Abstract
Introduction
Solitary pulmonary nodules (SPNs) are commonly found in imaging technologies, but are plagued by high false-positive rates.
Objective
We aimed to identify metabolic alterations in SPN etiology and diagnosis using less invasive plasma metabolomics and lipidomics.
Methods
In total, 1160 plasma samples were obtained from healthy volunteers (n = 280), benign SPNs (n = 157) and malignant SPNs (stage I, n = 723) patients enrolled from 5 independent centers. Gas chromatography-triple quadrupole mass spectrometry (GC‒MS) and liquid chromatography-Q Exactive Hybrid Quadrupole-Orbitrap mass spectrometry (LC‒MS) were used to analyze the samples for untargeted metabolomics and lipidomics.
Results and conclusion
GC‒MS-based metabolomics revealed 1336 metabolic features, while LC‒MS-based lipidomics revealed 6088 and 2542 lipid features in the positive and negative ion modes, respectively. The metabolic and lipidic characteristics of healthy vs. benign or malignant SPNs exhibited substantial pattern differences. Of note, benign and malignant SPNs had no significant variations in circulating metabolic and lipidic markers and were validated in four other centers. This study demonstrates evidence of early metabolic alterations that can possibly distinguish SPNs from healthy controls, but not between benign and malignant SPNs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Solitary pulmonary nodules (SPNs) are radiographic opacities that are smaller than or equal to 3 cm in diameter and are entirely surrounded by aerated lung parenchyma (Ost et al., 2003). More than four million Americans have undergone at least one chest CT scan, and approximately 1.5 million have a nodule identified (Gould et al., 2015). Benign SPNs are the consequence of lung inflammation or benign neoplasms (fibroma, hamartoma, neurofibroma and blastoma). Lung cancer, lymphoma, carcinoid, and sarcoma are the most common causes of malignant SPNs. Recent radiologic features and quantitative models have demonstrated that CT is useful for the early detection of malignant SPNs, although it suffers from significant false-positive rates. Overall, the majority of nodules are benign, the risk of lung cancer is approximately 1% (Horeweg et al., 2014), and a large number of nodules represent early, potentially curable lung malignancies (stage I).
To date, there is no consensus on how to manage these SPNs (Klaveren et al., 2009; Ost et al., 2003). After conventional assessment using current diagnostic techniques, up to 75% of patients will be classified as indeterminate (Ost et al., 2003). Depending on the probability of cancer, patients are managed with either follow-up CT imaging, additional diagnostic testing (including PET, CT-guided transthoracic needle biopsy, and/or bronchoscopy), or by definitive therapy with surgical excision (Au-Yong et al., 2020; Cruickshank et al., 2019; Nasim & Ost, 2019). Additional assays that can better stratify SPNs are desperately needed. A less invasive complementary test with good sensitivity and specificity is preferred. To decrease false-positive rates, blood biomarker assays could be developed to supplement CT screening and invasive methods.
A comprehensive understanding of benign and malignant SPNs could represent a large improvement towards the evaluation and management of SPNs. Metabolomics and lipidomics can reveal the downstream products of genes and proteins expressions, as reflects in phenotypic changes and alterations in pathophysiological states (Baharum et al., 2018; Nicholson & Lindon, 2008). Metabolomics is thus increasingly being used to diagnose diseases, unravel underlying disease mechanisms, as well as evaluate drug treatments and therapeutic outcomes (Wishart, 2016). With respect to SPNs, some studies have reported the use of metabolomics to elucidate metabolic profiles for SPNs mechanisms and to differentiate between benign and malignant SPNs, but these studies were limited by small sample sizes and a lack of multicenter validation (Fahrmann et al., 2016; Gao et al., 2013).
In our study, we used 1160 patients with benign and malignant SPNs as well as healthy subjects with the following underlying aims: (1) to differentiate between healthy individuals, benign SPNs, and malignant SPN patients with the aid of comprehensive metabolomic and lipidomic analyses, and (2) to provide probable guidance from the perspective of metabolomics and lipidomics for the management of malignant SPNs (stage I).
2 Material and methods
2.1 Participant characteristics
The discovery phase of the study included healthy volunteers as well as patients with benign SPNs and malignant SPNs from Center 1 (Jiangyin People’s Hospital, Wuxi, China). Patients who were enrolled from four other independent centers that formed the external validation phase cohort, including patients enrolled from Center 2 (Shanghai Ninth People's Hospital, Shanghai, China), Center 3 (Suzhou Municipal Hospital, Suzhou, China), Center 4 (Renmin Hospital of Wuhan University, Wuhan, China), and Center 5. (Xinhua Hospital, Shanghai, China). Healthy controls were not enrolled in the validation phase cohort. All participants were between the ages of 18 and 86 and were enrolled from March 2017 to June 2020. All participants were diagnosed using computed tomography scans, and individuals with SPNs were histologically verified as malignant or benign. Patients with co-morbid conditions concomitant illnesses such as other types of cancer, cardiovascular diseases, immunodeficiency diseases, and other nervous system diseases that may impact metabolism were excluded. Patients with a history of long-term drug use, as well as those who had received any type of cancer therapy, such as chemotherapy, radiation, or surgery three months prior to enrollment, were also excluded. Patients and healthy subjects gave written informed consent before enrollment. Prior to surgery, blood samples were obtained from all patients in EDTA-treated tubes. All blood samples were promptly centrifuged at 2000 × g for 15 min, and plasma was placed into clean Eppendorf tubes and kept at − 80 °C before analysis.
2.2 Untargeted metabolite profiling by GC‒MS and LC‒MS
Prior to comprehensive lipidomic profiling of the plasma samples, a sample preparation method based on liquid‒liquid MTBE extraction was utilized to cover diverse classes of lipids. Untargeted lipidomic analysis was performed using a Dionex UltiMate 3000 UHPLC system (Santa Clara, CA, USA) linked online via an electrospray ionization source with a Q ExactiveTM Hybrid Quadrupole-OrbitrapTM Mass Spectrometer (Thermo Fisher Scientific, Inc., MA, USA). Comprehensive metabolomic analysis was performed on a TRACE 1310 gas chromatograph equipped with an AS 1310 autosampler connected to a TSQ 8000 triple quadrupole mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) as described previously (Li et al., 2017). The detailed metabolomics and lipidomics methods are shown in the Supplementary methods.
The raw data files from GC‒MS and LC‒MS were converted to Analysis Base File (ABF) format by Abf Converter (http://www.reifycs.com/AbfConverter). Then, using the open-source program MS-Dial v.4.24 (Tsugawa et al., 2015), automatic peak selection, integration, retention time adjustment by the aforementioned database, and alignment were performed. The retention time index tolerance in GC‒MS was 3000, while the peak height threshold was 10,000. For LC‒MS, the peak height threshold was 1,000,000 for positive ion mode and 500,000 for negative ion mode. The resultant output data of high-quality time-aligned investigated metabolites, together with their related RT, m/z, and peak height acquired for each sample, were statistically analyzed. All metabolite identifications were manually double-checked.
2.3 Data analysis
The pretreated metabolomics, lipidomics data under positive ion mode and lipidomics data under negative ion mode the data were first normalized to internal standards, and then corrected using the quality control (QC) samples by LOESS (locally estimated scatterplot smoothing), Poly4 (fourth-order polynomial) and Poly6 (sixth-order polynomial) before multivariate analysis, respectively (Calderon-Santiago, 2021; Dunn et al., 2011; Wehrens et al., 2016). Methods for quality control-based robust LOESS, poly signal correction to provide signal correction and integration of data from multiple analytical batches are extensively applied for large-scale metabolic profiling of serum and plasma using GC‒MS and LC‒MS. Metabolic features detected in less than 50% of QC samples were removed. The Calculated RSD for all metabolic features in corrected QC samples was less than 20% using LC‒MS and less than 30% using GC‒MS (Calderon-Santiago, 2021; Dunn et al., 2011). The QC samples were obtained by pooling a 5 μL aliquot of each plasma sample, prepared using the above protocol, analyzed five times at the beginning of the run and injected in triplicate after every ten injections of the randomly sequenced samples. The RSD of the intrabatch metabolic features of the QC samples was less than 5%. The significance of each metabolite was analyzed by the Mann‒Whitney-Wilcoxon test with a false discovery rate (FDR) correction. The discrimination of variables was identified by orthogonal partial least-squares discriminant analysis (OPLS-DA) and partial least-squares discriminant analysis (PLS-DA). The quality of the OPLS-DA and PLS-DA models were evaluated by the R2Y and Q2 values, wherein R2Y shows the interpretation rate of the model, and Q2 indicates the prediction rate. Higher values of R2Y and Q2 indicate that the model is reliable and highly predictive. The Benjamini‒Hochberg FDR procedure was employed for the multiple test adjustments. Adjusted p values less than 0.05 were considered statistically significant. Differential metabolites were screened as those with variable importance in the projection (VIP) > 1.0 obtained from OPLS-DA and adjusted p values less than 0.05, where VIP indicates the contribution of each variable to group differences. Meanwhile, differential features for benign SPNs vs malignant SPNs were also screened using logistic regression and receiver operating characteristic (ROC) analysis. Pathway analyses were performed by MetaboAnalyst 5.0 (07/2020). PLS-DA was analyzed by SIMCA 14.1. Principal component analysis (PCA), OPLS-DA, and other statistical analyses were performed using R-4.1.3. OPLS-DA was analyzed by ropls package. QC correction was analyzed by MetaboQC. ROC was analyzed by pROC package.
3 Results
3.1 Participant characteristics and study design
In total, 1160 participants were enrolled in the study. The discovery phase included 280 healthy people, 42 benign SPN patients, and 118 malignant SPN patients. The validation group included patients from Center 2 (23 benign SPNs and 105 malignant SPNs), Center 3 (45 benign SPNs and 139 malignant SPNs), Center 4 (13 benign SPNs and 33 malignant SPNs), and Center 5 (34 benign SPNs and 328 malignant SPNs patients). The basic clinical characteristics of these participants are summarized in Table S1. There were no significant differences in the basic clinical characteristics between the normal and SPNs groups. The entire study is outlined in Fig. 1.
Comprehensive plasma metabolomic characterization of solitary pulmonary nodules. 280 health subjects (H) were included in center one. Patients diagnosed with indeterminate solitary pulmonary nodules (SPNs, n = 880) underwent thoracoscopic surgery and were divided into two groups based on their lung pathology: benign solitary pulmonary nodules (BSPNs, n = 157) and malignant solitary pulmonary nodules (MSPNs, n = 723) in five centers. The comprehensive metabolomic profiles of the two groups were determined by gas chromatography-triple quadrupole mass spectrometry (GC–MS) and Ultra-high performance liquid chromatography coupled with a Q Exactive hybrid quadrupole-orbitrap mass spectrometry (UPLC-MS/MS) to the fully capture metabolomic profiles of each group. Multivariate statistics were used to identify differential metabolites. Metabolic and lipidomic characteristics were observed in HC vs malignant or benign SPN patients and verified in four more centers
3.2 GC‒MS based metabolomics for SPNs
To examine metabolic alterations in SPNs, GC‒MS datasets were subjected to univariate and multivariate statistical analyses. Using the Mann‒Whitney-Wilcoxon test with FDR correction, we observed that 279 of 1336 metabolic characteristics showed significant difference (FDR < 0.05), with 146 metabolites greater in healthy controls than in benign SPNs, but 133 were the opposite (Fig. 2A and B). There was some overlap in the two groups using PCA, but clearly distinguished in the OPLS-DA model (Fig. 2C and D). The cumulative R2Y and Q2 values of the OPLSDA model were 0.837 and 0.582, respectively. Combined with variable importance in the projection (VIP > 1.0), 99 differential metabolites were found (Fig. 2F and Table S2), where perturbed metabolic pathways primarily included arginine and proline metabolism; propanoate metabolism; glutathione metabolism; butanoate metabolism; and glycine, serine and threonine metabolism (Fig. 2E). In healthy participants versus malignant SPNs patients, we observed 551 of 1336 metabolic characteristics with FDR < 0.05; the relative concentration levels of 353 metabolites were higher in healthy individuals than in malignant SPNs, while 198 were the opposite (Fig. 3 A and B). The OPLS-DA showed a significant difference, although the PCA did not (Fig. 3C and D), with an R2Y of 0.864 and Q2 of 0.695. Overall, 127 differential metabolites were identified by VIP > 1.0 and FDR < 0.05 (Fig. 3F and Table S3). The disturbed metabolic pathways mainly included glutathione metabolism, glycine, serine and threonine metabolism, valine, leucine and isoleucine biosynthesis and arginine and proline metabolism (Fig. 3E). When the three sets of data were combined, we detected 63 common differential metabolites in the healthy, benign, and malignant SPN groups. The enriched pathways were glutathione, butanoate, arginine and proline metabolism; D-glutamine and D-glutamate metabolism; and nitrogen metabolism (Figure S1).
GC–MS based metabolomic comparison of healthy subjects vs benign SPNs. A and B Cloud Plot of data set, 1057 features with p-value ≥ 0.05 and 279 features with p-value ≤ 0.05 includes visualization of the retention time, and the mass-to-charge ratio of features; C PCA score plots of healthy subjects vs benign SPNs; D Discriminative orthogonal projection to latent structure-discriminant analysis (OPLS-DA) score plots of healthy subjects vs benign SPNs; E Disturbed metabolic pathways identified from the comparison of healthy subjects vs malignant SPNs; F Heatmap of the differential metabolites from healthy subjects vs benign SPNs
GC–MS based metabolomic comparison of healthy subjects vs malignant solitary pulmonary nodules (SPNs). A and B Cloud Plot of data set, 785 features with p-value ≥ 0.05 and 551 features with p-value ≤ 0.05 includes visualization of the retention time, and the mass-to-charge ratio of features; C PCA score plots of healthy subjects vs malignant SPNs; D Discriminative OPLS-DA score plots of healthy subjects vs malignant SPNs; E Disturbed metabolic pathways identified from the comparison of healthy subjects vs malignant SPNs; F Heatmap of the differential metabolites from healthy subjects vs malignant SPNs
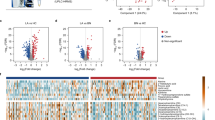
By comparing the metabolic features of benign SPNs with those of malignant SPNs using the Mann‒Whitney-Wilcoxon test with a FDR correction, there were no significant differential features (Fig. 4 A and B). The PCA of the two phenotypes revealed full overlap (Fig. 4C), and the OPLS-DA revealed no significant difference between the groups (Figure S2A) in the discovery phase, with R2Y of 0.671 and Q2 of -0.343. Multicenter samples were used for validation to corroborate this pattern of nonstatistical significance between the two groups. In validation Centers 2–5, there was no statistically significant difference in the sum of metabolic characteristics between the validation centers (Fig. 4D−G). Similarly, the PCA and OPLS-DA plots of GC‒MS-based metabolomics revealed no apparent separations but rather overlapped (Fig. 4H–K, Figure S2 B–E). The PLS-DA plots showed the similar trends of OPLS-DA plots (Figure S11). The areas under ROC curve AUROC of total features for benign vs malignant SPNs were around 0.5–0.6, accounting for 88%, indicated that the plasma metabolic profile in GC–MS of benign SPNs can not distinguish well from malignant SPNs.
GC–MS based metabolomics comparison of benign SPNs vs malignant SPNs. A and B Cloud Plot of data set, 1336 features with p-value ≥ 0.05 includes visualization of the retention time, and the mass-to-charge ratio of features in discovery set; C PCA score plots of benign SPNs vs malignant SPNs in discovery set; D–G Cloud plot analysis of benign SPNs vs malignant SPNs in validation sets (D: Center 2, E: Center 3, F: Center 4, G: Center 5); H–K PCA score plots of benign SPNs vs malignant SPNs in validation sets (H: Center 2, I: Center 3, J: Center 4, K: Center 5)
3.3 Lipidomics for SPNs using LC‒MS in the negative ion mode
Using negative ion mode for lipidomics, we discovered that 952 of 5136 metabolic characteristics had significant difference (FDR < 0.05) with the levels of 506 greater in healthy participants than in benign SPNs, while the converse was observed for 446 metabolic features (Figure S3 A and B). PCA and OPLS-DA models showed substantial differences between the healthy participants and patients with benign SPNs (Figure S3 C and D), with an R2Y of 0.872 and a Q2 of 0.508. Combined with VIP > 1.0, 48 differential metabolites were found (Figure S3 E and Table S4). In healthy controls and malignant SPNs, we found that 2075 of 4013 lipidomic features were significant difference (FDR < 0.05), in which the levels of 1187 features were higher in the healthy subjects than in malignant SPNs, while the converse was true for 888 features (Figure S4 A and B). The OPLS-DA model found significant discrimination between the two groups but not the PCA (Figure S4 C and D). R2Y and Q2 was 0.954 and 0.574, respectively. Finally, 100 distinct metabolites were identified using VIP > 1.0 and FDR < 0.05 (Figure S4 E and Table S5). Furthermore, by comparing differential metabolites in healthy participants, benign SPNs and malignant SPNs, we discovered 43 common differential metabolites (Figure S5).
By comparing benign SPNs to malignant SPNs, there was no statistically significant difference in their metabolic characteristics by the Mann‒Whitney-Wilcoxon test with a FDR correction (Fig. 5A and B). Using PCA, total overlap in their phenotypes was found (Fig. 5C). OPLS-DA model also revealed no significant difference between the groups in the discovery phase, with R2Y of 0.645 and Q2 of − 0.324 (Figure S6A). Multicenter samples were used to validate this pattern of nonstatistical significance between the two groups; there was no statistically significant variation in the totality of the metabolic variables between the validation centers (Fig. 5D–G). Similarly, there were no visible separations between Centers 2–5 in the PCA and OPLS-DA plots. (Figs. 5H–K, Figure S6 B–E). The PLS-DA plots showed the similar results of OPLS-DA plots (Figure S12). The AUROC values were around 0.5–0.6, accounting for 92% of total features, indicated that the plasma lipidic profile using LC–MS under negative ion mode of benign SPNs can not separate well from malignant SPNs.
LC–MS based lipidomics under negative ion mode for comparison of benign SPNs vs malignant SPNs. A and B Cloud Plot of data set, 6088 features with p-value ≥ 0.05 includes visualization of the retention time, and the mass-to-charge ratio of features in discovery set; C PCA score plots of benign SPNs vs malignant SPNs in discovery set; D–G Cloud plot analysis of benign SPNs vs malignant SPNs in validation sets (D: Center 2, E: Center 3, F: Center 4, G: Center 5); H–K PCA score plots of benign SPNs vs malignant SPNs in validation sets (H: Center 2, I: Center 3, J: Center 4, K: Center 5)
3.4 Lipidomics for SPNs using LC‒MS in the positive ion mode
Using the positive ion mode, we found that 250 of 2292 metabolic characteristics differed significantly (FDR < 0.05); the levels of 157 metabolites were higher in healthy subjects than in benign SPNs, while the converse was observed for 93 metabolites (Figure S7 A and B). The OPLS-DA showed a significant difference between the healthy participants and patients with benign SPNs, with an R2Y of 0.789 and a Q2 of 0.535. (Figure S7C and D). When VIP > 1.0 was used, 31 distinct metabolites were discovered (Figure S7 E and Table S6). In healthy subjects versus malignant SPNs, we found 633 of 1909 lipidomic features had significant difference (FDR < 0.05). The levels of 373 levels features were higher in the healthy subjects relative to the malignant SPNs, while the opposite was observed for 260 metabolites (Figure S8A and B). The OPLS-DA model demonstrated significant discrimination between the two groups (Figure S8 C and D). The cumulative R2Y and Q2 values were 0.914 and 0.671, respectively. Using the criterion, VIP > 1.0 and FDR < 0.05 revealed 52 differential metabolites (Figure S8 E and Table S7). Furthermore, by comparing differential metabolites in healthy controls vs. benign SPNs and malignant SPNs, we discovered 29 common differential metabolites (Figure S9).
By comparing the metabolic features of benign SPNs with those of malignant SPNs, there were no significant differential features by the Mann‒Whitney-Wilcoxon test with a FDR correction (Fig. 6A and B). Using the PCA model for the discovery phase, there was a total overlap in their phenotypes (Fig. 6C). The OPLS-DA model also showed no significant difference between the groups with R2Y of 0.666 and Q2 of − 0.37 (Figure S10A). To validate this pattern of nonstatistical significance between the two groups, multicenter samples were used. There was no statistically significant difference in the sum of the metabolic parameters between the validation centers (Fig. 6D–G). Similarly, PCA and OPLS-DA plots showed no discriminations for the 4 centers (Figs. 6H–K and S10 B–E). The results of PLS-DA plots were consistent with that of OPLS-DA plots (Figure S13). The AUROC values for 94% of total features is around 0.5–0.6 of benign vs malignant SPNs, indicated that the plasma lipidic profile of benign SPNs using LC–MS under positive ion mode can not differentiate well from malignant SPNs.
LC–MS based lipidomics under positive ion mode for comparison of benign SPNs vs malignant SPNs. A and B Cloud Plot of data set, 2542 features with p-value ≥ 0.05 includes visualization of the retention time, and the mass-to-charge ratio of features in discovery set; C PCA score plots of benign SPNs vs malignant SPNs in discovery set; D–G Cloud plot analysis of benign SPNs vs malignant SPNs in validation sets (D: Center 2, E: Center 3, F: Center 4, G: Center 5); H–K PCA score plots of benign SPNs vs malignant SPNs in validation sets (H: Center 2, I: Center 3, J: Center 4, K: Center 5)
4 Discussion
This study describes a less invasive and comprehensive plasma metabolomics and lipidomics approach for identifying metabolic changes in the pathogenesis and diagnosis of SPNs. In total, 1160 plasma samples were collected from 5 independent centers including 280 healthy volunteers and 880 SPN patients. The metabolic and lipidomic phenotypes of healthy subjects versus benign or malignant SPNs revealed significant pattern differences.
In summary, we found 224 of 1336 metabolites and 231 of 7428 lipids as differential metabolites and lipids (FDR adjusted p < 0.05, VIP > 1). Among them, both the benign and malignant SPN groups shared 63 metabolites and 72 lipids, and their change trends were similar. This indicates that the disease process is characterized by shared metabolic disorders. Based on the outcome of the comprehensive plasma metabolomics, 107 and 160 metabolic features were increased in benign and malignant SPNs, respectively, compared to healthy subjects, while 69 and 119 metabolic features showed a decreasing trend in the same comparison. We also discovered that metabolites associated with arginine and proline metabolism, such as glutamic acid, guanidinoacetate, creatine, proline, and spermidine, were elevated in patients with benign SPNs. Meanwhile, malignant SPNs had higher amounts of glutamic acid, glutathione, glycine, ornithine, spermidine, and guanidinoacetate, all of which are associated with glutathione metabolism. SPN individuals had lower levels of glycerophospholipids, which are known to play a protective role in the energy pathways of different respiratory illnesses such as lung infections and lung cancer (Issaq & Veenstra, 2013). Fatty acids and acylcarnitine, both of which may regulate β‑oxidation, have been found to be abnormally increased in non-small cell lung cancer (Harris et al., 2014). However, in our investigation, the aforementioned lipids were reduced in both benign and malignant SPNs, indicating that SPNs are physiologically different from malignant tumors.
Meanwhile, our findings demonstrate that benign and malignant SPNs are not significantly different but rather share common circulating metabolic and lipidomic signatures. This finding was validated in 5 centers, indicating that the metabolic features of both malignant and benign SPN patients are similar, but significantly different from those of healthy subjects.
In most cases, the malignant nodule is a potentially curable type of lung cancer that is in its early clinical stage (stage I) and biologically less aggressive (Klaveren et al., 2009). Malignant SPNs, on the other hand, reflect advanced lung cancer or metastatic malignancies that have migrated to the lung from other parts of the body, indicating a higher risk of lung cancer malignancy (Gao et al., 2013). The 5-year survival rate for advanced lung cancer remains disappointingly low at 15%, although early detection and treatment might raise this rate to 70–90% (Detterbeck et al., 2017). Previous research (Noreldeen et al., 2020) found that the metabolic levels of amino acids and lipids, as well as their derivatives, are considerably elevated in many forms of lung cancer. In our study, the amounts of amino acids, glycerolipids, and amino acids were reduced. As a result, we concluded that malignant SPNs are most likely an early stage of lung cancer that have little effect on blood circulation. In view of this presumption, the objective of diagnosing and managing operable malignant SPNs should be early identification and surgery, as well as preventing cancer cells from entering blood circulation.
The main limitation of this study lies in the composition of the study group. The malignant SPN patients (from the discovery and validation stages) were all diagnosed with stage I lung cancer. Thus, SPN patients at various stages of cancer should be recruited in future research to validate our findings. Metabolomic and lipidomic profoles of patients with malignant SPNs at other stages of lung cancer (stages II, III or IV) and benign SPNs need to be investigated by GC‒MS together with LC‒MS. The identified differential metabolites from the comparison of benign SPNs vs. healthy subjects and malignant SPNs vs. healthy subjects could be further validated using different cohorts. Finally, serum-plasma-tissue matched samples may be used for in-depth and comprehensive analysis.
5 Conclusions
We have reported a comprehensive plasma-based metabolomics and lipidomics profiles of benign and malignant solitary pulmonary nodules using GC‒MS and LC‒MS. Our findings indicate that the early metabolic alterations can distinguish SPNs from healthy controls, but not between benign and malignant SPNs.
Abbreviations
- GC–MS:
-
Gas chromatography-mass spectrometry
- LC–MS:
-
Liquid chromatography-mass spectrometry
- OPLS-DA:
-
Orthogonal partial least-squared discriminant analysis
- PCA:
-
Principal components analysis
- QC:
-
Quality control
- SPNs:
-
Solitary pulmonary nodules
- VIP:
-
Variable importance in the projection
References
Au-Yong, I. T. H., Hamilton, W., Rawlinson, J., & Baldwin, D. R. (2020). Pulmonary nodules. BMJ, 371, m3673. https://doi.org/10.1136/bmj.m3673
Baharum, S.N.; Azizan, K.A. (2018) Metabolomics in systems biology. In: Wan Mohd Aizat, Hoe-Han Goh, Syarul Nataqain Baharum (eds), Omics Applications for Systems Biologyed. Springer, cham https://doi.org/10.1007/978-3-319-98758-3_4
Calderon-Santiago, M. (2021). MetaboQC: Normalize metabolomic data using QC signal. r-project.org/web/packages/MetaboQC/MetaboQC.pdf.
Cruickshank, A., Stieler, G., & Ameer, F. (2019). Evaluation of the solitary pulmonary nodule. The Internal Medicine Journal, 49, 306–315. https://doi.org/10.1111/imj.14219
Detterbeck, F. C., Boffa, D. J., Kim, A. W., & Tanoue, L. T. (2017). The eighth edition lung cancer stage classification. Chest, 151(1), 193–203.
Dunn, W. B., Broadhurst, D., Begley, P., Zelena, E., Francis-McIntyre, S., Anderson, N., et al. (2011). Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nature Protocol, 6, 1060–1083. https://doi.org/10.1038/nprot.2011.335
Fahrmann, J. F., Grapov, D., DeFelice, B. C., Taylor, S., Kim, K., Kelly, K., et al. (2016). Serum phosphatidylethanolamine levels distinguish benign from malignant solitary pulmonary nodules and represent a potential diagnostic biomarker for lung cancer. Cancer Biomarkers, 16, 609–617. https://doi.org/10.3233/CBM-160602
Gao, L., Wen, Z., Wu, C., Wen, T., & Ong, C. N. (2013). Metabolic profiling of plasma from benign and malignant pulmonary nodules patients using mass spectrometry-based metabolomics. Metabolites, 3, 539–551. https://doi.org/10.3390/metabo3030539
Gould, M. K., Tang, T., Liu, I. L., Lee, J., Zheng, C., Danforth, K. N., et al. (2015). Recent trends in the identification of incidental pulmonary nodules. American Journal of Respiratory and Critical Care Medicine, 192, 1208–1214. https://doi.org/10.1164/rccm.201505-0990OC
Harris, F. T., Rahman, S. M., Hassanein, M., Qian, J., Hoeksema, M. D., Chen, H., et al. (2014). Acyl-coenzyme A-binding protein regulates Beta-oxidation required for growth and survival of non-small cell lung cancer. Cancer Prevention Research, 7, 748–757. https://doi.org/10.1158/1940-6207.CAPR-14-0057
Horeweg, N., van Rosmalen, J., Heuvelmans, M. A., van der Aalst, C. M., Vliegenthart, R., Scholten, E. T., et al. (2014). Lung cancer probability in patients with CT-detected pulmonary nodules: A prespecified analysis of data from the NELSON trial of low-dose CT screening. The Lancet Oncology, 15, 1332–1341. https://doi.org/10.1016/S1470-2045(14)70389-4
Issaq, H. J., & Veenstra, T. D. (Eds.). (2013). Proteomic and metabolomic approaches to biomarker discovery. Cambridge: Elsevier Academic Press.
Li, W. W., Shan, J. J., Lin, L. L., Xie, T., He, L. L., Yang, Y., et al. (2017). Disturbance in plasma metabolic profile in different types of human cytomegalovirus-induced liver injury in infants. Scientific Reports, 7, 15696. https://doi.org/10.1038/s41598-017-16051-8
Nasim, F., & Ost, D. E. (2019). Management of the solitary pulmonary nodule. Current Opinion in Pulmonary Medicine, 25, 344–353. https://doi.org/10.1097/01.mcp.0000130322.11513.c8
Nicholson, J. K., & Lindon, J. C. (2008). Systems biology: Metabonomics. Nature, 455, 1054–1056. https://doi.org/10.1038/4551054a
Noreldeen, H. A. A., Liu, X., & Xu, G. (2020). Metabolomics of lung cancer: Analytical platforms and their applications. Journal of Separation Science, 43, 120–133. https://doi.org/10.1002/jssc.201900736
Ost, D., Fein, A. M., & Feinsilver, S. H. (2003). The solitary pulmonary nodule. The New England Journal of Medicine., 348, 2535–2542. https://doi.org/10.1056/NEJMcp012290
Tsugawa, H., Cajka, T., Kind, T., Ma, Y., Higgins, B., Ikeda, K., et al. (2015). MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nature Methods., 12, 523–526. https://doi.org/10.1038/nmeth.3393
van Klaveren, R. J., Oudkerk, M., Prokop, M., Scholten, E. T., Nackaerts, K., Vernhout, R., et al. (2009). Management of lung nodules detected by volume CT scanning. The New England Journal of Medicine, 361, 2221–2229. https://doi.org/10.1056/NEJMoa0906085
Wehrens, R., Hageman, J. A., van Eeuwijk, F., Kooke, R., Flood, P. J., Wijnker, E., et al. (2016). Improved batch correction in untargeted MS-based metabolomics. Metabolomics, 12, 88. https://doi.org/10.1007/s11306-016-1015-8
Wishart, D. S. (2016). Emerging applications of metabolomics in drug discovery and precision medicine. Nature Reviews Drug Discovery, 15, 473–484. https://doi.org/10.1038/nrd.2016.32
Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant No. 81803694, 81904254), Fundamental Research Funds for the Central Universities (Grant No. 2632020ZD07); Natural Science Foundation of Jiangsu Province, China (Grant No. BK20190808); Natural Science Foundation of the Higher Education Institutions of Jiangsu Province, China (Grant No. 19KJB360002); Young Scientists Fund of the National Natural Science Foundation of Nanjing university of Chinese medicine (NZY81904254); Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), No. 87 [2018]; the Open Projects of the Discipline of Chinese Medicine of Nanjing University of Chinese Medicine Supported by the Subject of Academic priority discipline of Jiangsu Higher Education Institutions (Grant No. ZYX03KF52). The authors also thank Dr. Raphael N. Alolga from China Pharmaceutical University and American Journal Experts (an editing company) for the editorial services rendered.
Funding
National Natural Science Foundation of China,81803694,81904254,Fundamental Research Funds for the Central Universities,2632020ZD07,Natural Science Foundation of Jiangsu Province,BK20190808,Natural Science Foundation of the Higher Education Institutions of Jiangsu Province,19KJB360002,Young Scientists Fund of the National Natural Science Foundation of Nanjing university of Chinese medicine,NZY81904254,Priority Academic Program Development of Jiangsu Higher Education Institutions,87 [2018],the Open Projects of the Discipline of Chinese Medicine of Nanjing University of Chinese Medicine Supported by the Subject of Academic priority discipline of Jiangsu Higher Education Institutions, ZYX03KF52.
Author information
Authors and Affiliations
Contributions
WZ, L-yJ, LL, J-lW, JY, L-cD, Z-hZ and J-jS: conceptualization, Methodology and Writing- Original draft preparation. W-cX, YZ, M-jW, X-mC and H-qL: Data curation and Resources. WZ and J-jS: Investigation and Supervision. WZ and J-jS: Writing- Reviewing and Editing.
Corresponding authors
Ethics declarations
Conflict of interest
No potential conflict of interest declared.
Ethical approval
The Affiliated Jiangyin Hospital of Southest University Medical College approved this study (2017-036), in accordance with the Declaration of Helsinki, approved use of the patient plasma samples. Patients gave written informed consent before enrolment.
Data availability
The mass spectrometry data have been deposited to the Metabolomics Workbench (https://www.metabolomicsworkbench.org/). The study named ST001937. Reviewer account details: Username: jshan12345678. Password: Shan85811329 (http://dev.metabolomicsworkbench.org:22222/data/DRCCMetadata.php?Mode=Study&StudyID=ST001937). Further details and other data that support the findings of this study are available from the corresponding author upon request.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wei Zhou, Lili Lin, Lian-yong Jiang and Jin-long Wu are the co-first authors.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhou, W., Lin, L., Jiang, Ly. et al. Comprehensive plasma metabolomics and lipidomics of benign and malignant solitary pulmonary nodules. Metabolomics 18, 71 (2022). https://doi.org/10.1007/s11306-022-01929-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-022-01929-0