Abstract
In this study, the occurrence of 8 antibiotics [3 tetracyclines (TCs), 4 sulfonamides, and 1 trimethoprim (TMP)], 12 antibiotic resistance genes (ARGs) (10 tet, 2 sul), 4 types of bacteria [no antibiotics, anti-TC, anti-sulfamethoxazole (SMX), and anti-double], and intI1 in two wastewater treatment plants (WWTPs) were assessed and their influences in downstream lake were investigated. Both WWTPs’ effluent demonstrated some similarities, but the abundance and removal rate varied significantly. Results revealed that biological treatment mainly removed antibiotics and ARGs, whereas physical techniques were found to eliminate antibiotic resistance bacteria (ARBs) abundance (about 1 log for each one). UV disinfection did not significantly enhance the removal efficiency, and the release of the abundantly available target contaminants from the excess sludge may pose threats to human and the environment. Different antibiotics showed diverse influences on the downstream lake, and the concentrations of sulfamethazine (SM2) and SMX were observed to increase enormously. The total ARG abundance ascended about 0.1 log and some ARGs (e.g., tetC, intI1, tetA) increased due to the high input of the effluent. In addition, the abundance of ARB variation in the lake also changed, but the abundance of four types of bacteria remained stable in the downstream sampling sites.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Antibiotics have been widely used in human and veterinary medicines. The process of antibiotics’ transmission, with relevant antibiotic-resistance genes (ARGs) and antibiotic-resistance bacterias (ARBs) have been considered as emerging contaminants (Akinbowale et al. 2007; Kim et al. 2004). In the past, sulfonamides (SUs), tetracyclines (TCs), and trimethoprim (TMP) have been used to treat human and animal diseases, which could increase the antibacterial effect of sulfonamides as synergist (Batt et al. 2007; Pailler et al. 2009). In recent years, SU and TC resistance genes have received tremendous attention from researchers worldwide. So far, over 40 TC resistance genes and four sulfonamide resistance genes have been studied (Pei et al. 2006; Roberts 2005). IntI1 gene is considered to play a significant role in the spread of ARGs, as it can regulate the expression of exogenous genes and shows special relationship with sul (Baquero et al. 2008; Chen and Zhang 2013a; Sköld 2000; Wright et al. 2008). ARGs can be found on mobile elements such as transposons, integrons and plasmids, which would consequently transfer to other bacteria (Allen et al. 2010; Stokes and Gillings 2011). ARBs contain different ARGs, which may pose more risks to the environment due to proliferation and distribution. In addition, they can pose threats to human health through various pathways (Brooks et al. 2007; Pruden et al. 2006).
Globally, antibiotics, ARBs and ARGs in wastewater treatment plants (WWTPs) have attracted considerable researchers’ attention. Studies have shown that the WWTPs receive antibiotics, ARGs and ARBs from the sewage water of hospitals, residential areas, factories, and animal husbandry industry. Thus, these contaminants need to be removed in order to ensure public health safety (Huang et al. 2012; Szczepanowski et al. 2004; Zhang and Zhang 2011). However, traditional WWTP techniques are designed to remove conventional pollutants such as organic molecules and nutrients, but they may not be highly effective and efficient in removing antibiotics and ARGs and ARBs (Batt et al. 2006). Besides, effluents and excess sludge generated during the treatment processes may cause environmental health issues after being discharged or applied in agricultural fields (Luo et al. 2010; Verlicchi et al. 2012; Ye et al. 2007).
WWTPs usually consist of physical, chemical, and biological processes to eliminate conventional contaminants from the effluents without harming natural environments (Moura et al. 2012; Pescod 1992). Different stages play different functions in the reduction of target pollutants. Presently, WWTPs are also regarded as the important reservoirs of antibiotics and bacteria, where the antibiotic resistance organisms and determinants remained in the final effluents till it can be released to the environment (Ratola et al. 2012; Thomas and Nielsen 2005; Yang and Carlson 2003). The concentrations of antibiotics, ARGs and ARBs from the WWTPs effluents increase the spread and promotion of ARGs and ARBs into the natural environment, which transfer the antibiotic resistance into more pathogenic bacteria due to the abundance of ARGs from the high antibiotics concentration (Huang et al. 2012; Li et al. 2009; Livermore 2004; Pruden et al. 2006; Repice et al. 2013; Servais and Passerat 2009). Hence, it is essential to extensively investigate the fate of typical antibiotics and the corresponding resistance genes during the WWTP processes and their influences on the environment after being discharged.
The quality of surface water used as drinking water resource plays an important role for public health. Nowadays, the drinking water of millions of people from Zhejiang Province in China comes directly or indirectly from rivers and lakes. The effluents of WWTPs, which might contain hard-to-remove contaminants such as ARGs and ARBs, are usually discharged into nearby rivers and may turn into a new kind of pollution source. Obviously, it is considered as an important task to detect the hard-to-remove pollutants in the downstream water of WWTPs. Nevertheless, the knowledge in this area is still limited. So, monitoring the distribution of antibiotics, ARGs and ARBs would contribute to establishing adequate control measures and help reduce amount of these contaminants.
This study aimed to investigate the occurrence and removal of the targeted pollutants using different processes from two WWTPs and also in the downstream water (one lake basin) in eastern China. This study targeted 12 ARGs (10 tet, 2 sul), 8 relevant antibiotics (3 tetracyclines, 4 sulfonamides, and 1 trimethoprim), and 4 types of bacteria [no antibiotics, anti-TC, anti-sulfamethoxazole (SMX), and anti-double] for extensive analyses. In addition, intI1 was also monitored to indicate the horizontal gene transfer. The influences of the targeted pollutants at the final effluents in the downstream water body were also examined.
Materials and methods
Site description and sampling
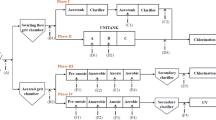
Samples were taken from two WWTPs (WWTP-Q and WWTP-L), and the downstream water adjacent to WWTP-L. The characteristics and the schematic flow diagram of the two WWTPs are given in Table 1 and Fig. 1. WWTP-Q is a plant receiving domestic wastewater and industrial effluents through an anaerobic-anoxic-oxic treatment (A/A/O) process. However, WWTP-L represents medium-sized plant using a triple oxidation process (OX). The downstream water is a lake spreading across 10 km2 and having the average water stage of 25 m throughout the year (Fig. S1). Samples were collected in September and October 2013.
Water samples of WWTP-Q were collected from the influent, primary clarifier effluent, biological reaction tank effluent, secondary clarifier effluent, and the final effluent. While samples of WWTP-L were collected separately from influent, anaerobic tank effluent, aerobic tank effluent, and the final effluent using water sampler. In addition, four sampling sites were chosen in the downstream lake including one upstream site (L1, 700 m from the WWTP-L influent) and three downstream sites (L2, 300 m; L3, 1200 m; L4, 2500 m from the WWTP-L effluent). The samples in the downstream lake were collected approximately 50 cm below the water surface. Mixed samples of WWTPs were collected during 24-h period after 1-h interval, while the lake samples were collected with the interval of 1 h during the daytime (8:00–18:00). Parallel samples were collected at the same time (n > 3). Fresh treated excess sludge samples were also collected from the two WWTPs. All Samples were stored in darkness at 4 °C and pretreated within 1 day for subsequent analyses.
Quantification of antibiotics
Eight antibiotics (three tetracyclines, four sulfonamides, and the trimethoprim) were chosen as our targeted compounds. All compound standards were generously provided by Dr. Ehrenstorfer (Germany). Methanol (LC/MS grade) and acetonitrile (LC/MS grade) were purchased from Fisher (USA), and the formic acid (LC grade) was purchased from Tedia Company. The detailed specifications of the selected antibiotics are given in Table S1.
For this study, we adopted the extraction and quantification methods from our previous study (Chen and Zhang 2013a). Water samples were first filtered through 0.45-μm glass fiber filters and then 0.80 g Na2EDTA was added into 1 L sample individually. Reaction was held for 1 h. Then, the pH of samples was adjusted to 4.8∼5.0 using 1 M HCl and NaOH. Oasis HLB cartridges (6 mL/500 mg, Waters, USA) were successively activated with 10.0 mL methanol, 10.0 mL ultra-pure water, and 5.0 mL of pH = 5.0 ± 0.2 ultra-pure water. The samples were then passed through cartridges at a flow rate of 5 mL/min. Cartridges were rinsed with 10 mL ultra-pure water and dried under gentle nitrogen gas for 30 min. Then, cartridges were eluted with 10 mL mixture of methanol/acetonitrile (1:1, v/v). The elutes were concentrated to 1 mL under gentle nitrogen gas at 40 °C and later diluted to a volume of 5 mL with methanol/water (1:1, v/v). Final treated samples were stored at −20 °C in the dark and analyzed by ultra-high performance liquid chromatography tandem mass spectrometry (UPLC-MS/MS).
After the freeze-drying process of sludge samples, 10 mL extractant (5 mL methanol + 5 mL Na2EDTA-McIlvaine) was added to 0.10 g treated solid sample, individually. Then, extract was sonicated for 10 min and centrifuged at 4000 rpm for another 10 min. This process was repeated twice, and the supernatants were collected and diluted to 800 mL using ultra-pure water.
UPLC-MS/MS was equipped with an Acquity™ UPLC and a Quattro Premier Micromass® MS (Waters/Micromass, Milford, MA). All target antibiotics were separated by a BEHC18 column (Waters Corp., 50 × 2.1 mm, 1.7 μm) and then identified and quantified by a MS/MS system via multi-reaction monitoring (MRM) mode. The MS/MS analysis was performed in the positive electrospray ionization (ESI) mode. The specific instrument operating parameters and conditions of the eight compounds are summarized in Tables S2 and S3.
The internal standards were used in this study. 13C6-sulfamethazine and 13C6-sulfamethoxazole (Cambridge Isotope Laboratories, USA) were used to quantify the sulfonamides. Thiabendazole-d6 and trimethoprim-d3 (Dr. Ehrenstorfer, Germany) were used to quantify tetracyclines and trimethoprim, respectively. The recoveries of targeted antibiotics in water and solid samples varied from 78 to 140 % and 71 to 150 %, respectively. Relative standard deviations (RSDs) of target compounds ranged from 1.0 to 3.5 % and showed good recoveries and instrumental precision. The correlation coefficients (R 2) of internal calibration curves were all higher than 0.999, indicating good linear correlation of curves.
Culturable count of antibiotic resistance bacteria
In this study, we used heterotrophic plate count (HPC) method as described by Gao and co-workers (2012). Four kinds of R2A agar medium were employed, comprising plates amended with no antibiotics, TC (TCI Company, 16.0 mg/L), sulfamethoxazole (SMX, Aladdin Company, 50.4 mg/L) and both two antibiotics (tetracycline and sulfamethoxazole) (Brooks et al. 2007; Pei et al. 2006). In order to inhibit the growth of fungi, 200 mg/L cycloheximide (Aladdin Company) was added into the plates.
Nearly 1.0 mL of water sample or 1.0 g of sludge sample was thoroughly diluted in 9.0 mL sterile physiological saline (0.85 %) by vortex. Then, tenfold serial dilutions were prepared for the subsequent tests. Approximately, 100 μL of each diluted sample was directly spread onto the four different plates. The plates were incubated at 35 °C for 72 h, and duplicate counting was also performed. Then, colony-forming units (CFUs) were calculated in per mL/g samples.
DNA extraction of samples
Water samples were concentrated and filtered using 0.22-μm filters via a vacuum filtration apparatus. After the clog, the filters were stored at −80 °C till DNA extraction. Total DNA of water and solid samples were extracted using Ultra-Clean Water DNA Kit (MoBio Laboratories, USA) and Fast DNA Spin Kit for Soil (MP Biomedicals, France), separately. The purified DNA concentrations were measured using spectrophotometric analysis (Nano Drop ND-2000c, Thermo, USA).
qPCR of antibiotic resistance genes
Thirteen antibiotic resistance genes, tet(A), tet(B), tet(C), tet(G), tet(L), tet(M), tet(O), tet(Q), tet(W), tet(X), sul1, sul2, and intI1, were quantified using real-time qPCR. 16S rRNA was also quantified in order to minimize the variance due to the differences among the bacterial abundance in the samples.
We followed the qPCR process as extensively described in our previous study (Chen and Zhang 2013a). The qPCR mixtures consisted of 0.3 μL of ROX reference dye, 0.2 μM concentration of each primer, 7.5 μL of SYBR premix Ex Taq™ (TaKaRa), 2 μL of template DNA, and 4.6 μL of dd H2O. StepOnePlus™ real-time PCR system (ABI, USA) was employed for amplification and quantification. The protocol was conducted as follows: 30 s at 95 °C, then 40 cycles of 5 s at 95 °C, 30 s at the annealing temperature, extension for another 30 s at 72 °C and a simultaneous fluorescence signal scanning at 72 °C, then a melt curve stage with temperature ramping from 60 to 95 °C. Details of the qPCR primers of the target genes and the annealing temperatures are given in Table S4.
Data analysis
The averages and standard deviations of all data were determined using Microsoft Excel 2010 and OriginPro 8.0 (Origin Lab Corporation, USA).
Results and discussion
Occurrence and concentrations of selected antibiotics
Figure 2 summarized the concentrations of target antibiotics in the sampling sites of the two WWTPs. Although the locations of WWTPs were different, both water samples of them presented the highest concentration at sulfamethoxazole (SMX, 426.0 and 136.7 ng/L, respectively), followed by oxytetracycline (OTC), TMP, and TC. No chlortetracycline (CTC) was detected in all WWTP-Q water samples. WWTP-Q had lower total antibiotic concentration (1409.9 μg/kg) than WWTP-L (3350.6 μg/kg) in sludge, among which OTC and TC had the highest concentrations, whether due to higher original amount in influent, or because TCs may enrich more in the sludge than sulfonamides (Kim et al. 2005). Besides, the magnitude order of total target antibiotics in sludge could be as high as mg/kg, which could pose potential risks such as ecotoxicity to the environment once being released (Zhu et al. 2013). However, the distribution of antibiotics between liquid and solid phase as well as their stability need to be further studied.
Concentrations of antibiotics in WWTPs (WWTP-Q: W-Q1, influent; W-Q2, primary clarifier effluent; W-Q3, biological reaction tank effluent; W-Q4, secondary clarifier effluent; W-Q5, the final effluent; W-QS, excess sludge. WWTP-L: W-L1, influent; W-L2, anaerobic tank effluent; W-L3, aerobic tank effluent; W-L4, final effluent; W-LS, excess sludge)
The removal rates of every antibiotic in each stage of two WWTPs are shown in Fig. S2. The total removal rates of seven antibiotics in WWTP-Q varied widely (TC 70.2, OTC 100, SD 46.7, SMX 82.2, SM2 41.4, SM1 76.6, and TMP 66.2 %) except CTC. Primary clarifier was usually used to remove suspended solids, and UV disinfection was used to eliminate microorganisms. Primary clarifier has lower removal rate of pharmaceuticals constituents (Sui et al. 2010), and in this study, it was found not effective in removing antibiotics (only 40 % removal was achieved). Kim and Tanaka (2009) once used UV to degrade antibiotics but dose was much higher (230 mJ/cm2) than the real UV dose (15 mJ/cm2) of WWTP-Q and only small proportions of antibiotics were degraded in this study (lower than 40 %). However, biological tank presented steady removal rates (all about 50 %), indicating that microbes could extensively biodegrade the antibiotics. Secondary clarifier removed most of OTC and SMX but not efficiently removed TC, sulfamerazine (SM1) and TMP, while sulfadiazine (SD) and sulfamethazine (SM2) increased in this process, and the reason could be the reenter of antibiotics from the sludge to water during the sludge-water separation, which needs further studies.
All eight antibiotics were detected in WWTP-L and the total removal rates of antibiotics were 100, 85.9, 87.9, 76.2, 75.0, 71.7, 54.7, and 58.0 %, respectively (Fig. S2). Anaerobic tank presented overall removal rates of about 30∼60 %, but different removal rates were observed in the aerobic tank. All the TC concentrations were eliminated, but only 10 % of CTC were removed indicating that the anaerobic tank presented more stable efficiency in antibiotic treatment as compared to aerobic tank. Moreover, as the previous two stages removed majority of antibiotics, thus UV disinfection played less important role in this process. McKinney and Pruden (2012) study indicated that UV disinfection has limited potential to damage ARGs. Apart from the treatment process, the differences between two WWTPs could also attribute to original influent loads and other operating parameters (aeration time, retention time of each stage, etc.). In addition, the results showed that the second clarifier has an advantage of removing OTC (100 %) and SMX (more than 60 %), and aerobic tank could eliminate TC (100 %) but has less efficient for CTC removal (only 10 %).
Abundance of target genes
Abundances of targeted 13 AGRs are summarized in Fig. 3. No significant differences of total ARGs abundance were noticed between the influents of the two WWTPs (1.38 × 1011 and 1.49 × 1011 copies/mL). However, the abundance of total targeted ARGs in excess sludge samples could be as high as 3.24 × 1010 and 9.68 × 1010 copies/g. Many researchers reported the occurrence of high abundance of ARGs in excess sludge, indicating the potential risks to the environment once they released (Auerbach et al. 2007; Munir et al. 2011). From Fig. 3, it could be found that tet(A), tet(C), tet(G), sul1, and int(I)1 accounted highest proportion of 67.95∼96.54 %, especially the intI1, which accounted for the 10.49∼30.19 % in the water samples and 8.10∼20.50 %, in the excess sludge. Some studies considered that the exogenous genes could be one of the significant factors for generating intracellular ARGs and intI1 played an important role in this process (Baquero et al. 2008; Sköld 2000). In this study, high abundance of intI1 may also contribute to the spread of ARGs in the environment.
Abundance of 13 target ARGs in WWTPs (WWTP-Q: W-Q1, influent; W-Q2, primary clarifier effluent; W-Q3, biological reaction tank effluent; W-Q4, secondary clarifier effluent; W-Q5, the final effluent; W-QS, excess sludge. WWTP-L: W-L1, influent; W-L2, anaerobic tank effluent; W-L3, aerobic tank effluent; W-L4, final effluent; W-LS, excess sludge)
The removal rates of ARGs in each stage of two WWTPs are shown in Fig. S3. Except for tetG (below 1.2 log), other ARGs could be removed more than 1.5 log (tetM, tetQ and tetW more than 2.4 log) in WWTP-Q. Biological tank was found highly effective for eliminating ARGs (about 50 %) and secondary clarifier could also removed ARGs 0.2∼1.2 log on account of sludge-water separation process. Nevertheless, primary clarifier (all blow 0.2 log) and UV disinfection (all below 0.4 log) played less essential part in reducing ARGs and the increase of tet(L) and tet(O) in primary clarifier may be due to the proliferation of genes. The decrease of ARGs in UV process may attribute to the fragmentation of genes or synthesis of pyrimidine dimer under UV (McKinney and Pruden 2012).
In WWTP-L, sul2 had the highest removal rate of about 2.26 log and others were between 1.1 and 1.8 log. ARGs declined relatively less in the anaerobic tank, but the aerobic tank could remove over 70 % of target genes except for tet(X) (15.3 %), demonstrating aerobic process was the key stage in removing ARGs. In addition, Chen and Zhang (2013b) also observed an increased of ARGs in anaerobic process and the increase of tet(W) (0.004 log) in this study also supported our findings. Similar to WWTP-Q, UV disinfection reduced relatively lower ARGs (all below 0.5 log except for tet(X)). Furthermore, two main genes tet(X) (0.37 log, 0.70 log) and tet(G) (0.29 log, 0.45 log) were declined among all ARGs in UV process, inferring that some ARGs could be removed more easily in this stage.
Culturable bacteria counts
Figure 4 presents the numbers of the total culturable bacteria and anti-TC, anti-SMX, and anti-double bacteria in the water and sludge samples of the two WWTPs. There was no significant difference of total detected bacteria abundance between the influents, and the removal rates of four types of bacteria were 1.72, 2.67, 2.10, and 2.23 log in WWTP-Q, and 1.81, 2.63, 1.76, and 2.66 log in WWTP-L, respectively. The abundance of bacteria in the excess sludge samples could be as high as 6.70 × 108, 9.50 × 106, 8.75 × 107, and 5.25 × 106 CFU/g in WWTP-L and nearly 1∼2 log higher than WWTP-Q. High abundance of ARBs existing in the excess sludge may pose potential risks to the environment (Brooks et al. 2007).
Abundance of 4 types of bacteria in WWTPs (WWTP-Q: W-Q1, influent; W-Q2, primary clarifier effluent; W-Q3, biological reaction tank effluent; W-Q4, secondary clarifier effluent; W-Q5, the final effluent; W-QS, excess sludge. WWTP-L: W-L1, influent; W-L2, anaerobic tank effluent; W-L3, aerobic tank effluent; W-L4, final effluent; W-LS, excess sludge)
In WWTP-Q treatment process, primary clarifier reduced bacteria about 1 log and indicated that the elimination of bacteria with suspended solids occurred under outer mechanical forces. Similarly, ARBs decreased nearly 1 log after the secondary clarifier and the physical forces such as gravity and flocculation were found to be the main approach to reduce ARBs. But, biological process exerted little influence on decreasing ARBs, which may attribute to the addition of original ARBs in the tank and the bacteria proliferation occurs during this process. In WWTP-L, the abundance of ARBs presented more obvious reduction in anaerobic process (about 1 log) than aerobic process. UV disinfection could reduce bacteria abundance 0.61, 0.91, 0.52, and 1.04 log, while all four types of bacteria reduced less than 0.5 log in WWTP-Q. The reason could be the difference in microbial loads, UV dose, and exposure retention time. Gao et al. (2012) also reported the much higher proportion of anti-SMX bacteria than anti-TC in samples. In this study, anti-SMX was found to be at the highest ratio of 50.0 % (anti-TC less than 3 %).
Influence of antibiotics, ARGs, and ARBs from effluent to the downstream water
The concentrations of antibiotics in the effluent and downstream lake sampling sites are presented in Fig. 5. Without the detection of TC in the effluent and CTC and SM1 in the lake water, all other seven antibiotics in effluent had higher concentration than L1. From L1 to L4, the concentrations of SD and TMP decreased gradually and were found slightly change from the effluent. The concentration of SMX increased from L1 (19.29 ng/L) to L2 (21.21 ng/L), and then decreased to 17.83 ng/L (L4). Thus, the SMX concentration variation in water is in agreement with the findings reported in early study (Zhu et al. 2013). SM2 presented a similar trend, which increased from L1 (1.11 ng/L) toL2 (1.75 ng/L) and reduced to 0.84 ng/L (L4), demonstrating the inflow of effluent increased the concentration of these two antibiotics in the lake water. However, the variation trends of TC and OTC were not obvious, as TC and OTC are the common medicines in the animal husbandry industry. The reason could be the flow of rain water from the nearby rural areas into the lake and it led to the uncertainty and non uniformity in their concentrations. Zhang et al. (2012) studied the abundance of antibiotics in Laizhou Bay and concluded that the upstream water could be the source of antibiotics. In addition, Kolpin et al. (2002) found about 80 % of 139 rivers in America had been polluted by antibiotics. Some studies also considered WWTP effluents as the main source of antibiotics to the surface water (Hirsch et al. 1999; Golet et al. 2002).
Figure 6 presents the abundance of targeted genes in the sampling sites. The abundance of total target genes increased from 7.91 × 108 copies/mL (L1) to 9.58 × 108 copies/mL (L2). The concentration in the effluent had 4.57 × 109 copies/mL, and then it decreased gradually, indicating that the inflow of effluent may contribute to the total abundance of ARGs in the downstream water. Similarly, int(I)1, tet(A), tet(G), tet(L), and tet(M) have the same trend. Besides, sul1 had the highest abundance in L1 (3.03 × 108 copies/mL), followed by tet(C) (1.79 × 108 copies/mL), whereas tet(C) dominated the most in L2 (3.56 × 108copies/mL), with sul1 followed (2.39 × 108copies/mL), demonstrating the effluent input may chang original ARGs abundance. For tet(X) and sul2, the abundance of ARGs reduced gradually from L1 to L4, which may attribute to less influence posed by relatively low ARGs abundance (104 to 106copies/mL) as compared to others (e.g., tet(C) and int(I)1) in the effluent. Czekalski et al. (2012) studied the detection and abundance of ARGs in the Lake Geneva and observed their different concentration levels. So, the influence of ARGs to the downstream water is a complicated process and involves with many physical factors and hydrological characteristics.
The abundance of bacteria in different sampling sites is presented in Fig. 7. It can be seen that the effluent had lower abundance (0.5 log) of total culturable bacteria than at L1 and had higher abundance with other three ARBs indicating of significantly higher removal efficiency of total culturable bacteria, but less effective for ARBs in WWTP. Both total culturable and anti-SMX bacterial numbers were found to decline about 0.5 log from L1 to L2, respectively, while the anti-TC bacteria remained stable (about 103 copies/mL). However, the anti-double bacteria increased about 0.5 log. Except for the proliferation of ARBs during the spread process, this phenomenon may also attribute to the input of the effluent and the dilution of lake water as well. Besides, it can be seen that the four types of bacteria remained steady from L2 to L4, which demonstrated that the environment in the downstream lake may be a stable biotope. Even so, it could be noticed that the abundance of bacteria was higher (e.g., 105 CFU/mL of anti-SMX bacteria and 103CFU/mL of anti-TC bacteria). The existence of high abundance of ARBs in the aquatic environment may spread the antibiotic resistance to other biotopes (Baquero et al. 2008). Lake and river waters are usually used for drinking water purpose in many areas. In recent study, research found most of residual pharmaceuticals could be efficiently removed by advanced drinking water treatment techniques (Dai et al. 2015). However, the water containing antibiotics and antibiotic resistance from the drinking water treatment plant may still not be thoroughly eliminated and the water from community supplies causes serious water borne diseases (Hirsch et al. 1999; Zhu et al. 2013).
Conclusions
In this study, the occurrence and abundance of 8 antibiotics, 13 corresponding ARGs, and 4 types of cultivable bacteria (including three ARBs) were evaluated in two WWTPs and the effluents were further purposively assessed in the downstream lake. Besides, their removal rates were also analyzed in different treatment stages of both WWTPs.
-
1.
In terms of overall removal, WWTP-Q had better treatment effect for the antibiotic OTC (100 %), while WWTP-L had the advantage of antibiotics TC, SD, and SM2 at the removal rate of 100, 76.2, and 71.7 %, respectively. In addition, removal rates of ARGs varied and WWTP-Q was found more efficient in reducing tet(M), tet(Q), and tet(W) (all above 2.4 log), while WWTP-L could only reduce sul2 genes remarkably (2.24 log). Bacteria removal rates achieved 7.21∼2.23 log (WWTP-Q) and 1.76∼2.66 log (WWTP-L).
-
2.
As for different treatment stages, biological tank showed better removal efficiency (about 50 % in WWTP-Q and 30∼60 % in WWTP-L) than any other treatment process for degrading antibiotics. Aerobic tank was found more effective in ARGs reduction than others (all above 50 % in WWTP-Q and more than 70 % except for tet(X) in WWTP-L). Physical method (primary settling tank and secondary clarifier) played more important roles in ARBs reduction (about 1 log in both WWTPs). UV treatment in both two WWTP had less effect in removing targets.
-
3.
Concentrations of SMX and SM2 increased in the downstream of WWTP-L. The total abundance of ARGs in downstream lake sampling sites increased about 0.1 log due to the effluents of WWTP-L, among which tet(C), int(I)1, and tet(A) abundance rose obviously. Besides, the abundance of four types of bacteria remained steady in the downstream sampling sites, demonstrating that the environment in the downstream lake may be a stable biotope.
-
4.
High concentrations of antibiotics and abundance of ARGs and ARBs were found in the excess sludge samples (levels of mg/kg, 1011 copies/g and 108 CFU/mL, respectively). The existence of targets in both effluent and sludge indicated they may pose potential risks to drinking water sources and soil, and then might harm human health, which should cause researcher and water industries’ attention.
References
Akinbowale OL, Peng H, Barton MD (2007) Diversity of tetracycline resistance genes in bacteria from aquaculture sources in Australia. J Appl Microbiol 103(5):2016–2025
Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J (2010) Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol 8:251–259
Auerbach EA, Seyfried EE, McMahon KD (2007) Tetracycline resistance genes in activated sludge wastewater treatment plants. Water Res 41(5):1143–1151
Baquero F, Martínez J, Cantón R (2008) Antibiotics and antibiotic resistance in water environments. Curr Opin Biotechnol 19:260–265
Batt AL, Bruce IB, Aga DS (2006) Evaluating the vulnerability of surface waters to antibiotic contamination from varying wastewater treatment plant discharges. Environ Pollut 142:295–302
Batt AL, Kim S, Aga DS (2007) Comparison of the occurrence of antibiotics in four full-scale wastewater treatment plants with varying designs and operations. Chemosphere 68:428–435
Brooks JP, Maxwell SL, Rensing C, Gerba CP, Pepper IL (2007) Occurrence of antibiotic resistant bacteria and endo toxin associated with the land application of biosolids. Can J Microbiol 53:616–622
Chen H, Zhang MM (2013a) Effects of advanced treatment systems on the removal of antibiotic resistance genes in wastewater treatment plants from Hangzhou, China. Environ Sci Technol 47(15):8157–8163
Chen H, Zhang MM (2013b) Occurrence and removal of antibiotic resistance genes in municipal wastewater and rural domestic sewage treatment systems in eastern China. Environ Int 55:9–14
Czekalski N, Berthold T, Caucci S, Egli A, Bürgmann H (2012) Increased levels of multiresistant bacteria and resistance genes after wastewater treatment and their dissemination into Lake Geneva, Switzerland. Front Microbiol 3:1–18
Dai S, Reiji K, Toshinari S, Michihiro A, Tetsuji N, Shoichi K (2015) Occurrence of selected pharmaceuticals at drinking water purification plants in Japan and implications for human health. Water Res 76:187–200
Gao P, Munir M, Xagoraraki I (2012) Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal wastewater treatment plant. Sci Total Environ 421–422:173–183
Golet E, Alder AC, Giger W (2002) Environmental exposure and risk assessment of fluoroquinolone antibacterial agents in wastewater and river water of the Glatt Valley watershed, Switzerland. Environ Sci Technol 36(17):3645–3651
Hirsch R, Ternes T, Haberer K, Kratz KL (1999) Occurrence of antibiotics in the aquatic environment. Sci Total Environ 225:109–118
Huang JJ, Hu HY, Lu SQ, Li Y, Tang F, Lu Y, Wei B (2012) Monitoring and evaluation of antibiotic-resistant bacteria at a municipal wastewater treatment plant in China. Environ Int 42:31–36
Kim I, Tanaka H (2009) Photodegradation characteristics of PPCPs in water with UV treatment. Environ Int 35:793–802
Kim SR, Nonaka L, Suzuki S (2004) Occurrence of tetracycline resistance genes tet(M) and tet(S) in bacteria from marine aquaculture sites. FEMS Microbiol Lett 237(1):147–156
Kim S, Eichhorn P, Jensen JN, Weber AS, Aga DS (2005) Removal of antibiotics in wastewater: effect of hydraulic and solid retention times on the fate of tetracycline in the activated sludge process. Environ Sci Technol 39:5816–5823
Kolpin DW, Furlong ET, Meyer MT, Thurman EM, Zaugg SD, Barber LB, Buxton H (2002) Pharmaceuticals, hormones, and other organic wastewater contaminants in U.S. streams, 1999−2000: a national reconnaissance. Environ Sci Technol 36(6):1202–1211
Li D, Yang M, Hu JY, Zhang J, Liu RY, Gu X (2009) Antibiotic-resistance profile in environmental bacteria isolated from penicillin production wastewater treatment plant and the receiving water. Environ Microbiol 11(6):1506–1517
Livermore D (2004) Can better prescribing turn the tide of resistance? Nat Rev Microbiol 2:73–78
Luo Y, Mao D, Rysz M, Zhou Q, Zhang H, Xu L et al (2010) Trends in antibiotic resistance genes occurrence in the Haihe River, China. Environ Sci Technol 44:7220–7225
McKinney CW, Pruden A (2012) Ultraviolet disinfection of antibiotic resistant bacteria and their antibiotic resistance genes in water and wastewater. Environ Sci Technol 46(24):13393–13400
Moura A, Pereira C, Henriques I, Correia A (2012) Novel gene cassettes and integrons in antibiotic resistant bacteria isolated from urban wastewaters. Res Microbiol 163(2):92–100
Munir M, Wong K, Xagoraraki I (2011) Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Res 45(2):681–693
Pailler JY, Krein A, Pfister L, Hoffmann L, Guignard C (2009) Solid phase extraction coupled to liquid chromatography-tandem mass spectrometry analysis of sulfonamides, tetracyclines, analgesics and hormones in surface water and wastewater in Luxembourg. Sci Total Environ 407(16):4736–4743
Pei R, Kim SC, Carlson KH, Pruden A (2006) Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res 40:2427–2435
Pescod MB (1992) Wastewater treatment and use in agriculture. FAO, Rome
Pruden A, Pei R, Storteboom H, Carlson KH (2006) Antibiotic resistance genes as emerging contaminants: studies in Northern Colorado. Environ Sci Technol 40(23):7445–7450
Ratola N, Cincinelli A, Alves A, Katsoyannis A (2012) Occurrence of organic microcontaminants in the wastewater treatment process. A mini review. J Hazard Mater 239:1–18
Repice C, Grande MD, Maggi R, Pedrazzani R (2013) Licit and illicit drugs in a wastewater treatment plant in Verona, Italy. Sci Total Environ 463–464:27–34
Roberts MC (2005) Update on acquired tetracycline resistance genes. FEMS Microbiol Lett 245(2):195–203
Servais P, Passerat J (2009) Antimicrobial resistance of fecal bacteria in waters of the Seineriver watershed (France). Sci Total Environ 408:365–372
Sköld O (2000) Sulfonamide resistance: mechanisms and trends. Drug Resist Updat 3(3):155–60
Stokes HW, Gillings MR (2011) Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol Rev 35:790–819
Sui Q, Huang J, Deng S, Yu G, Fan Q (2010) Occurrence and removal of pharmaceuticals, caffeine and DEET in wastewater treatment plants of Beijing, China. Water Res 44:417
Szczepanowski R, Krahn I, Linke B, Goesmann A, Pühler A, Schlüter A (2004) Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system. Microbiology 150:3613–3630
Thomas CM, Nielsen KM (2005) Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3:711–721
Verlicchi P, Al Aukidy M, Zambello E (2012) Occurrence of pharmaceutical compounds in urban wastewater: removal, mass load and environmental risk after a secondary treatment—a review. Sci Total Environ 429:123–155
Wright MS, Baker-Austin C, Lindell AH, Stepanauskas R, Stokes HW, McArthur JV (2008) Influence of industrial contamination on mobile genetic elements: class 1 integron abundance and gene cassette structure inaquatic bacterial communities. ISME J 2:417–428
Yang S, Carlson K (2003) Evolution of antibiotic occurrence in a river through pristine, urban and agricultural landscapes. Water Res 37(19):4645–4656
Ye ZQ, Weinberg HS, Meyer MT (2007) Trace analysis of trimethoprim and sulfonamide, macrolide, quinolone, and tetracycline antibiotics in chlorinated drinking water using liquid chromatography electrospray tandem mass spectrometry. Analyt Chem 79(3):1135
Zhang XX, Zhang T (2011) Occurrence, Abundance, and Diversity of Tetracycline Resistance Genes in 15 Sewage Treatment Plants across China and Other Global Locations. Environ Sci Technol 45:2598–2604
Zhang RJ, Zhang G, Zheng Q, Tang JH, Chen YJ, Xu WH, Zou YD, Chen XX (2012) Occurrence and risks of antibiotics in the Laizhou Bay, China: impacts of river discharge. Ecotox Environ Safe 80:208–215
Zhu S, Chen H, Li J (2013) Sources, distribution and potential risks of pharmaceuticals and personal care products in Qingshan Lake basin, Eastern China. Ecotox Environ Safe 6:154–159
Acknowledgments
The authors would to extends their sincerely thanks to the managers of wastewater treatment plants for providing the water samples and related information. This work was supported by the Major Science and Technology Program for Water Pollution Control and Treatment (2014ZX07101-012) and Natural Science Foundation of China (21277117).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Philippe Garrigues
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 1335 kb)
Rights and permissions
About this article
Cite this article
Li, J., Cheng, W., Xu, L. et al. Occurrence and removal of antibiotics and the corresponding resistance genes in wastewater treatment plants: effluents’ influence to downstream water environment. Environ Sci Pollut Res 23, 6826–6835 (2016). https://doi.org/10.1007/s11356-015-5916-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-015-5916-2