) were previously detected in P. vittata (Salatino and Prado 1998; Imperato 2000; Singh et al. 2008; Wahid et al. 2016). Moreover, the antioxidant properties of C-glycosyl flavones (Courts and Williamson 2015) and of some 3-O-glycosyl flavonols including rutin, kaempferol 3-O-rutinoside, and kaempferol 3-O-glucuronide were reported (Plumb et al. 1999; Yang et al. 2008; Leong et al. 2008; Ahmed et al. 2016). Besides, the preliminary 2,2-diphenyl-1-picrylhydrazyl (DPPH) scavenging assay showed high antioxidant effect of P. vittata root, stem, and leave extracts (inhibitory percentage twofold higher than ascorbic acid at 100 μg/mL and similar at 500 μg/mL—data not shown). This led to hypothesis that the intensive production of these compounds in P. vittata could be a part of the antioxidant defense mechanism that limits reactive oxygen species (ROS) production and oxidative stress generated by heavy metals. The link between oxidative injuries generated by Cu2+ and variations of metabolic pathways is well documented in literature, and phenolics are mainly involved in cell protection (Dixon and Paiva 1995; Ali et al. 2006).
Overall, these results showed the influence of heavy metal contamination on the secondary metabolites of Pteris vittata. These polyphenols are well known for their properties against oxidative stress and seem to be involved in metal tolerance of P. vittata. But the impact of these changes in metabolism (especially in roots) on the structure of bacterial community profile has also to be considered.
Effects of metal and plant on diversity and composition of soil bacterial communities
The Ha Thuong mining site is particularly contaminated in Cu, with Cu level in bulk soil reaching more than 3000 mg/kg, ten times the accepted limit for industrial soils in Vietnam. Considering bacterial abundance, our data highlighted differences in the total heterotrophic microflora and the numbers of 16S rDNA copies. The most polluted soil (BSP) showed the lowest density in both cultivable bacteria and qPCR counts. This observation agreed with previous reports indicating the decrease of bacterial biomass resulting from metal contamination (Konopka et al. 1999; Kelly et al. 1999; Ekelund et al. 2003; Guo et al. 2009). However, in the rhizosphere samples, as metal contamination levels were moderated (PVP), or low (PVC, DLP), these variations are more related to differences in the amount of organic carbon in soil (mainly originating from the root exudates) than to amount of metals. The higher difference between culturable counts and qPCR ones in the bulk soil compared to rhizosphere soil could be related to a higher amount of uncultivable bacteria in this very acidic and oligotrophic environment.
Regarding bacterial community structure, our results showed a very low diversity and richness index of the community from the bulk soil at the mining site (Table 5). The diversity indexes Np Shannon and Chao and the prevalence of phylum/class/genus/OTU revealed that diversity and richness increased in the presence of plants as expected; however, there were more differences between rhizosphere communities of two plants from the mining site rather than between communities associated to P. vittata at the mining site and at the control one. This could be a consequence of the phytoextraction process, which leads to a strong decrease of Cu and Pb levels in planted soil and were not high enough (i.e., <450 mg/kg Cu, <300 mg/kg Pb) and bioavailable to affect bacterial communities. Wakelin and collaborators (2010) studied the impact of Cu on bacterial communities in a Cu-polluted field site in Hygum, Danemark, using denaturing gel electrophoresis (DGGE) and reported that bacterial diversity increased with Cu gradient but dramatically decreased at total Cu level above 560 mg/kg (Wakelin et al. 2010). Another work at the same site using high-throughput DNA sequencing of the 16S rRNA gene highlighted that the increase of Cu pollution did not impact the OTU richness (Berg et al. 2012). No definitive conclusion can be drawn from our results due to the low number of treated samples but one can suggest that Cu at very high level decreased the bacterial diversity and richness, whereas at lower concentration (below 450 mg/kg), the differences in the diversity of rhizosphere community are explained by plant influence rather than metal pollution.
Regarding bacterial composition, our data revealed that the bacterial community of the mining bulk soil consisted mainly of two genera, Acidothiobacillus (Proteobacteria phylum) and Leptospirillum (Nitrospira phylum). These genera were not detected within the three other communities. These observations are in agreement with the capacity of these genera to be able to thrive in oligotrophic and extremely acidic environments contaminated with heavy metals (Sánchez-Andrea et al. 2011; Zhang et al. 2016). At the phyla level, previous studies showed the relative abundance of Proteobacteria to be negatively affected following the increase of Cu level, for both total community (Berg et al. 2012) and transcriptionally active community (Nunes et al. 2016). However, it was also shown that the modification of bacterial community in the presence of Cu led to the increase of Proteobacteria relative abundance (Turpeinen et al. 2004). The dominance of Proteobacteria was shown to be a characteristic of metal contaminated soil whereas pristine soils are usually dominated by Actinobacteria and Acidobacteria (Sheik et al. 2012). In our study, Nitrospira is the second dominant phylum in polluted bulk soil but its relative abundance was strongly reduced in rhizosphere samples from both plants (Fig. 5). This observation is in accordance with the result published by Nunes et al. (2016) indicating an increasing abundance of transcriptionally active Nitrospira along Cu gradient, suggesting that Nitrospira is a specific phylum dominating extreme Cu-polluted soil.
Our data showed that the OTU composition of the rhizosphere communities of P. vittata differed from that of the mine bulk soil. The dominant genera in the rhizosphere were Gp2, Gp3, Gp4, and Gp6 among Acidobacteria, Rhizomicrobium, Thibacter, Cupriavidus, Bacillus, and Pseudomonas. We also found that P. vittata select for different taxa in different soils. Some of these genera are known to prefer nutrient-limited environments (Hong et al. 2015) and are suggested to be bioindicators of Cu-related pollution in soils, i.e., Gp2, Gp3, Gp4, and Gp6 (Nunes et al. 2016). More interestingly, some genera as Cupriavidus were found at a higher level under metal pollution. Members of Cupriavidus genus are Gram-negative β-proteobacteria that are well known for their ecological role in heavy metal resistance, in particular C. metallidurans CH34 (Scherer and Nies 2009; von Rozycki and Nies 2009), C. necator N-1 (Poehlein et al. 2011), C. taiwanensis (Amadou et al. 2008), and C. gilardii CR3 (Wang et al. 2015). In addition, it has been demonstrated that some Cupriavidus species were also pathogenic such as C. gilardii (Karafin et al. 2010) and C. metallidurans (Langevin et al. 2011). Similarly, other genera known to harbor human opportunistic pathogens species including Ralstonia, Acinetobacter, Burkholderia, and Mycobacterium were also found to be enriched under metal pressure. Unfortunately, the taxonomic resolution achieved with our metagenomic strategy is not high enough to differentiate pathogenic species from non-pathogenic ones. Further studies are then needed to better understand the role of metal and/or plant secondary metabolites on the selection of specific taxa including opportunistic pathogens.
Conclusion
The present study showed a modification of P. vittata secondary metabolites, i.e., enhanced production of polyphenols known for their antioxidant properties, with heavy metal pollution, in particular Cu, and suggested the role of these discriminant metabolites in metal tolerance of this plant. Cu contamination level also altered the diversity and richness of bacterial communities in the mining bulk soil. In parallel, plant metabolism influenced the rhizosphere bacterial communities as some specific genera, of which Cupriavidus, Acinetobacter, Ralstonia, and Mycobacterium were increased in the rhizosphere of P. vittata at the polluted site. More detailed investigations are needed to understand whether the plant recruits beneficial bacteria to support the adaptation process to heavy metal stress, as supposed by Thijs et al., 2016. Similarly, our results suggested further work on the impact of secondary metabolites to soil bacterial communities including opportunistic pathogens.
Abbreviations
- UHPLC-DAD-ESI/QTOF-MS:
-
Ultrahigh-performance liquid chromatography with diode array detection coupled to electrospray ionization and quadrupole time-of-flight mass spectrometry
- UV:
-
Ultraviolet
- ESI:
-
Electrospray ionization
- MSMS:
-
Tandem mass spectrometry
- ESI/MS2 :
-
Electrospray ionization tandem mass spectrometry
- HRMS:
-
High-resolution mass spectrometry
- 1H-NMR:
-
Proton nuclear magnetic resonance
- RT:
-
Retention time
- SPE:
-
Solid phase extraction
- PVP:
-
Soil under P. vittata polluted
- BSP:
-
Bulk soil polluted
- DLP:
-
Soil under Dicranopteris linearis polluted
- PVC:
-
Soil under P. vittata control
- HSD:
-
Honest significant difference
- PCA:
-
Principal component analysis
- ANOVA:
-
Analysis of variance
- CFU:
-
Colony forming unit
- QPCR:
-
Quantitative real-time polymerase chain reaction
- OTU:
-
Operational taxonomic unit
- DGGE:
-
Denaturing gradient gel electrophoresis
- DPPH:
-
2,2-diphenyl-1-picrylhydrazyl
References
Ahmed SI, Hayat MQ, Tahir M et al (2016) Pharmacologically active flavonoids from the anticancer, antioxidant and antimicrobial extracts of Cassia angustifolia Vahl. BMC Complement Altern Med 16:460. doi:10.1186/s12906-016-1443-z
Ali MB, Singh N, Shohael AM et al (2006) Phenolics metabolism and lignin synthesis in root suspension cultures of Panax ginseng in response to copper stress. Plant Sci 171:147–154. doi:10.1016/j.plantsci.2006.03.005
Amadou C, Pascal G, Mangenot S et al (2008) Genome sequence of the β-rhizobium Cupriavidus taiwanensis and comparative genomics of rhizobia. Genome Res 18:1472–1483. doi:10.1101/gr.076448.108
Anh BTK, Kim DD, Tua TV et al (2011) Phytoremediation potential of indigenous plants from Thai Nguyen province, Vietnam. J Environ Biol 32:257–262
An ZZ, Huang ZC, Lei M et al (2006) Zinc tolerance and accumulation in Pteris vittata L. and its potential for phytoremediation of Zn- and As-contaminated soil. Chemosphere 62:796–802. doi:10.1016/j.chemosphere.2005.04.084
Azarbad H, Niklińska M, van Gestel CAM et al (2013) Microbial community structure and functioning along metal pollution gradients. Environ Toxicol Chem 32:1992–2002. doi:10.1002/etc.2269
Bardon C, Piola F, Bellvert F et al (2014) Evidence for biological denitrification inhibition (BDI) by plant secondary metabolites. New Phytol 204:620–630. doi:10.1111/nph.12944
Berg J, Brandt KK, Al-Soud WA et al (2012) Selection for Cu-tolerant bacterial communities with altered composition, but unaltered richness, via long-term Cu exposure. Appl Environ Microbiol 78:7438–7446. doi:10.1128/AEM.01071-12
Clifford MN, Johnston KL, Knight S, Kuhnert N (2003) Hierarchical scheme for LC-MSn identification of chlorogenic acids. J Agric Food Chem 51:2900–2911. doi:10.1021/jf026187q
Courts FL, Williamson G (2015) The occurrence, fate and biological activities of C-glycosyl flavonoids in the human diet. Crit Rev Food Sci Nutr 55:1352–1367. doi:10.1080/10408398.2012.694497
Crupi P, Genghi R, Antonacci D (2014) In-time and in-space tandem mass spectrometry to determine the metabolic profiling of flavonoids in a typical sweet cherry (Prunus avium L.) cultivar from Southern Italy. J Mass Spectrom 49:1025–1034. doi:10.1002/jms.3423
Danh LT, Truong P, Mammucari R, Foster N (2014) A critical review of the arsenic uptake mechanisms and phytoremediation potential of Pteris vittata. Int J Phytoremediation 16:429–453. doi:10.1080/15226514.2013.798613
Dixon RA, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7:1085–1097. doi:10.1105/tpc.7.7.1085
Edgar RC, Haas BJ, Clemente JC et al (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi:10.1093/bioinformatics/btr381
Ekelund F, Olsson S, Johansen A (2003) Changes in the succession and diversity of protozoan and microbial populations in soil spiked with a range of copper concentrations. Soil Biol Biochem 35:1507–1516. doi:10.1016/S0038-0717(03)00249-9
Epelde L, Martín-Sánchez I, González-Oreja JA et al (2012) Impact of sources of environmental degradation on microbial community dynamics in non-polluted and metal-polluted soils. Sci Total Environ 433:264–272. doi:10.1016/j.scitotenv.2012.06.049
Gong X-L, Chen Z-H, Liang N-C (2007) Advances in study on chemical constituents and pharmacological activities of plants of genus Pteris. Chin J Chin Mater Medica 32:1382–1387
Gracelin DHS, Britto AJD, Kumar PBJR (2012) Qualitative and quantitative analysis of phytochamicals in five Pteris species. Int J Pharm Pharm Sci 5:105–107
Gu L, Kelm MA, Hammerstone JF et al (2003) Screening of foods containing proanthocyanidins and their structural characterization using LC-MS/MS and thiolytic degradation. J Agric Food Chem 51:7513–7521. doi:10.1021/jf034815d
Guo Z, Megharaj M, Beer M et al (2009) Heavy metal impact on bacterial biomass based on DNA analyses and uptake by wild plants in the abandoned copper mine soils. Bioresour Technol 100:3831–3836. doi:10.1016/j.biortech.2009.02.043
Hartmann A, Schmid M, Tuinen D van, Berg G (2008) Plant-driven selection of microbes. Plant Soil 321:235–257. doi: 10.1007/s11104-008-9814-y
Hong C, Si Y, Xing Y, Li Y (2015) Illumina MiSeq sequencing investigation on the contrasting soil bacterial community structures in different iron mining areas. Environ Sci Pollut Res Int 22:10788–10799. doi:10.1007/s11356-015-4186-3
Imperato F (2000) Kaempferol and quercetin 3-O-(X″,X″-di-protocatechuoyl)-glucuronides from Pteris vittata. Am Fern J 90:141–144. doi:10.2307/1547491
Jaishankar M, Tseten T, Anbalagan N et al (2014) Toxicity, mechanism and health effects of some heavy metals. Interdiscip Toxicol 7:60–72. doi:10.2478/intox-2014-0009
Jaishee N, Chakraborty U (2015) Comparative assessment of phytochemicals and HPLC analyses of phenolics present in Dicranopteris linearis (N. Burm.) Underw and Pteris vittata L. Int J Pharm Pharm Sci Res 5:1–7
Kachenko AG, Singh B, Bhatia NP (2007) Heavy metal tolerance in common fern species. Aust J Bot 55:63–73. doi:10.1071/BT06063
Kajdžanoska M, Gjamovski V, Stefova M (2010) HPLC-DAD-ESI-MSn identification of phenolic compounds in cultivated strawberries from Macedonia. Maced J Chem Chem Eng 29:181–194
Karafin M, Romagnoli M, Fink DL et al (2010) Fatal infection caused by Cupriavidus gilardii in a child with aplastic anemia. J Clin Microbiol 48:1005–1007. doi:10.1128/JCM.01482-09
Karamać M (2009) Chelation of Cu(II), Zn(II), and Fe(II) by tannin constituents of selected edible nuts. Int J Mol Sci 10:5485–5497. doi:10.3390/ijms10125485
Kelly JJ, Häggblom M, Tate RL III (1999) Changes in soil microbial communities over time resulting from one time application of zinc: a laboratory microcosm study. Soil Biol Biochem 31:1455–1465. doi:10.1016/S0038-0717(99)00059-0
Konopka A, Zakharova T, Bischoff M et al (1999) Microbial biomass and activity in lead-contaminated soil. Appl Environ Microbiol 65:2256–2259
Kozich JJ, Westcott SL, Baxter NT et al (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi:10.1128/AEM.01043-13
Langevin S, Vincelette J, Bekal S, Gaudreau C (2011) First case of invasive human infection caused by Cupriavidus metallidurans. J Clin Microbiol 49:744–745. doi:10.1128/JCM.01947-10
Lenart-Boroń A, Boroń P (2014) The effect of industrial heavy metal pollution on microbial abundance and diversity in soils—a review. In: Hernandez Soriano MC (ed) Environmental risk assessment of soil contamination. InTech. p759–783. doi: 10.5772/57406
Leong CNA, Tako M, Hanashiro I, Tamaki H (2008) Antioxidant flavonoid glycosides from the leaves of Ficus pumila L. Food Chem 109:415–420. doi:10.1016/j.foodchem.2007.12.069
Lin L-Z, Harnly JM (2010) Phenolic component profiles of mustard greens, yu choy, and 15 other brassica vegetables. J Agric Food Chem 58:6850–6857. doi:10.1021/jf1004786
Lou Z, Wang H, Zhu S et al (2011) Antibacterial activity and mechanism of action of chlorogenic acid. J Food Sci 76:M398–M403. doi:10.1111/j.1750-3841.2011.02213.x
Ma LQ, Komar KM, Tu C et al (2001) A fern that hyperaccumulates arsenic. Nature 409:579. doi:10.1038/35054664
Mabry TJ, Markham KR, Thomas MB (1970) The systematic identification of flavonoids. Springer Berlin Heidelberg, Berlin, Heidelberg doi:10.1007/978-3-642-88458-0
Maldonado PD, Rivero-Cruz I, Mata R, Pedraza-Chaverrí J (2005) Antioxidant activity of A-type proanthocyanidins from Geranium niveum (Geraniaceae). J Agric Food Chem 53:1996–2001. doi:10.1021/jf0483725
Marques V, Farah A (2009) Chlorogenic acids and related compounds in medicinal plants and infusions. Food Chem 113:1370–1376. doi:10.1016/j.foodchem.2008.08.086
Martucci MEP, De Vos RCH, Carollo CA, Gobbo-Neto L (2014) Metabolomics as a potential chemotaxonomical tool: application in the genus Vernonia schreb. PLoS One 9:e93149. doi:10.1371/journal.pone.0093149
Mendoza-Wilson AM, Castro-Arredondo SI, Espinosa-Plascencia A et al (2016) Chemical composition and antioxidant-prooxidant potential of a polyphenolic extract and a proanthocyanidin-rich fraction of apple skin. Heliyon 2:e00073. doi:10.1016/j.heliyon.2016.e00073
Michalet S, Rohr J, Warshan D et al (2013) Phytochemical analysis of mature tree root exudates in situ and their role in shaping soil microbial communities in relation to tree N-acquisition strategy. Plant Physiol Biochem 72:169–177. doi:10.1016/j.plaphy.2013.05.003
Nguyen THH, Sakakibara M, Sano S, Mai TN (2011) Uptake of metals and metalloids by plants growing in a lead-zinc mine area, Northern Vietnam. J Hazard Mater 186:1384–1391. doi:10.1016/j.jhazmat.2010.12.020
Nunes I, Jacquiod S, Brejnrod A, et al (2016) Coping with copper: legacy effect of copper on potential activity of soil bacteria following a century of exposure. FEMS Microbiol Ecol 92:fiw175. doi: 10.1093/femsec/fiw175
Paul T, Das B, Apte K, Suchitra (2012) Hypoglycemic activity of Pteris vittata L., a fern on alloxan induced diabetic rats. Inventi Impact Planta active 2:88–91.
Plumb GW, Price KR, Williamson G (1999) Antioxidant properties of flavonol glycosides from green beans. Redox Rep Commun Free Radic Res 4:123–127. doi:10.1179/135100099101534800
Poehlein A, Kusian B, Friedrich B et al (2011) Complete genome sequence of the type strain Cupriavidus necator N-1. J Bacteriol 193:5017. doi:10.1128/JB.05660-11
Qiang L, Luo F, Zhao X et al (2015) Identification of proanthocyanidins from litchi ( Litchi chinensis Sonn .) pulp by LC-ESI-Q-TOF-MS and their antioxidant activity. PLoS One 10:e0120480. doi:10.1371/journal.pone.0120480
Quast C, Pruesse E, Yilmaz P et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:590–596. doi:10.1093/nar/gks1219
R Core Team (2014) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/
Salatino MLF, Prado J (1998) Flavonoid glycosides of Pteridaceae from Brazil. Biochem Syst Ecol 26:761–769. doi:10.1016/S0305-1978(98)00032-5
Sánchez-Andrea I, Rodríguez N, Amils R, Sanz JL (2011) Microbial diversity in anaerobic sediments at Río Tinto, a naturally acidic environment with a high heavy metal content. Appl Environ Microbiol 77:6085–6093. doi:10.1128/AEM.00654-11
Scherer J, Nies DH (2009) CzcP is a novel efflux system contributing to transition metal resistance in Cupriavidus metallidurans CH34. Mol Microbiol 73:601–621. doi:10.1111/j.1365-2958.2009.06792.x
Schloss PD, Westcott SL, Ryabin T et al (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi:10.1128/AEM.01541-09
Sharma SS, Dietz K-J (2009) The relationship between metal toxicity and cellular redox imbalance. Trends Plant Sci 14:43–50. doi:10.1016/j.tplants.2008.10.007
Sheik CS, Mitchell TW, Rizvi FZ et al (2012) Exposure of soil microbial communities to chromium and arsenic alters their diversity and structure. PLoS One 7:e40059. doi:10.1371/journal.pone.0040059
Singh BK, Quince C, Macdonald CA et al (2014) Loss of microbial diversity in soils is coincident with reductions in some specialized functions. Environ Microbiol 16:2408–2420. doi:10.1111/1462-2920.12353
Singh M, Govindarajan R, Rawat AKS, Khare PB (2008) Antimicrobial flavonoid rutin from Pteris vittata L. against pathogenic gastrointestinal microflora. Am Fern J 98:98–103
Singh S, Parihar P, Singh R et al (2016) Heavy metal tolerance in plants: role of transcriptomics, proteomics, metabolomics, and ionomics. Front Plant Sci 6:1143. doi:10.3389/fpls.2015.01143
Thijs S, Sillen W, Rineau F et al (2016) Towards an enhanced understanding of plant–microbiome interactions to improve phytoremediation: engineering the metaorganism. Front Microbiol doi. doi:10.3389/fmicb.2016.00341
Tu C, Ma LQ (2002) Effects of arsenic concentrations and forms on arsenic uptake by the hyperaccumulator ladder brake. J Environ Qual 31:641–647
Tu C, Ma LQ, Bondada B (2002) Arsenic accumulation in the hyperaccumulator Chinese brake and its utilization potential for phytoremediation. J Environ Qual 31:1671–1675
Turpeinen R, Kairesalo T, Häggblom MM (2004) Microbial community structure and activity in arsenic-, chromium- and copper-contaminated soils. FEMS Microbiol Ecol 47:39–50. doi:10.1016/S0168-6496(03)00232-0
von Rozycki T, Nies DH (2009) Cupriavidus metallidurans: evolution of a metal-resistant bacterium. Antonie Van Leeuwenhoek 96:115–139. doi:10.1007/s10482-008-9284-5
Vukics V, Guttman A (2010) Structural characterization of flavonoid glycosides by multi-stage mass spectrometry. Mass Spectrom Rev 29:1–16. doi:10.1002/mas.20212
Wahid F, Khan T, Shehzad O et al (2016) Phytochemical analysis and effects of Pteris vittata extract on visual processes. J Nat Med 70:8–17. doi:10.1007/s11418-015-0930-8
Wakelin SA, Chu G, Lardner R et al (2010) A single application of Cu to field soil has long-term effects on bacterial community structure, diversity, and soil processes. Pedobiologia 53:149–158. doi:10.1016/j.pedobi.2009.09.002
Wang J, Zhao F-J, Meharg AA et al (2002) Mechanisms of arsenic hyperaccumulation in Pteris vittata. Uptake kinetics, interactions with phosphate, and arsenic speciation. Plant Physiol 130:1552–1561. doi:10.1104/pp.008185
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi:10.1128/AEM.00062-07
Wang X, Chen M, Xiao J et al (2015) Genome sequence analysis of the naphthenic acid degrading and metal resistant bacterium Cupriavidus gilardii CR3. PLoS One 10:e0132881. doi:10.1371/journal.pone.0132881
Whitaker BD, Stommel JR (2003) Distribution of hydroxycinnamic acid conjugates in fruit of commercial eggplant (Solanum melongena L.) cultivars. J Agric Food Chem 51:3448–3454. doi:10.1021/jf026250b
Yang J, Guo J, Yuan J (2008) In vitro antioxidant properties of rutin. LWT - Food Sci Technol 41:1060–1066. doi:10.1016/j.lwt.2007.06.010
Zhang X, Niu J, Liang Y et al (2016) Metagenome-scale analysis yields insights into the structure and function of microbial communities in a copper bioleaching heap. BMC Genet 17:21. doi:10.1186/s12863-016-0330-4
Acknowledgments
Hoang Nam Pham wish to gratefully thank the Vietnam Ministry of Education and Training, the University of Sciences and Technologies of Hanoi, and team “Environmental resistance and bacterial efflux,” UMR 5557 CNRS Microbial Ecology, for the financial support. We thank the platforms CESN (Centre d’Etudes des Substances Naturelles) and PARMIC (Plateau d’Analyse du Risque MICrobiologique, UMR Ecologie Microbienne, Université Lyon1) for the equipment facilities, and Institut of Marine Biochemistry (Vietnam Academy of Science and Technology, Hanoi) is also acknowledged for providing valuable assessment during the development of this subject.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Philippe Garrigues
Electronic supplementary material
Fig. S1
Typical UHPLC/DAD chromatogram at 280 nm of P. vittata root extracts. PVP: From plants grown on contaminated soil (Ha Thuong); PVC: From plants grown on non-polluted soil (USTH) (DOCX 157 kb).
Fig. S2
Typical UHPLC/DAD chromatogram at 280 nm of P. vittata stems extracts. PVP: From plants grown on contaminated soil (Ha Thuong); PVC: From plants grown on non-polluted soil (USTH) (DOCX 182 kb).
Fig. S3
Typical UHPLC/DAD chromatogram at 280 nm of P. vittata leaves extracts. PVP: From plants grown on contaminated soil (Ha Thuong); PVC: From plants grown on non-polluted soil (USTH) (DOCX 185 kb).
Table S1
(DOCX 11 kb).
Rights and permissions
About this article
Cite this article
Pham, H.N., Michalet, S., Bodillis, J. et al. Impact of metal stress on the production of secondary metabolites in Pteris vittata L. and associated rhizosphere bacterial communities. Environ Sci Pollut Res 24, 16735–16750 (2017). https://doi.org/10.1007/s11356-017-9167-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-017-9167-2