Abstract
Background
Inflammatory bowel disease is a group of pathologies characterised by chronic inflammation of the intestine and an unclear aetiology. Its main manifestations are Crohn’s disease and ulcerative colitis. Currently, biopsies are the most used diagnostic tests for these diseases and metabolomics could represent a less invasive approach to identify biomarkers of disease presence and progression.
Objectives
The lipid and the polar metabolite profile of plasma samples of patients affected by inflammatory bowel disease have been compared with healthy individuals with the aim to find their metabolomic differences. Also, a selected sub-set of samples was analysed following solid phase extraction to further characterise differences between pathological samples.
Methods
A total of 200 plasma samples were analysed using drift tube ion mobility coupled with time of flight mass spectrometry and liquid chromatography for the lipid metabolite profile analysis, while liquid chromatography coupled with triple quadrupole mass spectrometry was used for the polar metabolite profile analysis.
Results
Variations in the lipid profile between inflammatory bowel disease and healthy individuals were highlighted. Phosphatidylcholines, lyso-phosphatidylcholines and fatty acids were significantly changed among pathological samples suggesting changes in phospholipase A2 and arachidonic acid metabolic pathways. Variations in the levels of cholesteryl esters and glycerophospholipids were also found. Furthermore, a decrease in amino acids levels suggests mucosal damage in inflammatory bowel disease.
Conclusions
Given good statistical results and predictive power of the model produced in our study, metabolomics can be considered as a valid tool to investigate inflammatory bowel disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Inflammatory bowel disease (IBD) is a group of pathologies characterised by a chronic phlogosis, and a not specified etiology (Baumgart and Carding 2007). The main clinically defined forms of IBD are Crohn’s disease (CD) and ulcerative colitis (UC) (Kaser et al. 2010; Kumar and Clark 2016). The incidence of IBD fluctuates from country to country but the main two typologies affect 1.5 million Americans, 2.2 million Europeans, and several hundred thousands more worldwide (Kumar and Clark 2016; Cosnes et al. 2011). Diagnosis of IBDs is particularly challenging, as other diseases causing similar signs and symptoms need to be excluded first through a combination of tests, but the ultimate diagnostic tool remains endoscopic examination coupled with biopsies. Furthermore, discrimination between the two manifestations, UC and CD, is particularly complicated given the similarity of the symptoms, resulting in 10–15% of cases lacking a defined diagnosis (undefined colitis) (Kumar and Clark 2016). Different causative agents have been proposed in the past for CD and UC diagnosis. One of these theories is based on the T-cell pathway, that proposes the idea that T cells activation is due to a confluence of genetic and environmental factors that causes an immune imbalance, ending with the characteristic inflammation, prerogative of these diseases (Maul et al. 2005). Moreover, change in the intestinal flora, has been suggested as a crucial factor that can trigger the emergence of IBD (Weinstock et al. 2002; Summers et al. 2003). However, it is clear that most of the symptoms that occur during the progression of disease are common for both pathologies.
Metabolomics and lipidomics could both represent useful tools in elucidating the mechanisms and suggesting diagnostic and therapeutic approaches for IBDs. Metabolomics is the study of metabolites in a biofluid, cell, tissue or organism to understand the mechanism behind the response to a given stimulus/condition. By extension lipidomics aims to understand how lipid metabolism is perturbed by a biological stimulus. A number of studies have recently used metabolomics and lipidomics to examine IBD, demonstrating differences in the metabolic profile of IBD patients when compared with healthy individuals (Lu et al. 2012) or, in a smaller number of cases, discriminating between IBD subtypes (Storr et al. 2013; Lin et al. 2011). Lipid species are also altered in these diseases, with Fan et al. (2015) demonstrating that triaclyglycerol and cholesterol levels were significantly higher and lower in CD when compared with UC, respectively. These alterations of lipid profiles, including suggested changes in plasmalogens may contribute to the pathogenesis of IBD (Balasubramanian et al. 2009). Moreover, Agouridis et al. (2011) highlighted lower total cholesterol and high density lipoprotein cholesterol levels in IBD samples when compared with healthy individuals, while low density lipoprotein cholesterol levels were higher when compared with healthy patients. Santoru et al. 2017 have reported higher levels of several lipids like diacylglycerol (18:0/22:2), diacylglycerol (16:0/18:2) and n-acylphosphatidylethanolamines (18:1/16:1/18:0) in IBD patient when compared with healthy individuals, while urobilin, phosphatidylcholine (16:0/3:1), urobilinogen, phosphatidic acid (19:0/16:1), phosphatidylserine (22:2/18:0), phosphatidylcholine (22.2/14:1) and ceramide (18:1/22:0) were decreased. Recently, Scoville et al. (2018), studying the metabolomic profile of the serum of IBD patients, showed variations in a number of lipid, amino acid, and tricarboxylic acid cycle related in IBD patients: for example deoxycholate, sphingomyelin, glycochenodeoxycholate glucuronide, taurolithocholate 3-sulfate, dehydroepiandrosterone sulfate, as well as long chain, polyunsaturated, branched chain, and monohydroxy fatty acids were significantly decreased in CD when compared with healthy individuals. Also several compound related to the tricarboxylic acid cycle, like citrate, aconitate, α-ketoglutarate, succinate, fumarate and malate were found decrease in CD and UC when compared with healthy individuals.
A major challenge in the field of lipidomics is the identification of individual lipid species, in part reflecting the similar chemical structure across the class of compounds as well as the high proportion of isobaric species. To address these issues, several analytical methods have been proposed including tailored condensed phase separations coupled to MS or tandem mass spectrometry strategies (Han and Gross 2005, Fahy et al. 2007; Sud et al. 2006, Niemela and; Sjöström 1986). In this respect ion mobility-mass spectrometry (IM-MS) has been suggested as one such technique that could improve the separation of a wide range of lipid species (Kliman et al. 2011; Hinz et al. 2018). In addition, drift tube ion mobility (DTIM) allows calculation of the ion’s collision cross section (CCS), a physicochemcal parameter, when travelling through an inert gas such as nitrogen. In this study, with the aim to create a metabolomic fingerprinting profile for the diagnosis of IBD, metabolomic variations of the compound’s levels in CD, UC and healthy plasma samples were investigated by either ultra high performance liquid chromatography coupled with a DTIM time of flight mass spectrometer for lipidomics, or a high performance liquid chromatography coupled with a triple quadrupole mass spectrometer system for polar metabolomics.
2 Materials and methods
2.1 Patients
Patients were recruited at the Department of Gastroenterology of the Policlinico Universitario di Monserrato (CA), Italy. All patients were fasted from 8 to 12 h before the blood test. The whole blood samples were collected into tubes containing ethylenediaminetetraacetic acid to avoid blood coagulation. Samples were then centrifuged to obtain the plasma fraction which was transferred into an eppendorf tube, delivered to the laboratory within 3 h and stored at − 80 °C until use. All clinical information of patients are reported in supplementary materials (Table S1). Briefly, the 78 UC patients (31 females, 47 males) showed ranges of age and body mass index (BMI) of 20–76 and 15.7–35.6, respectively (averages of age and BMI were 48 ± 13 and 24 ± 4, respectively), while the same ranges for the 50 CD patients (28 females and 22 males) were 21–78 and 16–31, respectively (averages of age and BMI were 47 ± 12 and 24 ± 4, respectively). Furthermore, patients had their diagnosis confirmed by endoscopic, histological and radiographic data (Agouridis 2011; Nikolaus 2007; Schroeder 1987; Harvey 1980) resulting in 21 CD and 39 UC samples to be in a quiescent or inactive phase of the disease, 8 CD and 25 UC samples to have a moderate activity, and 21 CD and 14 UC samples to show a severe activity.
Also, patients were under a different course of therapies: 14 CD and 6 UC were under the Adalimumab treatment, 9 CD and 12 UC were under the Azathioprine treatment, 4 CD and 7 UC were using Infliximab, 16 CD and 48 UC were under a traditional therapy (corticosteroids, 5-aminosalicylates), while 7 CD and 5 UC were not under a therapy.
The healthy volunteers (n = 60, 21 females and 39 males) were recruited locally (Sardinia, Italy). These individuals showed ranges of age and body mass index (BMI) of 24–66 and 18.9–31.2, respectively (averages of age and BMI were 40 ± 13 and 23 ± 3, respectively) Exclusion criteria were age above 80 or below 20 years, recent use of antibiotics or probiotics and pregnancy.
2.2 Materials
Methanol, chloroform, dichloromethane, isopropanol, acetonitrile and analytical standards were purchased from Sigma Aldrich (Milan, Italy).
2.3 Internal standards
A mixture of 26 deuterated lipids (methanol solution) containing phosphatidic acid (PA), phosphatidylcholines (PC), phosphatidylethanolamines (PE),phosphatidylinositols (PI), phosphatidylserines (PS), sphingomyelins (SM), ceramides (Cer), triacylglycerols (TAG) and fatty acids (FA) at the conenctration of 28.57 µM was used for the plasma lipid profile analysis (Avanti Polar Lipids, Inc., Alabama 35007-9105).
A mixture of seven standards containing D3-proline, D8-valine, D10-leucine, U-13C lysine, U-13C Glutamic acid, D5-phenylalanine and D4-serotonin (2018 Merck KGaA, Darmstadt, Germany; Cambridge Isotope Laboratories, Inc., Cambridge,United Kingdom), at the concentration of 10 µM, dissolved in a 0.1 M solution of HCl, was used for the plasma polar profile analysis.
2.4 Sample preparation for untargeted lipidomics
Plasma samples were centrifuged at 5139g for 10 min at 4 °C and 400 µL of supernatant was transferred to eppendorf tubes. A modified Folch method was used to extract the lipophilic compounds. Quality control (QC) samples, which contained 20 µL of each plasma sample analysed, were also prepared using the same method. Briefly, 400 µL of each plasma sample was mixed with 600 µL of methanol, 600 µL of chloroform and 175 µL of Milli-Q water and centrifuged at 5139g for 30 min. at 4 °C. The high volume of plasma was chosen with the aim to use a single extraction protocol to analyse also the aqueous metabolic profile designated for the gas chromatography-MS and 1H NMR analysis (experiments from an unpublished study). The lipid chloroform phase was separated and dried under a gentle stream of nitrogen. Samples were suspended in 1 mL of methanol, vortexed for 3 min and sonicated for 5 min. After sonication, 50 µL of the samples was transferred into vials and diluted with 50 µL of internal standard and 75 µL of Chromasol ultrapure water.
2.4.1 Solid phase extraction of the lipid fraction
Ten folch extracts from each class (CD, UC and C) were submitted to chromatographic separation by solid phase extraction (SPE) to obtain three different fractions each containing different lipid classes: neutral lipids, fatty acids and phospholipids. Each class was formed of five males and five females without other pathologies, no smokers and with a normal BMI (18.5–24.9 kg/m2). The dried organic phases from the Folch extracts were reconstituted in 800 µL of chloroform and subjected to ultrasound for 5 min to dissolve and mix the lipids. After sonication, 20 µL of samples was transferred into a vial and diluted with 100 µL of internal standard lipid mixture. The solution was sequentially dried under a gentle stream of nitrogen and then suspended in 800 µL of chloroform. Chromabond aminopropyl-modified silica NH2 (1 mL/100 mg) columns from Macherey–nagel were conditioned with 3 mL of hexane. After conditioning, 800 µL of samples were added and eluted using: 1 mL of chloroform: isopropanol 2:1 (v/v) solution for the neutral lipids extraction; 1 mL of diethyl ether: acetic acid 98:2 (%) solution for the fatty acids and 1 mL of methanol for the phospholipids. All the samples were then dried and suspended with 50 µL of a mixture of isopropanol: acetonitrile: water (2:1:1 v/v).
2.5 Sample preparation for the polar phase analysis of plasma samples
From the plasma sample 20 µL were transferred into an Eppendorf tube and 100 µL of theinternal standard mix solution for the polar metabolites analysis was added. Samples were sonicated for 5 min and centrifuged for 5 min at 5139g. The obtained solution was dried under a gentle stream of nitrogen. Samples were then suspended in 100 µL of a 10 mM ammonium acetate solution, vortexed, sonicated for 5 min and centrifuged 5 min at 5139g. Samples were then transferred to a 96 multi-well plate for LC/MS analysis.
2.6 LC/DTIM-QTOF analysis
An Agilent Infinity II UPLC coupled to a 6560 Ion mobility QTOF was used with an Agilent Jetstream source, operated in the positive and negative ion mode while, the capillary potential was set to 60 V and the needle at 20 kV. High purity Nitrogen (99.999%) was used as a drift gas with a trap fill time and a trap release time of 2000 and 500 µs, respectivelly. Before the analysis, the instrument was calibrate using an Agilent tuning solution at the mass range of m/z 100–1700. Samples were evaporated with Nitrogen at the pressure of 48 mTorr and at the temperature of 375 °C, while an Agilent reference mass mix for mass re-calibration was continuously infused during the run schedule. An Agilent tuning mix solution was injected every ten samples to perform DTCCSN2 re-calibration. Full-scan spectra were obtained in the ranges of 100–1700 amu for the lipid plasma extracts, setting the detector at 1500 V. The organic fractions were analysed using a Waters Acquity CSH C18 column (100 × 2.1 mm 1.7 µm). The mobile phase for positive ionization mode consisted of (A) 10 mM ammonium formate solution in 60% of milliQ water and 40% of acetonitrile and (B) 10 mM ammonium formate solution containing 90% of isopropanol, 10% of acetonitrile. The mobile phase was pumped at a flow rate of 400 µL/min as follows: initially 60% of A, then a linear decrease from 60 to 50% of A in 2 min then at 1% in 5 min staying at this percentage for 1.9 min and then brought back to the initial conditions in 1 min. The Agilent MassHunter LC/MS Acquisition console from The MassHunter suite was used for data acquisition. The mobile phase for negative ionization mode differed only for the use of 10 mM ammonium acetate instead of ammonium formate. Identical DTIM parameters were used for the SPE extracts analysis, while the mobile phase was pumped at a flow rate of 400 µL/min using the following gradient: initially 60% of A, then a linear decrease to 50% of A in 2.10 min and decreased to 30% of A in 10 min and to 1% of A in 6 min maintaining these condition for 0.1 min, back to the initial conditions in 2 min. Identifications of the most important compounds were confirmed by the use of a targeted MS/MS using different collision energies (20, 30 and 40 V) and comparing the fragmentation pattern with the LIPID MAPS and METLIN libraries (The Scripps Research Institute, La Jolla, CA). An overlay of the extracted ion chromatograms of all the identified species is shown in Fig. 1, while their retention times, DTCCSN2 and fragmentations are reported in Table 1.
2.7 LC/MS/MS QqQ analysis of plasma samples
Samples were analysed by a Thermo scientific UHPLC + series coupled with a TSQ Quantiva mass spectrometer (Thermos Fisher Scientific, Waltham, Massachusetts, United States). The electrospray voltage was set to 3500 V for positive ionisation mode and to 2500 V for the negative ionisation mode. The aqueous phases were analysed with an ACE Excel 2 C18 PFP (100A. 150 × 2.1 mm 5 µ) column. The mobile phase contained 0.1% of formic acid water solution as solvent A and 0.1% of A formic acid acetonitrile solution as solvent B. The mobile phase was pumped at a flow rate of 500 µL/min initially at 100% of A for 1.60 min, then a linear decrease from 100 to 70% of A in 2.4 min and to 10% in 0.5 min, then constant for 0.5 min and brought back to initial condition after 0.1 min. The Xcalibur software (Thermos Fisher Scientific, Waltham, Massachusetts, United States) was used for data acquisition. Putative recognition of all detected metabolites was performed using a targeted MS/MS analysis and relative comparisons of fragmentations with analytical standards. Calculated masses and mass fragments of the measured compounds are reported in supplementary materials (Table S2).
2.8 Data processing
Data acquired with the Agilent 6560 DTIM Q-TOF LC-MS in LC/DTIM-QTOF mode was pre-processed with a set of software from the MassHunter Workstation suite (Agilent Technologies, Santa Clara, USA) to perform mass re-calibration, DTCCSN2 re-calibration, and deconvolution, yielding a matrix containing all features present across all samples. This matrix was further processed with KniMet (Liggi et al. 2017, 2018), a pipeline based on the KNIME analytic platform (Berthold et al. 2007) for the post-processing of metabolomics MS-based data. Features were filtered based on their presence in blanks and QC samples (threshold = 40%) (Dunn et al. 2011, 2012). The remaining features were subjected to missing values imputation with Random Forest and Probabilistic Quotient Normalisation (Dieterle et al. 2006) based on QCs. Finally, data was annotated based on accurate mass match with the LIPID MAPS database (Fahy et al. 2007; Schmelzer et al. 2007).
2.9 Multivariate statistical data analysis
All the multivariate statistical analysis (MVA) were performed using the SIMCA software 14.0 (Umetrics, Umeå, Sweden). Principal component analysis (PCA) was used along Hotelling’s T2 and DmodX tests to evaluate distribution of the samples and evaluate the presence of any possible outliers. This was followed by partial least square-discriminant analysis (PLS-DA) with its orthogonal extension (OPLS-DA), which was used as classificatory model to visualize and evaluate the differences between samples. Statistical power of the models, as well as the possible presence of overfitting, were evaluated by considering the R2X, R2Y and Q2 parameters along with permutation test and cross validation analysis. From the PLS-DA, the variable importance in the project (VIP) and the coefficient analysis were used to provide, respectively, the contribution of the components to the separation of sample groups and the expression of how strongly the variable class is correlated to the systematic part of each of the metabolites. Further, the discriminant compounds highlighted by the VIP and loadings analyses were compared with those found in the S-plot from the OPLS-DA that combines the modelled covariance and correlation in a scatter plot (Eriksson 2013).
2.10 Univariate statistical analysis
GraphPad Prism software (version 7.01, GraphPad Software, Inc., CA, USA) was used to perform univariate statistical analysis of the data. To verify the significance of metabolites obtained using multivariate statistical analysis, a Mann–Whitney U test was performed (Ruxton 2006). For the analysis of the samples subjected to SPE extraction, all the univariate statistical results were obtained using a one way non parametric test with the Bonferroni correction (Steinbach et al. 1994). Variables with a p value lower than 0.05 were considered statistically significant.
3 Results and discussion
3.1 Untargeted lipid analysis of IBD samples revealed changes in the levels of phospholipids
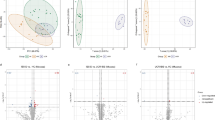
The lipid profile of IBD and healthy samples was studied by LC/DTIM-QTOF. Data processing yielded 1038 (498 annotated as lipids) and 607 (104 annotated as lipids) features for positive (PIA) and negative ionization analysis (NIA), respectively, which were subjected to MVA. The extract composition consists of lipids from the following classes: 7 free FA, 23 lysophosphatidylcholines (LPC), 106 PC, 45 PS, 10 PE, 92 diacylglycerols (DAG), 26 cholesteryl esters (CE), 49 SM, 10 Cer and 234 TAG. The unsupervised analysis of both (PIA) (PCA R2X = 0.69, Q2 = 0.44) and (NIA) (PCA R2X = 0.5 and Q2 = 0.4) ion mode features did not indicate any clusters related to gender, age, BMI, therapies, state of disease and localization of inflammation associated with the samples (Fig. S1). Also, trying to forcing classifications through the use of a supervised analysis no one of the models based on the above cited classifications showed significant statistical result.DmodX and Hotelling’s T2 analysis showed 7 outliers for PIA, which were removed from the subsequent analysis, while no outliers were identified for NIA. Three different OPLS-DA were performed: CD vs C, UC vs C and CD vs UC. Models displayed a clear separation between healthy subjects and both pathological classes in the two-way comparisons (Fig. 2, OPLS-DA model for PIA data of CD and healthy patients, R2Y = 0.67, Q2 = 0.51 (A); OPLS-DA model for PIA data of UC and healthy patients R2Y = 0.71, Q2 = 0.33 (B); both models passed permutation test with Q2 of − 0.48 and − 0.14, respectively). For the NIA, OPLS-DA also discriminated between the healthy controls and both disease groups (Fig. 2, OPLS-DA model for NIA of CD and healthy patients, R2Y = 0.7, Q2 = 0.7 (C), OPLS-DA model of NIA of UC and healthy patients, R2Y = 0.7, Q2 = 0.5 (D); both models passed permutation test with Q2 = − 0.20 and − 0.31, respectively). In contrast, the comparison between the two pathological classes did not show separation between groups for both ionization modes (data not shown). The most discriminant compounds highlighted by the VIP list, the loadings and the S-plot where subjected to MS/MS analysis and cross-referenced to the LIPID MAPS and METLIN (The Scripps Research Institute, La Jolla, CA) libraries to further confirm their identities (Table 1; Fig. 3.) Additional confidence in these identifications was obtained by comparing the DTCCSN2 values of these compounds with those available in recent literature (Paglia et al. 2015; Stow et al. 2017; Zhou et al. 2017; Kyle et al. 2018), resulting in an average relative standard deviation of 0.6% (Table S3). The statistical significance of the discriminant compounds from the MVA analyses were confirmed by the Mann–Whitney test. Regarding the results obtained for CD samples in PIA, LysoPC 18:0, LysoPC 18:1, LysoPC 18:2, PC 18:0/18:1, PC 18:1/18:1, PC 18:2/18:2 and PC 18:3/20:4 were decreased in CD samples, while PC16:0/22:6, PC18:0/22:6, PC 18:3/20:3, SM 16:1/20:1, TAG 18:1/18:2/20:4 and TAG 16:0/18:1/18:2 were increased in CD samples. Similar results were obtained for UC samples in PIA where LysoPC 18:1, LysoPC 18:2, PC 18:0/18:1, PC 18:1/18:1, PC 18:2/18:2, PC 18:3/20:4, TAG 14:0/16:0/18:2 and TAG 16:0/18:0/18:1 were decreased for UC samples, while PC 16:0/20:4, PC 16:0/22:6, SM 18:2/24:0 and TAG 18:1/18:2/20:4 were increased in UC samples. On the other hand, analysis of the NIA data showed that PC 18:1/18:2, PC 16:0/22:6 and PC 18:0/18:2 were decreased in CD samples, while free fatty acid including oleic acid, linoleic acid, palmitic acid and palmitoleic acid were increased in CD and UC samples. Only PC 18:1/18:2 was decreased in UC samples (Fig. 4, Table S4).
Scores plot from the LC/DTIM-QTOF analysis. a Scores plot from the PIA OPLS-DA of CD samples (red filled circles) vs control samples (grey filled circles): R2Y = 0.7, Q2 = 0.5; b Scores plot from the PIA OPLS-DA of UC samples (black filled circles) vs healthy patient samples (grey filled circles). R2Y = 0.7 Q2 = 0.3. c Scores plot from the NIA OPLS-DA of CD samples (red filled circles) vs control samples (grey filled circles): R2Y = 0.70 Q2 = 0.5; d Scores plot from the NIA OPLS-DA of UC samples (black filled circles) vs control samples (grey filled circles): R2Y = 0.7 Q2 = 0.5
Among the most discriminant compounds, LysoPC (18:0, 18:1 and 18:2) were found to be decreased in pathological samples. The major pathway for the production of LysoPC, such as 18:0, 18:1 and 18:2, all of which decreased in the disease state, is the hydrolysis of fatty acids from PCs by phospholipases such as phospholipase A2 (PLA2). PLA2 has been reported to be relevant during different inflammatory processes, including IBD pathogenesis (Schaloske et al. 2006), and the expression of its activator antigen was found to be increased in the monocytes and granulocytes of IBD intestinal sections (Peterson et al. 1996). In addition, one of our previous studies showed a modification of the PLA2 metabolic pathways in IBD faecal samples (Santoru et al. 2017). Therefore, the decrease in LysoPC in plasma derived from IBD patients observed in this study may suggest a higher PLA2 activity. In addition, levels of saturated and monounsaturated PCs containing 18:0, 18:1 and 18:2 fatty acids were decreased in plasma samples of UC and CD patients, where increased levels of PCs containing polyunsaturated fatty acids such as arachidonic and docosahexaenoic acid were also found. As these fatty acids represent substrates for the generation of eicosanoids and other oxylipins, both of which are important during inflammatory responses, this result may indicate a higher demand for these lipids in IBD patients. In support, Ehehalt et al. (2004) also highlighted an increase of PC and LysoPC (PC 16:0/18:1, PC 16:0/18:2, PC 18:0/18:1, PC 18:0/18:2, LysoPC 16:0 and LysoPC 18:0) in the rectal mucus of UC patients compared to CD and healthy individuals. On the other hand, some PC levels were found decreased in the pathological samples when compared with controls, and this trend can be explained by a change of the lipid composition in the intestinal mucosa as already suggested by Bischoff et al. (2014).
3.2 Class-specific analysis of lipids showed changes in the lipid profile
Analysis of the lipidome did not highlight any difference between CD and UC samples, which could be related to the high intra-class variability as well as the similarity between these two pathological conditions which could reflect also in their lipid profile. To increase the class specificity of our untargeted approach and provide a more in-depth analysis of potential differences between the UC and CD, we performed SPE on a sub-set of the samples matched for age, gender, BMI and therapy. This separated lipids into three fractions, fatty acids, neutral lipids and phospholipids prior to LC-MS analysis.
The LIPID MAPS database was used to annotate compounds based on accurate mass match, resulting in 98 TAG, 44 glycerophosphocolines, 43 free FA, 28 SM, 42 DAG, 32 Cer, 11 LysoPC and 9 CE. One-way ANOVA test was performed for the 3 classes of samples CD, UC and C by expressing each lipid as the ratio of the total signal for that particular class of lipids and applying a Bonferroni correction. Statistical analysis of the results from SPE of the phospholipid fraction showed an increase in PCs in UC samples when compared with CD samples, while unsaturated PCs were decreased in CD samples when compared with UC. Furthermore, saturated DAGs were increased in samples from CD patients compared to UC. MVA of this fraction did not show any outliers or clusters (PCA R2X = 0.50, Q2 = 0.3), and OPLS-DA comparing IBD samples vs healthy individuals reported the same results of the untargeted analysis described above (data not shown). However, unlike the analysis of the untargeted lipidomics data, the model comparing CD vs UC for the phospholipid fraction reported good validation parameters (R2Y = 0.7, Q2 = 0.3, and passed permutation test). Twelve lipids were identified as significantly increased by both multivariate statistics and the univariate Mann–Whitney test for UC samples when compared to healthy individuals, while only one was increased in CD samples when compared with UC. For the UC group the identified compounds were: PC18:2/18:0, PC18:1/18:0, PC22:5/16:0, PC20:3/18:0, PC16:0/18:2, PC16:0/18:1, PC 18:1/18:1, PC16:0/20:3 and PC16:1/16:0 (Table 2). From the analysis of the neutral lipid fraction, the only category that was found significantly altered was CE, which was increased in UC compared with CD. At last, the analysis of the fatty acid fraction showed a significant increase of unsaturated FA in CD compared to controls (Fig. 5). MVA was performed also for the neutral lipid and FA fractions, however, no significant differences were found comparing CD vs UC (data not shown). Among these findings, variations in CE levels can be correlated with a change in the lipoprotein system (Bruce et al. 1998). In fact, lipoprotein levels are known to be affected by a range of physiological and pathological conditions (Ansell et al. 2003), and these findings were confirmed by a study of Biyyani et al. (2010) where, in a lipoprotein profile comparison between IBD patients and controls, lower total cholesterol and high-density lipoprotein cholesterol levels were detected in IBD patients. Furthermore hypocholesterolaemia is a common symptom in patients with various types of acute disease, including surgery, trauma, burn injury and sepsis, with it suggested that chemokine action might drive this variation (Fraunberger et al. 2000). Chemokines are chemotactic cytokines that stimulate leukocytes migration to areas of inflammation and lead cell activation events (Banks et al. 2003). It has been reported that the local inflammation and tissue damage in UC and CD is caused by a local expression of specific chemokines in IBD tissues (Banks et al. 2003). Further experiments are recommended to understand why the levels of CEs were increased in UC when compared with CD.
Graphs indicating the levels of lipid categories from the SPE plasma samples examined by one-way ANOVA test between CD vs UC vs C samples with a Bonferroni correction. Variable intensities are shown in the y-axis. *, **, and *** indicates levels of significance with a p value < 0.05, < 0.01, < 0.001, respectively
3.3 Alterations in the amino acid metabolism
Fifty-five polar compounds from 166 plasma samples were identified and measured from the LC/MS/MS analysis. Data was processed by first dividing the detected area of the most abundant ion by that of the related ion from the internal standard. Group means for these ratios were compared using the Mann–Whitney U test. Overall, 16 and 21 compounds were found to be significantly changed when samples from the CD vs healthy groups and UC vs healthy groups were compared, respectively. β-amino isobutyric acid, α-hydroxy butyric acid, histidine, creatine, isocitric acid and citric acid levels were increased in CD samples, while alanine, betaine, citrulline, threonine, hydroxyproline, proline, valine, methionine, tyrosine, methionine sulfoxide where decreased in CD when compared with healthy samples(Fig. 2S). For the UC group cysteine, β-amino isobutyric acid, AMP, α-hydroxybutyric, cAMP, creatine, isocitric acid, GMP, citric acid, uridine, GABA, β-hydroxy isovaleric acid and ketoleucine were increased, while arginine, alanine, betaine, citrulline, threonine, proline, methionine, tyrosine and tryptophan were decreased when compared with healthy samples (Figs. 3S, 4). The levels of cAMP, found to be increased in UC are consistent with the results of Moore et al. (1995), who described how the production of cAMP in leucocytes is due to 3-adrenergic catecholamines, histamine and the E series prostaglandins by a receptor coupled activation of adenylate cyclase. In vitro studies have also reported that the agents responsible for the increase of cAMP levels were implicated in the reduction of several inflammatory metabolic pathways, including the release of histamine as well as leukotrienes and the proliferation of lymphocytes (Harvarth 1991, Rivkin et al. 1977). Moreover, a profound decrease of amino acid levels was found in IBD patients, confirming the results already reported by Hisamatsu et al. (2012) that suggested a multivariate analysis comprising plasma amino acid profiles as a novel, non-invasive approach for the diagnosis and monitoring of IBD. Similarly, Scoville et al. (2018) reported alteration on the levels of several amino acids in the serum of CD subjects compared to both control and UC samples, while no variation occurred when comparing UC with control samples. These amino acid changes in IBD samples could be linked to the mucosa damage, characteristics of these disease, and is corroborated by the decreased plasma levels of citrulline, a marker of extensive mucosal barrier injury of the small intestine (Blijlevens et al. 2004). Moreover, these amino acid variations could reflect the use of proteins as a catabolic energy source in the inflammatory state (Hong et al. 2009). In fact, T and B cells utilize amino acids, glucose and lipids as energy sources during oxidative phosphorylation and mitogenic stimulation (Kominsky et al. 2010.). In addition, it has been noticed how a specific protein intake in IBD patients can cause changes in the intestinal health by modulating the quantity and the nature of both amino acids absorbed and undigested proteins delivered to the large intestine (Vidal-Lletjós et al. 2017).
4 Conclusions
In this study, both metabolomics and lipidomics provided good classification not only between healthy and diseased samples but also in separating samples from UC and CD patients. From this study, the most discriminatory metabolites associated with UC and CD were several amino acids such as arginine, alanine, betaine, citrulline, threonine, proline, methionine, tyrosine and tryptophan and several lysophosphatidylcholines (18:2, 16:0, 18:1, 18:0), phosphatidylcholines (36:4, 38:7, 34:3, 34:2, 36:2, 38:6, 40:6, 42:2, 32:1, 38:5, 36:3,, 36:1, 34:1, 38:3) triacylglycerol (56:7, 52:3, 52:1) and free fatty acids (16:0, 18:2, 24:2, 16:1, 18:0, 22:1, 24:3, 18:1). However, to validate these potential biomarkers would require a validation cohort of comparable size and further consideration of any confounding factors.
References
Agouridis, A. P., Elisaf, M., & Milionis, H. J. (2011). An overview of lipid abnormalities in patients with inflammatory bowel disease. Annals of Gastroenterology, 24(3), 181–187.
Ansell, B. J., Navab, M., Hama, S., Kamranpour, N., Fonarow, G., Hough, G., et al. (2003). Inflammatory/antiinflammatory properties of high-density lipoprotein distinguish patients from control subjects better than high-density lipoprotein cholesterol levels and are favorably affected by simvastatin treatment. Circulation, 108(22), 2751–2756.
Balasubramanian, K., Kumar, S., Singh, R. R., Sharma, U., Ahuja, V., Makharia, G. K., et al. (2009). Metabolism of the colonic mucosa in patients with inflammatory bowel diseases: An in vitro proton magnetic resonance spectroscopy study. Magnetic Resonance Imaging, 27(1), 79–86.
Banks, C., Bateman, A., Payne, R., Johnson, P., & Sheron, N. (2003). Chemokine expression in IBD. Mucosal chemokine expression is unselectively increased in both ulcerative colitis and Crohn’s disease. The Journal of Pathology, 199(1), 28–35.
Baumgart, D. C., & Carding, S. R. (2007). Inflammatory bowel disease: Cause and immunobiology. The Lancet, 369(9573), 1627–1640.
Berthold, M. R., & Hand, D. J. (2007). Intelligent data analysis: An introduction. Springer, Berlin.
Bischoff, S. C., Barbara, G., Buurman, W., Ockhuizen, T., Schulzke, J. D., Serino, M., et al. (2014). Intestinal permeability—a new target for disease prevention and therapy. BMC Gastroenterology, 14(1), 189.
Biyyani, R. S. R. S., Putka, B. S., & Mullen, K. D. (2010). Dyslipidemia and lipoprotein profiles in patients with inflammatory bowel disease. Journal of Clinical Lipidology, 4(6), 478–482.
Blijlevens, N. M. A., Lutgens, L. C. H. W., Schattenberg, A. V. M. B., & Donnelly, J. P. (2004). Citrulline: A potentially simple quantitative marker of intestinal epithelial damage following myeloablative therapy. Bone Marrow Transplantation, 34(3), 193.
Bruce, C., Chouinard, R. A. Jr., & Tall, A. R. (1998). Plasma lipid transfer proteins, high-density lipoproteins, and reverse cholesterol transport. Annual Review of Nutrition, 18(1), 297–330.
Cosnes, J., Gower–Rousseau, C., Seksik, P., & Cortot, A. (2011). Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology, 140(6), 1785–1794.
Dieterle, F., Ross, A., Schlotterbeck, G., & Senn, H. (2006). Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Analytical Chemistry, 78(13), 4281–4290.
Dunn, W. B., Broadhurst, D., Begley, P., Zelena, E., Francis-McIntyre, S., Anderson, N., et al. (2011). Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nature Protocols, 6(7), 1060.
Dunn, W. B., Wilson, I. D., Nicholls, A. W., & Broadhurst, D. (2012). The importance of experimental design and QC samples in large-scale and MS-driven untargeted metabolomic studies of humans. Bioanalysis, 4(18), 2249–2264.
Ehehalt, R., Wagenblast, J., Erben, G., Lehmann, W. D., Hinz, U., Merle, U., et al. (2004). Phosphatidylcholine and lysophosphatidylcholine in intestinal mucus of ulcerative colitis patients. A quantitative approach by nanoelectrospray-tandem mass spectrometry. Scandinavian Journal of Gastroenterology, 39(8), 737–742.
Eriksson, L., Byrne, T., Johansson, E., Trygg, J., & Vikström, C. (2013). Multi-and megavariate data analysis basic principles and applications. Malmö: Umetrics Academy.
Fahy, E., Sud, M., Cotter, D., & Subramaniam, S. (2007). LIPID MAPS online tools for lipid research. Nucleic Acids Research, 35, W606–W612.
Fan, F., Mundra, P. A., Fang, L., Galvin, A., Moore, X. L., Weir, J. M., et al. (2015). Lipidomic profiling in inflammatory bowel disease: Comparison between ulcerative colitis and Crohn’s disease. Inflammatory Bowel Diseases, 21(7), 1511–1518.
Fraunberger, P., Nagel, D., Walli, A. K., & Seidel, D. (2000). Serum cholesterol and mortality in patients with multiple organ failure. Critical Care Medicine, 28, 3574–3575.
Han, X., & Gross, R. W. (2005). Shotgun lipidomics: Multidimensional MS analysis of cellular lipidomes. Expert Review of Proteomics, 2(2), 253–264.
Harvath, L., Robbins, J. D., Russell, A. A., & Seamon, K. B. (1991). cAMP and human neutrophil chemotaxis. Elevation of cAMP differentially affects chemotactic responsiveness. The Journal of Immunology, 146(1), 224–232.
Harvey, R. F., & Bradshaw, M. J. (1980). Measuring Crohn’s disease activity. Lancet, 1(8178), 1134–1135.
Hinz, C., Liggi, S., & Griffin, J. L. (2018). The potential of ion mobility mass spectrometry for high-throughput and high-resolution lipidomics. Current Opinion in Chemical Biology, 42, 42–50.
Hisamatsu, T., Okamoto, S., Hashimoto, M., Muramatsu, T., Andou, A., Uo, M., et al. (2012). Novel, objective, multivariate biomarkers composed of plasma amino acid profiles for the diagnosis and assessment of inflammatory bowel disease. PloS one, 7 (1), e31131.
Hong, S. K. S., Maltz, B. E., Coburn, L. A., Slaughter, J. C., Chaturvedi, R., Schwartz, D. A., et al. (2009). Increased serum levels of L-arginine in ulcerative colitis and correlation with disease severity. Inflammatory bowel diseases, 16(1), 105–111.
Kaser, A., Zeissig, S., & Blumberg, R. S. (2010). Inflammatory bowel disease. Annual Review of Immunology, 28, 573–621.
Kliman, M., May, J. C., & McLean, J. A. (2011). Lipid analysis and lipidomics by structurally selective ion mobility-mass spectrometry. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids, 1811(11), 935–945.
Kominsky, D. J., Campbell, E. L., & Colgan, S. P. (2010). Metabolic shifts in immunity and inflammation. The Journal of Immunology, 184(8), 4062–4068.
Kumar, P., & Clark, M. L. (2016) Kumar and Clark’s clinical medicine (9th ed.). Amsterdam: Elsevier, pp. 309–319.
Kyle, J. E., Aly, N., Zheng, X., Burnum-Johnson, K. E., Smith, R. D., & Baker, E. S. (2018). Evaluating lipid mediator structural complexity using ion mobility spectrometry combined with mass spectrometry. Bioanalysis, 10(5), 279–289.
Liggi, S. (2018). sonial/KniMet: First release of KniMet (Version v1.2.0). Zenodo. https://doi.org/10.5281/zenodo.1196407.
Liggi, S., Hinz, C., Hall, Z., Santoru, M. L., Poddighe, S., Fjeldsted, J., et al. (2017). KniMet: A pipeline for the processing of chromatography–mass spectrometry metabolomics data. Metabolomics, 14(4), 52.
Lin, H. M., Helsby, N. A., Rowan, D. D., & Ferguson, L. R. (2011). Using metabolomic analysis to understand inflammatory bowel diseases. Inflammatory Bowel Diseases, 17(4), 1021–1029.
Lu, K., Knutson, C. G., Wishnok, J. S., Fox, J. G., & Tannenbaum, S. R. (2012). Serum metabolomics in a Helicobacter hepaticus mouse model of inflammatory bowel disease reveal important changes in the microbiome, serum peptides, and intermediary metabolism. Journal of Proteome Research, 11(10), 4916–4926.
Maul, J., Loddenkemper, C., Mundt, P., Berg, E., Giese, T., Stallmach, A., et al. (2005). Peripheral and intestinal regulatory CD4+ CD25 high T cells in inflammatory bowel disease. Gastroenterology, 128(7), 1868–1878.
Moore, A. R., & Willoughby, D. A. (1995). The role of cAMP regulation in controlling inflammation. Clinical & Experimental Immunology, 101(3), 387–389.
Niemelä, K., & Sjöström, E. (1986). Simultaneous identification of aromatic and aliphatic low molecular weight compounds from alkaline pulping liquor by capillary gas-liquid chromatography-mass spectrometry. Holzforschung-International Journal of the Biology, Chemistry, Physics and Technology of Wood, 40(6), 361–368.
Nikolaus, S., & Schreiber, S. (2007). Diagnostics of inflammatory bowel disease. Gastroenterology, 133(5), 1670–1689.
Paglia, G., Kliman, M., Claude, E., Geromanos, S., & Astarita, G. (2015). Applications of ion-mobility mass spectrometry for lipid analysis. Analytical and Bioanalytical Chemistry, 407(17), 4995–5007.
Peterson, J. W., Dickey, W. D., Saini, S. S., Gourley, W., Klimpel, G. R., & Chopra, A. K. (1996). Phospholipase A2 activating protein and idiopathic inflammatory bowel disease. Gut, 39(5), 698–704.
Rivkin, I., & Neutze, J. A. (1977). Influence of cyclic nucleotides and a phosphodiesterase inhibitor on in vitro human blood neutrophil chemotaxis. Archives internationales de pharmacodynamie et de therapie, 228(2), 196–204.
Ruxton, G. D. (2006). The unequal variance t-test is an underused alternative to Student’s t-test and the Mann–Whitney U test. Behavioral Ecology, 17(4), 688–690.
Santoru, M. L., Piras, C., Murgia, A., Palmas, V., Camboni, T., Liggi, S., et al. (2017). Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Scientific Reports, 7(1), 9523.
Schaloske, R. H., & Dennis, E. A. (2006). The phospholipase A2 superfamily and its group numbering system. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids, 1761(11), 1246–1259.
Schmelzer, K., Fahy, E., Subramaniam, S., & Dennis, E. A. (2007). The lipid maps initiative in lipidomics. Methods in Enzymology, 432, 171–183.
Schroeder, K. W., Tremaine, W. J., & Ilstrup, D. M. (1987). Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. New England Journal of Medicine, 317(26), 1625–1629.
Scoville, E. A., Allaman, M. M., Brown, C. T., Motley, A. K., Horst, S. N., Williams, C. S., et al. (2018). Alterations in lipid, amino acid, and energy metabolism distinguish Crohn’s disease from ulcerative colitis and control subjects by serum metabolomic profiling. Metabolomics, 14(1), 17.
Steinbach, G., Morotomi, M., Nomoto, K., Lupton, J., Weinstein, I. B., & Holt, P. R. (1994). Calcium reduces the increased faecal 1, 2-sn-diacylglycerol content in intestinal bypass patients: A possible mechanism for altering colonic hyperproliferation. Cancer Research, 54(5), 1216–1219.
Storr, M., Vogel, H. J., & Schicho, R. (2013). Metabolomics: Is it useful for IBD? Current Opinion in Gastroenterology, 29(4), 378.
Stow, S. M., Causon, T. J., Zheng, X., Kurulugama, R. T., Mairinger, T., May, J. C., & Hann, S. (2017). An interlaboratory evaluation of drift tube ion mobility—mass spectrometry collision cross section measurements. Analytical Chemistry, 89(17), 9048–9055.
Sud, M., Fahy, E., Cotter, D., Brown, A., Dennis, E. A., Glass, C. K., et al. (2006). Lmsd: Lipid maps structure database. Nucleic Acids Research, 35(suppl_1), D527–D532.
Summers, R. W., Elliott, D. E., Qadir, K., Urban Jr, J. F., Thompson, R., & Weinstock, J. V. (2003). Trichuris suis seems to be safe and possibly effective in the treatment of inflammatory bowel disease. The American Journal of Gastroenterology, 98(9), 2034–2041.
Vidal-Lletjós, S., Beaumont, M., Tomé, D., Benamouzig, R., Blachier, F., & Lan, A. (2017). Dietary protein and amino acid supplementation in inflammatory bowel disease course: What impact on the colonic mucosa? Nutrients, 9(3), 310.
Weinstock, J. V., Summers, R. W., Elliott, D. E., Qadir, K., Urban, J. F., & Thompson, R. (2002). The possible link between de-worming and the emergence of immunological disease. The Journal of Laboratory and Clinical Medicine, 139(6), 334–338.
Zhou, Z., Tu, J., Xiong, X., Shen, X., & Zhu, Z. J. (2017). LipidCCS: Prediction of collision cross-section values for lipids with high precision to support ion mobility–mass spectrometry-based lipidomics. Analytical Chemistry, 89(17), 9559–9566.
Acknowledgements
We thank John Fjeldsted and Christine Miller for their support in the Ion Mobility analyses. This study was funded by Agilent Technologies, Regione Autonoma della Sardegna (L.R.7/2007, Grant Number F71J12001180002), and the Medical Research Council UK (Grant Number MR/P011705/1).
Author information
Authors and Affiliations
Contributions
PC, LA, JLG, AM and PU conceived the study, directed the project and designed the experiments. AM, MLS, CP and SL, performed the lipid metabolite profile extraction of the plasma samples. AM and CM performed the polar metabolite profile extraction of plasma samples. AM, CH, JW, JD and SL performed metabolomics and lipidomics experiments and data analysis. AM, CH and ZH, contributed on the lipid targeted analysis. AM wrote the first draft of the manuscript, PC, LA, SL, CH, and JLG contributed to the final version. AM, SL, CH, JLG, PC and LA, critically reviewed the data and the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee (Comitato Etico Indipendente della A.O.U. di Cagliari via Ospedale, 54 - 09124 – Cagliari reference number: PG/2014/11480) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Murgia, A., Hinz, C., Liggi, S. et al. Italian cohort of patients affected by inflammatory bowel disease is characterised by variation in glycerophospholipid, free fatty acids and amino acid levels. Metabolomics 14, 140 (2018). https://doi.org/10.1007/s11306-018-1439-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-018-1439-4