Abstract
Nonylphenol polyethoxylates (NPEOs), although banned for decades, are still widely used in manufactories and thus affect human lives. In this study, a highly efficient NPEO-degrading bacterium, Sphingomonas sp. Y2, was isolated from sewage sludge by enrichment culture. Strain Y2 ensured the complete removal of NPEO in 48 h and degraded 99.2 % NPEO (1,000 mg L−1) within 30 h at a specific growth rate of 0.73 h−1 in minimum salt medium. To date, this degradation efficiency is the highest reported for NPEO metabolism by a pure bacterium under this condition. Furthermore, the application of this bacterium to wastewater treatment demonstrated that it metabolized 98.5 % NPEO (1,000 mg L−1) within 5 days with a specific growth rate of 2.03 day−1. The degradation intermediates, identified as nonylphenol, short-chain NPEOs and short-chain nonylphenol polyethoxycarboxylates by high-performance liquid chromatography and gas chromatography-mass spectrometry, indicated the sequential exo-cleavage of the EO chain. Additionally, the enzymes involved in the biodegradation were inducible rather than constitutive. Considering that strain Y2 exhibits prominent biodegradation advantages in industrial wastewater treatment, it might serve as a promising potential candidate for in situ bioremediation of contamination by NPEOs and other structurally similar compounds.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Surfactants are a large class of surface-active compounds widely used in industry, which adversely affect human health. Alkylphenol ethoxylates (APEs) are a major group of nonionic surfactants widely used in the production of industrial and domestic detergents, among which the C8- and C9-APEs are the most prevalent. Nonylphenol polyethoxylates (NPEOs) constitute approximately 80 % of total APEs (Dokianakis et al. 2006); their low biodegradability, estrogenic and mutagenic activities, and degradation products have received increasing attention. Many applications of NPEOs have been banned by the European Commission because of the environmental and health problems caused by these surfactants and their corresponding breakdown products (Frassinetti et al. 2011). However, despite the pollution caused by NPEOs (Brooke and Thursby 2005; Hayashi et al. 2005; Karahan et al. 2010; Montgomery-Brown et al. 2003), these compounds are still utilized because they cannot be completely substituted by alternative chemicals owing to technical and economic reasons. Furthermore, commercial NPEOs are complex mixtures of isomers and oligomers (Lu et al. 2008), which increases the complexity of the degradation process and the difficulty in elucidating the underlying theoretical mechanisms.
To date, there have been no detailed reports regarding the mineralization of NPEOs. It is generally accepted that nonylphenol (NP) and short-chain NPEOs are the main biodegradation products of NPEOs under anaerobic conditions without the formation of short-chain nonylphenol polyethoxycarboxylates (NPECs) (Chang et al. 2004; Lu et al. 2007, 2008; Paterakis et al. 2012). The ethoxylate (EO) chain-shortening aerobic pathway consists of exo-cleavage of the EO chain by hydroxyl shift or oxidation to produce short-chain NPEOs and NPECs by releasing acetaldehyde or glyoxylic acid at each step, respectively (Gu et al. 2010; John and White 1998; Lu et al. 2008; Montgomery-Brown et al. 2003). Furthermore, NPEOs might also be transformed to the corresponding NPECs by direct oxidation of the EO chain with neither EO chain cleavage nor short-chain product formation (Di Corcia et al. 1998; Gu et al. 2010; Hayashi et al. 2005; Jonkers et al. 2001). Additionally, Franska et al. (2003) noted that alkylphenol (Triton X-100) was degraded by central fission to produce neutral, mono-, and di-carboxylated polyethylene glycols (PEGs).
Surfactant-laden wastewaters generally have high chemical oxygen demand (COD) levels owing to the high levels of insoluble and soluble organic materials and surfactants contained therein. Before being introduced into the environment, the COD must be reduced to acceptable levels. Recently, biological methods of COD reduction have attracted more attention and their application in practice is being attempted. Bioaugmentation remains an economically and ecologically friendly methodology for the in situ bioremediation of contaminated sites (Mrozik and Piotrowska-Seget 2010). Furthermore, certain applications of disposing xenobiotics have been well documented (Gratia et al. 2009; Kristanti et al. 2014; Łebkowska et al. 2011; Manu and Chaudhari 2002; Nopcharoenkul et al. 2011; Wang et al. 2014a, b). It follows that the application of efficient microorganisms to decontaminate NPEOs and other similar pollutants in wastewater appears to be both essential and promising.
The acclimatization of NPEOs-degrading organisms and studies of their degradation characteristics have been frequently reported. Different genera such as Pseudomonas (John and White 1998; Tasaki et al. 2006); Sphingomonas, and Cupriavidus (Gu et al. 2010) are capable of metabolizing NPEOs. However, to our knowledge, their practical utilization for NPEOs bioremediation in wastewater in subsequent trials has not yet been documented. This might be desirable, as application of a judicious consortium of growing cells with effective biodegradation ability might ensure more effective removal of contaminants from wastewater in situ. In this study, we isolated and acclimated the microorganism Sphingomonas sp. Y2, which is capable of effectively degrading NPEO and certain other structurally related compounds; furthermore, this strain exhibited remarkable adaptation ability and remediation efficiency for practical use in industrial wastewater treatment. We proposed the mechanism for the degradation of NPEO by this bacterium based on subsequent metabolite detection, which was important to assess the environmental risk and might provide a theoretical basis for practical application of Y2 or its degradation process in wastewater management.
Materials and methods
Reagents and chemicals
Commercially available NPEOs with an average EO chain length of 9 (CAS no. 9016-45-9, ∼10 %) was purchased from Aladdin Industrial Corporation (Shanghai, China) for bacterial biodegradation testing. All organic solvents were high-performance liquid chromatography (HPLC) grade. The ultrapure water used in this study was filtered using a 0.22-μm filter membrane if necessary. All general compounds were of analytical grade.
The wastewater used in this research was sampled from the wastewater treatment plant (120.9E, 30.2 N) in Shaoxing, Zhejiang Province, China, which receives multiple types of wastewater from all of the industries and living quarters in the city. Wastewater was collected in plastic bottles from the influent wastewater of the sewage and stored at 4 °C.
Screening and identification of the degrading bacterium
The NPEO-degrading bacterium was isolated from activated sludge by serial dilution. The liquid minimum salt medium (MSM, pH 7.2) contained (per L): 7.04 g Na2HPO4.12H2O, 1.00 g KH2PO4, 0.50 g (NH4)2SO4, 0.10 g MgCl2.6H2O, 0.05 g Ca(NO3)2.4H2O, and 1 mL trace element solution (Schmidt et al. 1992). If needed, 2.0 % agar was added to the medium. Cells were grown in 200 mL liquid medium in 500-mL Erlenmeyer flasks at 30 °C in a rotary shaker at 200 rpm. An aliquot of the enrichment was transferred to fresh medium each week for approximately two months. The enrichment was then diluted and spreaded onto solid plates with 1,000 mg L−1 NPEO as the sole carbon source.
One yellow-pigmented colony, Y2, was isolated and then further identified by amplifying the 16S rDNA gene using the universal bacterial primers: 27F: 5′-AGA GTT TGA TCA TGG CTC AG-3′ and 1492R: 5′-TAC GGT TAC CTG TTA CGA CTT-3′. The PCR products were subjected to electrophoresis on 0.8 % agarose gels and then sequenced by Sangon Biotech Co. Ltd. (Shanghai, China). The obtained 16S rDNA gene sequence was blasted against other previously reported microorganisms capable of degrading non-ionic surfactants contained in the National Center for Biotechnological Information database. Complete sequence alignment was carried out using CLUSTAL X 2.0 software and then a phylogenetic tree was constructed using the neighbor-joining method by MEGA 6.0 software (Tamura et al. 2013).
The Y2 isolate was cultured on 0.5× Luria-Bertani broth (0.5 × LB) (2.5 g L−1 yeast extract, 5 g L−1 peptone, and 5 g L−1 NaCl) agar to observe its morphological characteristics and further monitored under a transmission electron microscope. The assimilation and enzyme tests were performed using API 20NE and API ZYM systems (BioMérieux, Marcy-l’Étoile, France) according to the manufacturers’ instructions. BIOLOG GN2 aerobic microplates (Biolog, Inc., Hayward, CA) were used to test the carbon source utilization pattern. Catalase and oxidase activities were determined by 5.0 % hydrogen peroxide and 10.0 % N, N-dimethyl-p-phenylenediamine, respectively.
NPEO biodegradation in MSM
A carbon source is used to maintain cell growth, form cell structures, and produce metabolites. Different concentrations of NPEO (100, 1,000, and 10,000 mg L−1) were added to a series of 500-mL flasks containing 200 mL MSM. 20-mL samples were withdrawn under aseptic condition to measure cell growth and residual NPEO concentrations every 12-h interval. Growth curves under different substrate concentrations were evaluated by optical density (OD) at λ = 600 nm using a 7230G spectrophotometer (MAPADA, Shanghai, China), and the specific growth rate was calculated by ORIGIN 9.0 software using a Gompertz model (Winsor 1932; Zwietering et al. 1990). Flasks were incubated in the shaker at 200 rpm and 30 °C.
The bacterial strain, incubated in 0.5 × LB liquid medium until late exponential phase, was used as the inoculum. The precipitates were resuspended after having been washed twice with sterile water, of which 0.2 mL was inoculated into 100-mL Erlenmeyer flasks containing 20 mL MSM with 1,000 mg L−1 NPEO as the sole carbon source. Samples were withdrawn every 6 h to monitor the bacterial biomass and NPEO consumption in triplicate sets. Control flasks without inocula were cultured under the same conditions.
Biodegradation of NPEO in the wastewater medium
We carried out additional work to exploit the potential for NPEO biodegradation in industrial wastewater by Sphingomonas sp. Y2. The wastewater in this experiment contained visible suspended substances. The main physical and chemical characteristics were analyzed and listed in Table 1. The COD and metal ions were measured by a COD monitoring instrument (HACH, USA) and inductively coupled plasma optical emission spectrometry (Optima 8000DV, Perkin-Elmer, USA), respectively. The total nitrogen (TN) and total phosphorus (TP) were determined by a flow analyzer (San plus system, SKALAR, Breda, The Netherlands). After natural precipitation without any further disposal, the wastewater was sterilized under 115 °C for 30 min to avoid the Maillard reaction and other interactions between different substances and ions (Zhang et al. 2012), which constituted the wastewater medium (WWM). Sustainable treatment of NPEO-contaminated wastewater by strain Y2 was evaluated at the flask-scale. Cultures were grown in 100-mL Erlenmeyer flasks containing 20 mL WWM with dissolved 1,000 mg L−1 NPEO. Flasks without inocula were used to examine its bioavailability by strain Y2. Cell growth and biodegradation analysis were performed every day. The change curve of COD was also monitored at specific times.
Analysis procedures
For analysis the NPEO and the corresponding metabolites concentrations in MSM, cultures were firstly extracted by isovolumetric trichloromethane in a JY92 IIN ultrasonic cleaner (SCENTZ, Ningbo, China) for 30 min, and then organic phase was separated with water phase by centrifugation at 10,000×g by 5 min. In the next step, the aqueous phase was acidified to pH 2.0 by 2 M HCl before being subjected to an additional extraction. The organic phases were subsequently collected together, dried over anhydrous Na2SO4, and evaporated under reduced pressure. Remaining residues were re-dissolved in 5 mL methanol and then the methanolic samples were filtered (0.22 μm) for HPLC and gas chromatography-mass spectrometry (GC-MS). As for in WWM, sample preparation was performed as mentioned above except for the third time extraction conducted by the same volume of ethyl acetate. All treatment processes were performed in triplicate for accuracy.
During the experiment, the concentration of NPEO was determined by HPLC using an Eclipse C18 column (250 × 4.6 mm × 5 μm; Agilent Technologies, Santa Clara, CA, USA) and a UV-vis detector. Reverse phase HPLC was performed as follows: the isocratic elution solution was comprised of methanol and water at the volume ratio of 95:5 with a 1-mL min−1 flow rate. An aliquot of 20 μL of the sample was injected automatically and detected at 225-nm wavelength at 30 °C.
The degradation products were determined with the help of a gas chromatograph interfaced with a DB-5 fused-silica capillary column and a mass spectrometer. Samples were derivatized with N,O-bis (trimethylsilyl) trifluoroacetamide and trimethylchlorosilane, and then subjected to GC-MS analysis as previously described (Lu et al. 2007) with slight modification. A 10-μL sample was injected into the injector in the splitless mode whose temperature was set to 280 °C. The GC column temperature was programmed to increase from 80 to 280 °C via a ramp of 10 °C, after which it was maintained at 280 °C for 5 min. The helium carrier gas was maintained at a constant flow rate of 1 mL min−1. A 40–600 m/z mass range was recorded. Compounds were identified by comparison with authentic substances and previously published results.
Enzyme assays
Sphingomonas sp. Y2 was grown in 0.5 × LB and MSM until the early stationary phase. Culture was centrifuged at 7,000×g for 10 min and washed twice with cold 0.1 M Tris-HCl buffer (pH 7.2); then approximately 1.0 g cells were resuspended in 15 mL buffer. The cells were disrupted by ultrasonication at 300 W under 0 °C (6 s on and 9 s off for 30 times). After centrifugation at 10,000×g for 30 min, the enzyme activity was estimated by the concentration decrease of NPEO. Reactions were performed in a 10-mL flask system, which contained the appropriate amount of cell-free extract and 1,000 mg L−1 NPEO in 0.1 M Tris-HCl buffer (pH 7.2). The enzymatic reaction mixture was incubated in a water bath shaker at 60 rpm at 30 °C. Flasks were withdrawn at 0, 6, and 12 h, and then immediately subjected to extraction. A reaction mixture without enzyme was set as the blank control. Statistical analysis was performed to evaluate the analysis of variance (ANOVA) using SPSS software (Chaîneau et al. 2005).
Once crude enzyme obtained by Y2 grown in MSM as described above, an initial purification of combination of ammonium sulfate graded precipitation and dialysis was performed. Enzyme of treatment groups transformed NPEO in the shortest time of period was further measured its dehydrogenase activity as Liu et al. (2006) mentioned. The 2-mL reaction mixture included an appropriate amount of enzyme, 0.1 mM phenazine methosulfate, 0.1 mM 3-(4,5-dimethylthiazolyl-2)- 2,5-diphenyltetrazolium bromide (MTT), 5 mM NPEOs in 0.1 M Tris-HCl buffer (pH 7.2) with and without 0.1 M nicotinamide adenine dinucleotide (NAD) or 0.1 M flavin adenine dinucleotide (FAD). A reaction mixture without substrate was used as the control test. The dehydrogenase activity was measured by the increase in the absorbance at 570 nm of MTT.
Results and discussion
Isolation and identification of an NPEO-degrading strain
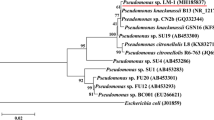
In this study, with NPEO as the sole carbon and energy source, a bacterium capable of degrading NPEO at high efficiency was isolated from sewage sludge after continuous enrichment cultivation. It was found to be phylogenetically closely related to Sphingomonas sp. LB126 by 16S rDNA gene sequencing and BLAST homology search (Fig. 1); and the degrading bacterium was designed as Sphingomonas sp. Y2. The accession number in the NCBI database is KT957299. The characteristics of Y2 were as follows: Gram-negative and rod-shaped (1.0–2.5 × 0.4–0.8 μm) with lateral flagella. This aerobic microorganism was catalase positive, oxidase negative, protease negative, β-glucosidase positive, and nitrogenase negative. Furthermore, it was capable of utilizing more than 30 kinds of carbon sources as determined by a BIOLOG GN2 system test including α-cyclodextrin, bromosuccinic acid, and proline. One of the most extraordinary features possessed by members of Sphingomonas is the ability to degrade environmental refractory pollutants. Strain Y2 was found to be efficient in degrading medium-length ethoxylates and some phenolic substances, while being unable to effectively degrade long-length ethoxylates, monohydric alcohols, and alkanes (Table 2). Thus, it appeared as if the underlying process used for degrading the EO chain by strain Y2 had no relationship with utilization of the linked bulky hydrophobic groups as reported for Pseudomonas sp. TR01 (Maki et al. 1994).
Phylogenetic distributions of strain Y-2 and other relative bacteria species based on the 16S rDNA gene sequences. The phylogenetic tree was constructed using neighbor-joining algorithm with the kimura-2-parameter model in the MEGA 6.0 software. Bootstrap values (percentages of 1,000 replications) are shown at the branch points. The scale bar represents evolutionary distance (2 substitutions per 100 nucleotides)
Growth and biodegradation of NPEO in MSM
In Fig. 2a, a positive correlation can be observed between growth rate of Y2 and the concentration of NPEO. It was suggested by the results that a significant growth could be obtained with NPEO as the sole carbon source at initial concentrations of 1,000 and 10,000 mg L−1. Strain Y2 effected removal of these substrates to approximately 60–100 % of the original levels within 48 h. When the initial concentration of NPEO was 100 mg L−1, the cells kept growing albeit slowly owing to deficiency of a sufficient carbon source. Furthermore, it seemed that high concentrations of NPEO had little effect on the growth of strain Y2, since it could degrade 20,000 mg L−1 NPEO (data not shown).
a Growth curve and degradation kinetics of NPEO under different concentrations. The filled symbols: the bacterial growth at three tested concentrations; the open symbols: the NPEO degradation profile. b Degradation kinetics of NPEO by strain Y2 and cell growth curve in MSM with the initial concentration of 1,000 mg L−1. Flasks incubated with Sphingomonas sp. Y2 (inverted triangle) were compared to controls without bacterium (triangle). Growth of Y2 was determined by OD600 (square). Data are expressed as the means ± SD of three independent experiments
The growth curve of strain Y2 in the presence of 1,000 mg L−1 NPEO as the sole carbon source is presented in Fig. 2b. The level of reduction of NPEO was on a reverse parallel with the OD600, which reflects the cell density. After a short lag phase, the Y2 strain grew rapidly and then metabolized 68.3 % NPEO from 12–18 h. The substrate concentration decreased sharply during the plateau phase, and was barely detectable in the stable phase. Approximately 99.2 % substrates were removed after 30 h, and the biodegradation efficiency reached 99.3 % at 36 h. The pollutant was completely removed by Y2 in 48 h. The significant decrease in the NPEO degradation rate during the later stage might be caused by substrate exhaustion or an accumulation of short-chain intermediates, which might be expected to affect the bacterial growth (Frassinetti et al. 2011). The level of NPEO in the control flasks remained constant throughout the experiment, suggesting negligible volatilization and abiotic degradation. The specific growth rate in MSM was 0.73 h−1, which was much higher than the highest specific growth rate (0.47 h−1) reported from the biodegradation of Triton X-100 by another isolated bacterial community (Wyrwas et al. 2013), and the biodegradation efficiency by Y2 was more notable as well (99.2 % in 30 h vs. approximately 90.0 % after 32 h). Notably, compared to Sphingomonas sp. NP42a (Gu et al. 2010), which is capable of degrading >95.0 % 200 mg L−1 NPEO within 2 days, Y2 could dispose 100.0 % 1,000 mg L−1 NPEO within 2 days. The performance of strain Y2 in 1,000 mg L−1 NPEO was also better than Pseudomonas sp. TX1 (Chen et al. 2006), which could degrade 1,000 mg L−1 OPEO. The extraction recovery rate of the NPEO was 99.4 ± 1.7 % and the detection limit using a UV-Vis detector was 1.00 mg L−1.
NPEO biodegradation in WWM
Synthetic wastewater or treated sewage has often been used for in situ restoration research, whereas the choice of influent raw wastewater as an experimental subject has been rare. Slightly disproportionate biodegradability behaviors of the different types of wastewaters (synthetic vs. domestic) might exist despite the same COD/TN/TP ratios (Bracklow et al. 2007). Therefore, synthetic wastewater cannot be entirely returned to its original condition. Furthermore, the wastewater in this study contained relatively high COD and low nitrogen and phosphorus; therefore, the average COD/TN/TP ratio (Table 1) was inferior compared with the optimum ratio of 100:5:1 that was necessary for efficient bacterial growth (Yang et al. 2009). The presence of even a small amount of surfactant in wastewater is highly visible because they induce foaming and subsequently lower the oxygen concentration in water, thus exerting remarkably large effects on aerobic processes (Yang et al. 2009). In situ bioremediation with microorganisms capable of metabolizing organic pollutants is a promising method for destroying these unwanted chemicals. However, successful bioaugmentation relies on various biotic and abiotic factors, among which the availability of the specialized consortia is the most significant (Ma et al. 2009).
Bacteria affiliated with α- and β-proteobacteria are dominant in NPEOs-amended reactors, suggesting that these types of bacteria play specific roles in the degradation of NPEOs (Lozada et al. 2006). Accordingly, in this research, we elevated the biodegradation of NPEO in industrial wastewater on a laboratory scale. The COD in WWM declined to less than 30.0 % after 5 day degradation by Y2, and in the first 3 days, the COD decreased sharply (Fig. 3c), emphasizing the potential of wastewater treatment with Sphingomonas sp. Y2. In Fig. 3a, strain Y2 can be seen to have degraded 74.0 % NPEO during the plateau phase, and metabolized 98.5 % NPEO in 5 with a 2.03 day−1 specific growth rate in WWM. The OD600 slightly decreased in the last 2 days, which might be due to deteriorating circumstances for growth. In contrast, little growth of Y2 occurred in WWM without NPEO even prolonged culturing time (Fig. 3b). Though a longer lag phase and relatively more time were needed to catabolize NPEO, Y2 removed 93.8 % NPEO at the end of the second day. With generally similar tendencies, strain Y2 removed 100.0 and 98.5 % 1,000 mg L−1 NPEO in MSM and WWM within in 48 h and 5 days, respectively. These delays were speculated to be due to either a requirement of more time for the induction of specific metabolic enzymes or minimal nutrients in WWM or the poor conditions arising from the presence of potential hazardous substances, as the complex compositions in natural wastewater might affect the biodegradation process. However, the substantial degradation efficiency of strain Y2 in WWM (the recovery of NPEO in WWM was tested as 82.1 ± 6.7 %) indicated that this strain serves as a candidate for environmental in situ restoration. This is the first study reporting the remediation of NPEOs pollution in natural wastewater by a single strain.
a Degradation kinetics of NPEO by strain Y2 and cell growth curve in WWM. 1,000 mg L−1 substrate was degraded by Sphingomonas sp. Y2 in 5 days (triangle) and the growth curve was determined by OD600 (square). The control flasks without inocula were incubated under the same condition (inverted triangle). b Cell growth curve (square) of strain Y2 in WWM without NPEO as the sole carbon source. c The variation curve of COD (square) with NPEO degradation in WWM. Error bars indicate standard deviation among three replicates
Wastewater treatment plants play an important role in the disposal of refractory pollutants before their discharge into the aquatic environment. Biological treatments, particularly the activated sludge process, have been widely used for the removal of organic compounds from wastewater; however, their performance depends on various parameters (Shokrollahzadeh et al. 2008). Compared to the activated sludge reported by González et al (2007), strain Y2 decomposed as much as 98.5 % NPEOs in less time. Thus, the results indicated that Sphingomonas sp. Y2 was successful as an inoculant to improve the in situ remediation of NPEOs and other structurally similar pollutants. Furthermore, the mineralization of NPEOs by a combination of Y2 and other microorganisms capable of metabolizing the intermediates produced by Y2 appear to be an exceptional method, as organisms that degrade short-chain NPEOs have been described in multiple reports (Liu et al. 2006), NPECs (Montgomery-Brown et al. 2008; Zhang et al. 2007), and NPs (Wang et al. 2014a, b). Alternatively, a subsequent chemical treatment step could also be integrated for the removal of NPEOs in industrial wastewater.
Identification and analysis of biodegradation intermediates
Complete mass balancing during biodegradation studies is difficult, and the ultimate fate of APEs and their metabolites has not been adequately understood (Montgomery-Brown et al. 2003). Here, the degradation products were extracted and analyzed by HPLC and GC-MS. We found that NPEO decreased (Fig. 4a) and two new peaks appeared on HPLC with increased cultivation time (Fig. 4b). The relative retention time of NPEO, product 1, and product 2 were approximately 3.6, 2.1, and 4.4 min, respectively. Therefore, it was suitable to detect and analyze the biodegradation of NPEOs. The reduction of the sum of substrate and intermediates indicated an incomplete mass balance (data not shown), which was probably due to the poor extraction efficiency of the metabolites in cells or absorbed on flasks or of other undetected substances.
Reliable identification of the low molecular weight breakdown products was conducted by GC-MS. As shown in Fig. 5a, total ion chromatography (TIC) identified many peaks for each compound, which each consisted of many isomers (Lee et al. 1997). The mass spectrograms are listed in Fig. 5b. NP, NP1EO, NP2EO, NP3EO, NP1EC, and NP2EC were identified as the main biodegradation intermediates, whereas PEGs were absent. The mass spectrograms of NP, NP1EO, NP1EC, and NP2EC were similar to those of the authentic compounds and as previously reported (Liu et al. 2006). Furthermore, NP2EO and NP3EO were compared with the authentic compounds. Short-chain NPEOs constituted about 90.0 % of the metabolites at 48 h (data not shown). The rapid decrease of NPEO and the increase of its intermediates suggested that EO chains were rapidly removed by Sphingomonas sp. Y2. Product 2, detected by preparative HPLC and GC-MS, was suggested to consist of short-chain NPEOs, 81.8 % of which tended to distribute in the precipitate, whereas product 1 was thought to represent short-chain NPECs, 76.1 % of which were dispersed in the supernatant. These distributions might be explained by the hydrophilicity of the NPEC carboxyl groups. Additionally, we monitored the formation of metabolites every 2 h for 24 h, which determined that intermediate product 1 accumulated subsequent to product 2 (data not shown). Hence, it was speculated that the short-chain NPEOs were further oxidized to the corresponding carboxylate acids to a slight degree (Fig. 6), which was similar to the hydroxyl shift cleavage followed by the terminal oxidation of EO chains reported by Gu et al. (2010).
In general, carboxylated intermediates are the main products generated under aerobic conditions whereas NPs and short-chain NPEOs are the ultimate products generated during anaerobic conditions (Lu et al. 2007). However, an inconsistent phenomenon was observed for strain Y2, which accumulated NP, short-chain NPEOs, and NPECs by step removal of either unit. Notably, Y2 showed a relatively high evolutionary similarity with the NPEO-degrading Sphingmonas sp. NP42a (98 %) (Gu et al. 2010), but they were found to contain different metabolic pathways, which might be explained by the metabolic versatility of Sphingomonas. S. sp. NP42a primarily produced NP1EO and NP2EO without NPECs. It was generally acknowledged that acetaldehyde and glyoxylic acid formation would occur during NPEOs degradation according to the non-oxidative hydroxyl shift model and the oxidative model, respectively (Gu et al. 2010). However, this anticipated release of acetaldehyde or glyoxylic acid was not observed during the degradation period in this study, even by the resting cells and the cell extractions (data not shown).
Biodegradation enzyme inductivity and alcohol dehydrogenase analysis
Little activity was found with cell-free crude supernatant prepared from cells grown in 0.5 × LB medium, suggesting that the enzyme required for metabolizing NPEO was inductively expressed. The medium had a significant impact on the decrease of NPEO (p < 0.05), whereas different incubation time did not (p > 0.05). The results were consistent with those of previous studies (Chen et al. 2006; Liu et al. 2006), demonstrating that secondary metabolism needed to be modulated in order for cells to exhibit the best possible biodegradation efficiencies.
Alcohol dehydrogenase (ADH) has been supposed to be involved in NPEO removal on the basis of short-chain NPEC formation, as it could oxidize the terminal hydroxyl to a carboxyl group. ADH1 oxidation of OPEO in Pseudomonas putida S-5 (Tasaki et al. 2006) and EO chain nonylphenol dehydrogenase (NPEO-DH) from Ensifer sp. AS08 (Liu et al. 2013) have been validated by heterogeneous expression. In this study, an alternative approach was adopted because of the formation of inclusion bodies in E. coli. The enzyme activity increased twofold by further addition of NAD or FAD compared to that without the cofactors. These results suggested that NPECs were formed by strain Y2 attacking the terminal alcohol group of the EO chain by inducible ADH. It is being carried out to construct mutant libraries to explore the corresponding NPEO-degrading gene clusters.
Conclusion
In this study, an efficient NPEO-degrading bacterium was isolated from sewage sludge by enrichment cultivation. Sphingomonas sp. Y2 could metabolize 1,000 mg L−1 NPEO in both MSM and WWM up to 100.0 % and 98.5 % within 48 h and 5 days, respectively. This is the first trial assay of isolated bacteria to examine biodegradation in industrial wastewater. In addition, this bacterium was shown to metabolize not only NPEO and other structurally similar compounds but also some phenolics. Furthermore, Y2 was suggested to degrade NPEO by an exo-cleavage of EO units with inducible metabolizing enzymes, generating NP, short-chain NPEOs, and short-chain NPECs as the ultimate products. Therefore, Sphingomonas sp. Y2 represents a potential tool for the treatment of wastewater contaminated by NPEOs and other similar pollutants in situ.
References
Bracklow U, Drews A, Vocks M, Kraume M (2007) Comparison of nutrients degradation in small scale membrane bioreactors fed with synthetic/domestic wastewater. J Hazard Mater 144:620–626
Brooke L, Thursby G (2005) Ambient aquatic life water quality criteria for nonylphenol. Report for the United States EPA, Office of Water, Office of Science and Technology, Washington DC
Chaîneau CH, Rougeux G, Yéprémian C, Oudot J (2005) Effects of nutrient concentration on the biodegradation of crude oil and associated microbial populations in the soil. Soil Bio Biochem 37:1490–1497
Chang BV, Yu CH, Yuan SY (2004) Degradation of nonylphenol by anaerobic microorganisms from river sediment. Chemosphere 55:493–500
Chen H-J, Guo G-L, Tseng D-H, Cheng C-L, Huang S-L (2006) Growth factors, kinetics and biodegradation mechanism associated with Pseudomonas nitroreducens TX1 grown on octylphenol polyethoxylates. J Environ Manage 80:279–286
Di Corcia A, Costantino A, Crescenzi C, Marinoni E, Samperi R (1998) Characterization of recalcitrant intermediates from biotransformation of the branched alkyl side chain of nonylphenol ethoxylate surfactants. Environ Sci Technol 32:2401–2409
Dokianakis SN, Kornaros M, Lyberatos G (2006) Impact of five selected xenobiotics on isolated ammonium oxidizers and on nitrifying activated sludge. Environ Toxicol 21:310–316
Franska M, Franski R, Szymanski A, Lukaszewski Z (2003) A central fission pathway in alkylphenol ethoxylate biodegradation. Water Res 37:1005–1014
Frassinetti S, Barberio C, Caltavuturo L, Fava F, Di Gioia D (2011) Genotoxicity of 4-nonylphenol and nonylphenol ethoxylate mixtures by the use of Saccharomyces cerevisiae D7 mutation assay and use of this text to evaluate the efficiency of biodegradation treatments. Ecotoxicol Environ Saf 74:253–258
González S, Petrovic M, Barceló D (2007) Removal of a broad range of surfactants from municipal wastewater—comparison between membrane bioreactor and conventional activated sludge treatment. Chemosphere 67:335–343
Gratia E, Weekers F, Margesin R, D’Amico S, Thonart P, Feller G (2009) Selection of a cold-adapted bacterium for bioremediation of wastewater at low temperatures. Extremophiles 13:763–768
Gu X, Zhang Y, Zhang J, Yang M, Tamaki H, Kamagata Y, Li D (2010) Isolation of phylogenetically diverse nonylphenol ethoxylate-degrading bacteria and characterization of their corresponding biotransformation pathways. Chemosphere 80:216–222
Hayashi S, Saito S, Kim J-H, Nishimura O, Sudo R (2005) Aerobic biodegradation behavior of nonylphenol polyethoxylates and their metabolites in the presence of organic matter. Environ Sci Technol 39:5626–5633
John DM, White GF (1998) Mechanism for biotransformation of nonylphenol polyethoxylates to xenoestrogens in Pseudomonas putida. J Bacteriol 180:4332–4338
Jonkers N, Knepper TP, de Voogt P (2001) Aerobic biodegradation studies of nonylphenol ethoxylates in river water using liquid chromatography–electrospray tandem mass spectrometry. Environ Sci Technol 35:335–340
Karahan Ö, Olmez-Hanci T, Arslan-Alaton I, Orhon D (2010) Modelling biodegradation of nonylphenol ethoxylate in acclimated and non-acclimated microbial cultures. Bioresour Technol 101:8058–8066
Kristanti RA, Toyama T, Hadibarata T, Tanaka Y, Mori K (2014) Bioaugmentation involving a bacterial consortium isolated from the rhizosphere of Spirodela polyrhiza for treating water contaminated with a mixture of four nitrophenol isomers. RSC Adv 4:1616–1621
Łebkowska M, Zborowska E, Karwowska E, Miaśkiewicz-Pęska E, Muszyński A, Tabernacka A, Naumczyk J, Jęczalik M (2011) Bioremediation of soil polluted with fuels by sequential multiple injection of native microorganisms: field-scale processes in Poland. Econ Geol 37:1895–1900
Lee H-B, Peart TE, Bennie DT, Maguire RJ (1997) Determination of nonylphenol polyethoxylates and their carboxylic acid metabolites in sewage treatment plant sludge by supercritical carbon dioxide extraction. J Chromatogr A 785:385–394
Liu X, Ohta T, Kawabata T, Kawai F (2013) Catalytic mechanism of short ethoxy chain nonylphenol dehydrogenase belonging to a polyethylene glycol dehydrogenase group in the GMC oxidoreductase family. Int J Mol Sci 14:1218–1231
Liu X, Tani A, Kimbara K, Kawai F (2006) Metabolic pathway of xenoestrogenic short ethoxy chain-nonylphenol to nonylphenol by aerobic bacteria, Ensifer sp. strain AS08 and Pseudomonas sp. strain AS90. Appl Microbiol Biotechnol 72:552–559
Lozada M, Figuerola ELM, Itria RF, Erijman L (2006) Replicability of dominant bacterial populations after long-term surfactant-enrichment in lab-scale activated sludge. Environ Microbiol 8:625–638
Lu J, Jin Q, He Y, Wu J (2007) Biodegradation of nonylphenol polyethoxylates under Fe(III)-reducing conditions. Chemosphere 69:1047–1054
Lu J, Jin Q, He Y, Wu J, Zhao J (2008) Biodegradation of nonylphenol polyethoxylates under sulfate-reducing conditions. Sci Total Environ 399:121–127
Ma F, Guo J-b, Zhao L-j, Chang C-c, Cui D (2009) Application of bioaugmentation to improve the activated sludge system into the contact oxidation system treating petrochemical wastewater. Bioresour Technol 100:597–602
Maki H, Masuda N, Fujiwara Y, Ike M, Fujita M (1994) Degradation of alkylphenol ethoxylates by Pseudomonas sp. strain TR01. Appl Environ Microbiol 60:2265–2271
Manu B, Chaudhari S (2002) Anaerobic decolorisation of simulated textile wastewater containing azo dyes. Bioresour Technol 82:225–231
Montgomery-Brown J, Drewes JE, Fox P, Reinhard M (2003) Behavior of alkylphenol polyethoxylate metabolites during soil aquifer treatment. Water Res 37:3672–3681
Montgomery-Brown J, Li Y, Ding W-H, Mong GM, Campbell JA, Reinhard M (2008) NP1EC degradation pathways under oxic and microxic conditions. Environ Sci Technol 42:6409–6414
Mrozik A, Piotrowska-Seget Z (2010) Bioaugmentation as a strategy for cleaning up of soils contaminated with aromatic compounds. Microbiol Res 165:363–375
Nopcharoenkul W, Pinphanichakarn P, Pinyakong O (2011) The development of a liquid formulation of Pseudoxanthomonas sp. RN402 and its application in the treatment of pyrene-contaminated soil. J Appl Microbiol 111:36–47
Paterakis N, Chiu TY, Koh YKK, Lester JN, McAdam EJ, Scrimshaw MD, Soares A, Cartmell E (2012) The effectiveness of anaerobic digestion in removing estrogens and nonylphenol ethoxylates. J Hazard Mater 199–200:88–95
Schmidt S, Wittich RM, Erdmann D, Wilkes H, Francke W, Fortnagel P (1992) Biodegradation of diphenyl ether and its monohalogenated derivatives by Sphingomonas sp. strain SS3. Appl Environ Microbiol 58:2744–2750
Shokrollahzadeh S, Azizmohseni F, Golmohammad F, Shokouhi H, Khademhaghighat F (2008) Biodegradation potential and bacterial diversity of a petrochemical wastewater treatment plant in Iran. Bioresour Technol 99:6127–6133
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729
Tasaki Y, Yoshikawa H, Tamura H (2006) Isolation and characterization of an alcohol dehydrogenase gene from the octylphenol polyethoxylate degrader Pseudomonas putida S-5. Biosci Biotechnol Biochem 70:1855–1863
Wang M-Z, He H-Z, Zheng X, Feng H-J, Lv Z-M, Shen D-S (2014a) Effect of Pseudomonas sp. HF-1 inoculum on construction of a bioaugmented system for tobacco wastewater treatment: analysis from quorum sensing. Environ Sci Pollut Res 21:7945–7955
Wang Z, Yang Y, Sun W, Xie S (2014b) Biodegradation of nonylphenol by two alphaproteobacterial strains in liquid culture and sediment microcosm. Int Biodeter Biodegr 92:1–5
Winsor CP (1932) The Gompertz curve as a growth curve. Proc Natl Acad Sci U S A 18:1–8
Wyrwas B, Dymaczewski Z, Zgoła-Grześkowiak A, Szymański A, Frańska M, Kruszelnicka I, Ginter-Kramarczyk D, Cyplik P, Ławniczak Ł, Chrzanowski Ł (2013) Biodegradation of Triton X-100 and its primary metabolites by a bacterial community isolated from activated sludge. J Environ Manage 128:292–299
Yang X, Xue Y, Wang W (2009) Mechanism, kinetics and application studies on enhanced activated sludge by interior microelectrolysis. Bioresour Technol 100:649–653
Zhang J, Liu L, Li J, Du G, Chen J (2012) Enhanced glucosamine production by Aspergillus sp. BCRC 31742 based on the time-variant kinetics analysis of dissolved oxygen level. Bioresource Technol 111:507–511
Zhang J, Yang M, Qiao Y, Zhang Y, Chen M (2007) Biodegradation of nonylphenoxy carboxylates mixtures in two microcosms. Sci Total Environ 388:392–7
Zwietering MH, Jongenburger I, Rombouts FM, van’t Riet K (1990) Modeling of the bacterial growth curve. Appl Environ Microbiol 56:1875–1881
Acknowledgement
This study was supported by the National Natural Science Foundation of China (41271335; 31470191), the Major State Basic Research Development Program of China (973 program) (2015CB150502), the High Technology Research and Development Program of China (863 Program) (2012AA06A203), and the National Key Technology Rand D Program (2012BAC17B04), the Science and Technology Project of Zhejiang Province (2013C3303; 2014C33019), and the Environmental Science Project of Zhejiang Province (2012B006).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
There is no conflict of interest to report.
Additional information
Responsible editor: Gerald Thouand
Rights and permissions
About this article
Cite this article
Bai, N., Wang, S., Abuduaini, R. et al. Isolation and characterization of Sphingomonas sp. Y2 capable of high-efficiency degradation of nonylphenol polyethoxylates in wastewater. Environ Sci Pollut Res 23, 12019–12029 (2016). https://doi.org/10.1007/s11356-016-6413-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-016-6413-y