Abstract
Introduction
In natural product research, bioassay-guided fractionation was previously widely employed but is now judged to be inadequate in terms of time and cost, particularly if only known compounds are ultimately isolated. The development of metabolomics, along with improvements in analytical tools, allows comprehensive metabolite profiling. This enables dereplication to target unknown active compounds early in the purification workflow.
Objectives
Starting from an ethanolic extract of violet leaves, this study aims to predict redox active compounds within a complex matrix through an untargeted metabolomics approach and correlation analysis.
Methods
Rapid fractionation of crude extracts was carried out followed by multivariate data analysis (MVA) of liquid chromatography–high resolution mass spectrometry (LC–HRMS) profiles. In parallel, redox active properties were evaluated by the capacity of the molecules to reduce 2,2-diphenyl-1-picrylhydrazyl (DPPH·) and superoxide (O2 ·−) radicals using UV–Vis and electron spin resonance spectroscopies (ESR), respectively. A spectral similarity network (molecular networking) was used to highlight clusters involved in the observed redox activities.
Results
Dereplication on Viola alba subsp. dehnhardtii highlighted a reproducible pool of redox active molecules. Polyphenols, particularly O-glycosylated coumarins and C-glycosylated flavonoids, were identified and de novo dereplicated through molecular networking. Confirmatory analyses were undertaken by thin layer chromatography (TLC)–DPPH–MS assays and nuclear magnetic resonance (NMR) spectra of the most active compounds.
Conclusion
Our dereplication strategy allowed the screening of leaf extracts to highlight new biologically active metabolites in few steps with a limited amount of crude material and reduced time-consuming manipulations. This approach could be applied to any kind of natural extract for the study of various biological activities.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
In natural product research, identification and characterisation of bioactive compounds from crude extracts remains challenging. To identify active molecules, conventional treatments include extraction, bio-guided fractionation, isolation, and characterisation steps. However, these procedures are time consuming as the composition of natural extracts can be complex. There is also a risk of redundant results, such as isolating known compounds or loss of bioactivity during manipulations (Ayouni et al. 2016). Holistic methods such as metabolomics can enhance the classical reductionist workflow based on iterative bioassays and paralleled fractionation to reach active compounds. This approach, referred to as untargeted metabolite profiling, and subsequent statistical analysis was revealed as a valuable tool in natural product research (Cox et al. 2014).
Ultra-high performance liquid chromatography–high resolution mass spectrometry (UHPLC–HRMS) has been used to study complex mixtures, taking advantage of sensitive detection and high chromatographic and mass resolutions. This approach consists of the pre-identification of known compounds in a single analytical run, which is of particular interest in the dereplication process (Wolfender et al. 2015). As a preliminary step, chemical compound databases are interrogated based on accurate mass of all detected features (m/z-retention time (RT) pairs), which produces molecular formula hits. However, several possibilities may be obtained, mainly due to isobaric matches. Therefore, additional filters are needed to rank database hits, such as cross-research with chemotaxonomic data or tandem mass spectrometry (MS/MS) fragmentation patterns. Indeed, development of in silico tools to mirror predicted fragmentation behaviour with experimental data limits the number of matches for a given feature (Xiao et al. 2012; Kind and Fiehn 2016). Additionally, a mass spectral similarity network (e.g., molecular networking) was developed to help interpret LC–MS/MS spectra (Yang et al. 2013; Grapov et al. 2015), complementing this dereplication strategy with the propagation of assigned peaks (Allard et al. 2016).
MS/MS-based dereplication or de novo identification strategies are necessary to target active compounds early in the purification process as untargeted metabolite profiling methods generate tens to thousands of variables, depending of the analytical workflow (Wolfender et al. 2010). Multivariate data analysis (MVA) is commonly used to exploit the variable space in order to rank the features most involved in sample separation. Orthogonal partial least square (OPLS) regression models enable the prediction of features involved in a given activity for each sample (Worley and Powers 2013; Bylesjö et al. 2006).
Violets contain various natural products with diverse biological activities (Muhammad et al. 2012). These include anti-inflammatory, antimicrobial (Witkowska-Banaszczak et al. 2005) and, predominantly, antioxidant activity (Vukics et al. 2008). Previous work describes over 200 compounds in this genus, mainly flavonoids, terpenoids and phenylpropanoids (Zhu et al. 2015). However, only 30 of the 500 species were studied. Thus, a potential supply of new biologically active compounds remains to be discovered, particularly non-volatile metabolites. Many interesting activities are related to the redox active functions of the molecules. Monitoring the ability to reduce free radicals by a single electron reaction is straightforward and automatable in vitro (López-Alarcón and Denicola 2013). This makes it possible to identify reducing agents (also known as antioxidants), which are useful in liquid or soft medium as preservatives. In vivo, the antioxidant character of a compound is not predominantly linked to its ability to scavenge free radicals due to kinetic constraints. The redox property of the molecule, in its reversibility, is involved. It is the oxidised form that paradoxically activates the Nrf2 (NF-E2-related factor 2) signalling pathway and allows the expression of antioxidant enzymes and proteins (Forman et al. 2014). Thus evaluating in vitro reductants is the first step in identifying new structures capable of having active oxidised forms in vivo. The aim of this work was to identify metabolite clusters most probably involved in reduction reactions starting from a complex extract of leaves of violet of Toulouse (Viola alba subsp. Dehnhardtii, Violaceae).
2 Materials and methods
2.1 Plant material
In spring 2016, seven distinct flowerpots of violet of Toulouse (Viola alba subsp. dehnhardtii) were collected from the Toulouse Municipal Greenhouses, France. Samples were washed before separation of flowers, aerial parts and roots. To stabilise the vegetable matter, each plant part was lyophilised and ground into powder.
2.2 Leaf extraction
Metabolites were extracted by adding 10 volumes of 80% ethanol (EtOH) to the powdered leaf (10.0 ± 0.2 g). The solutions were then sonicated in a bath (Fisher Scientific) at room temperature for 30 min and filtered. This procedure was repeated once with fresh solvent. Each extract was evaporated under reduced pressure (Buchi rotavapor R-114) and 100 mg of the crude extracts were finally suspended in 1 mL of methanol prior to solid phase extraction (SPE) (1 g Sep-Pak® C18 cartridge, Waters, Milford, MA, USA). Rapid fractionation was achieved using five aqueous methanolic solutions of increasing organic solvent concentrations (F1: 95/5 H2O/MeOH; F2: 75/25; F3: 50/50; F4: 25/75; F5: 0/100). The five collected fractions of each extract were dried under reduced pressure. One part was put aside for redox properties analysis and the other part was dissolved to 1 mg/mL for UHPLC–HRMS analysis.
2.3 Redox properties assay
The reductive properties of the crude extracts and their respective fractions were firstly evaluated by the DPPH radical scavenging assay (Nguyen et al. 2013). Each sample was analysed in triplicate and five concentrations were tested in order to graphically determine the half maximal inhibitory concentration (IC50). Trolox (Fluka, purity >98%) and rutoside (Sigma, purity >94%) were used as positive controls. Secondly, the capacity of the molecules to reduce the superoxide radical (O2 ·−) was analysed by electron spin resonance (ESR) (Hubert et al. 2008). Rutoside was employed as a positive control. The results were analysed with WinEPR software (v. 2.11b, Bruker): a baseline correction was firstly done before second order integration of the signal.
2.4 UHPLC–HRMS profiling
All extracts were profiled using a UHPLC–DAD–LTQ Orbitrap XL instrument (Ultimate 3000, Thermo Fisher Scientific, Hemel Hempstead, UK). The UV detection was performed by a diode array detector (DAD) from 210 to 400 nm. Mass detection was performed using an electrospray source in positive ionisation (PI) and negative ionisation (NI) modes at 15,000 resolving power (full width at half maximum (FWHM) at 400 m/z). The mass scanning range was m/z 100–1500 Da. The capillary temperature was 300 °C and ISpray voltage was fixed at 4.2 kV (positive mode) and 3.0 kV (negative mode). Mass measurement was externally calibrated before starting the experiment. Each full MS scan was followed by data dependant MS/MS on the three most intense peaks using stepped collision-induced dissociation (CID) (35% normalised collision energy, isolation width 2 Da, activation Q 0.250). The LC–MS system was run in binary gradient mode using a BEH C18 Acquity column (100 × 2.1 mm i.d., 1.7 µm, Waters, MA, USA) equipped with a guard column. Mobile phase A (MPA) was 0.1% formic acid (FA) in water and mobile phase B (MPB) was 0.1% FA in acetonitrile. Gradient conditions were: 0 min, 95% MPA; 0.5 min 95% MPA; 12 min, 5% MPA; 15 min, 5% MPA, 15.5 min, 95% MPA; 19 min, 95% MPA. The flow rate was 0.3 mL/min, column temperature 40 °C and injection volume was 2 µL.
2.5 Data processing
The UHPLC–HRMS raw data were converted to abf files (Reifycs Abf Converter) and processed with MS-DIAL version 2.54 (Tsugawa et al. 2015) for mass signal extraction between 100 and 1500 Da from 0 to 12.5 min. Respective MS1 and MS2 tolerance were set to 0.01 and 0.4 Da in centroid mode. The optimised detection threshold was set to 2 × 104 for MS1 and 10 for MS2. Adducts and complexes were identified to exclude them from the final peak list. Finally, the peaks were aligned on a quality control (QC) reference file with a retention time tolerance of 0.1 min and a mass tolerance of 0.025 Da. The resulting peak list was then exported to comma-separated value (CSV) format prior to MVA using SIMCA-P+ (version 14.0, Umerics, Umea, Sweden).
2.6 Statistical analysis
Comma-separated value files were directly imported into SIMCA-P+ (version 14.0, Umerics, Umea, Sweden). For MVA, all data were log transformed and pareto scaled. The OPLS regression analysis was done with DPPH IC50 values as Y input. Coefficient scores were used to rank variables according to their reductive potential. For each model, a leave-one-subject-out cross-validation was performed to assess the model fit. The validity of the discriminant model was verified using permutation tests (Y-scrambling).
2.7 Identification of significant features
Molecular formulae of significant features were calculated with MS-FINDER 2.10 (Tsugawa et al. 2016). Various parameters were used in order to reduce the number of potential candidates, such as the element selection exclusively including C, H, O; mass tolerance fixed to 10 ppm and the isotopic ratio tolerance set to 20%. Only natural product databases focused on plants were selected from Universal Natural Products Database (UNPD), KNApSAc, PlantCyc, Dictionary of Natural Products (DNP, CRC press, v25:2) and CheBI. The results were presented as a list of compounds sorted according to the score value of the match. This value encompassed uncertainty on accurate mass, the isotopic pattern score and the experimental MS/MS fragmentation mirrored to in silico matches. Chemical classes were retained for identified features with a score above 5 and only structures with a score above 7 were retained for thorough analysis.
2.8 Mass spectral similarity network
The text file format exported from MS-DIAL was cleaned-up by eliminating the identified adducts before importing into MetamapR (version 1.4.0) (Grapov et al. 2015). A mass spectral similarity network was created with a maximum of ten connections between nodes, a cut off fixed at 0.3 and a retention time filter at 1.5 min. The calculated edge list was then downloaded and processed with Cytoscape 2.8.3 (Shannon et al. 2003). An attribute file containing all processed information, in particular m/z values, OPLS coefficients and chemical classes of identified features was imported to improve network visualisation.
2.9 Validation of the model by TLC–DPPH–MS
A TLC method was developed to confirm the statistical results. TLC separation was undertaken for fractions F2 and F3 with respective migration solvent composition: ethyl acetate, formic acid, acetic acid, water (50:5.5:5.5:13.5) and ethyl acetate, formic acid, water (60:6.5:6.5). 675 µL of a 10 mg/mL solution was deposed in band of 180 mm in a 20 cm TLC plate. The plate was then placed in an oven at 70 °C for 15 min and then observed under UV at 254 nm for F2 and 366 nm for F3. A small part of the plate was revealed with a purple DPPH solution at 600 mol/L. Then, yellow active spots were collected on the non-treated surface with the use of a TLC–MS interface (Camag, Muttenz, Switzerland) and directly injected into the LTQ-Orbitrap instrument.
2.10 Purification of compounds of interest
Purification of active compounds was undertaken starting from 200 mg of F2. LC–UV separation was performed using a XBridge™ C18 prep column (4.6 × 150 mm i.d., 5 µm, Waters, MA, USA) on an HPLC-DAD–QTOF-MS instrument (HPLC Alliance 2695- QTof Premier, Waters, MA, USA). Mobile phase A (MPA) was 0.1% formic acid (FA) in water and mobile phase B (MPB) was 0.1% FA in acetonitrile. The linear gradient program was as follows: 0 min, 95% MPA; 15 min 75% MPA; 16 min, 50% MPA; 18 min, 50% MPA, 18.5 min, 95% MPA; 23.5 min, 95% MPA. The flow rate was 20 mL/min and injection volume was 250 µL. For all analyses, detection was performed by UV at 325 and 254 nm and collection was automatically done by filling 30% of the tube. Samples were kept at ambient temperature during the whole analysis.
All collected fractions were dried using a Speedvac (SpeedVac plus, Thermo Savant™) and dissolved in DMSO-d6 for NMR analysis (Bruker cryo 500 MHz, Germany) (see Supplementary Table S1).
3 Results
3.1 Radical scavenging properties in vitro
Redox properties of extracts were evaluated by studying their ability to scavenge 2,2-diphenyl-1-picrylhydrazyl (DPPH·) radical. The analyses were first carried out on ethanolic extracts of flowers, leaves, and roots. The results showed that roots presented weak radical scavenging activity, with an IC50 of 1525 ± 122 mg/L. Flowers and leaves had greater activity, with IC50 values of 475 ± 15 and 467 ± 14 mg/L, respectively (see Supplementary Data S1). Leaves were chosen for further investigations as flowers are more fragile and less abundant.
Radical scavenging activity was measured for the crude ethanolic leaf extracts of seven individual plants of violet of Toulouse and respective fractions (N = 42) of decreasing polarity (F1–F5) (Fig. 1A; plain bar plots). While the seven crude extracts displayed an IC50 value of 467 ± 14 mg/L, high IC50 values were obtained for SPE fractions F1 and F5 (2605 ± 380 and 1337 ± 26 mg/L, respectively) demonstrating weak radical scavenging activity in vitro in these cases. Significant activities were measured for fractions F2, F3 and F4, with IC50 values of 53 ± 5, 153 ± 9 and 429 ± 10 mg/L, respectively. For comparison, the positive controls Trolox and rutoside possess IC50 values of approximately 7 and 14 mg/L, respectively. Therefore, fractions F2 and F3 seemed promising for the discovery of antioxidant compounds in vitro as preservatives.
MVA workflow: A comparison of radical scavenging properties of leaf crude extracts and respective fractions of violet of Toulouse obtained by C18-SPE using DPPH assays (plain bar plots) and ESR (striped bar plots). B PCA score plot of the ESI–NI dataset (QC: quality controls, F1–F5 denote respective C18-SPE fractions). C (a) Coefficient plot obtained by OPLS regression; (b) emphasis of the first loadings; (c) ranking of the first seven loadings estimated as radical scavenger compounds according to the OPLS regression
To confirm these results, ESR experiments were conducted to measure the capacity of extracts (final concentration of 50 mg/L) to trap superoxide radical (O2 ·−). The corresponding values of the ESR signal double integrations are presented in Fig. 1A (striped bar plots). As for DPPH assay, F2 and F3 appeared to have the most redox potential as demonstrated by the low intensity of the remaining signal.
3.2 UHPLC–HRMS-based metabolomics approach
UHPLC–HRMS profiles of all the 46 extracts (7 crude extracts, 35 SPE fractions, and 6 QC samples prepared by pooling an aliquot of all fractions) afforded 434 and 527 features (m/z-RT pairs) in NI and PI modes, respectively.
The C18-SPE procedure is clearly effective at resolving the extracts according to polarity, with polar fractions 1–3 eluting early (<6 min in our UHPLC conditions) and fractions 4 and 5, containing apolar compounds, eluting at later retention times (see Supplementary Data S2).
As a preliminary step, principal component analysis (PCA) was applied as an exploratory data analysis to provide an unsupervised overview of the LC–MS fingerprints. PCA clustered all independent biological replicates from the same fraction. Thus, one fraction was related to one position on the plot with slight variability highlighting a reproducible response. As expected, PCA grouped crude extracts and QC near the plot centre. By contrast, single fractions were well distributed (Fig. 1B). These results indicated that SPE fractionation and LC–MS workflows highlighted variability in the data set, and were highly reproducible. Moreover, it demonstrated a stable chemical composition within aerial parts of the seven independent plants under study. After PCA, we applied OPLS regression analysis to obtain a classification of the loadings (i.e. m/z-RT pairs) regarding the DPPH IC50 value (input Y). The quality of model prediction was good (R2Y = 0.977, Q2Y = 0.952) and a permutation test assessed its validity (see Supplementary Data S3). This supervised method allowed the ranking of potential redox-active compounds according to their regression coefficient values (Fig. 1C(a, b)): negative coefficients were correlated to potential redox-active compounds and positive coefficients to less-active redox compounds. From the list containing the first hits (Fig. 1C(c)), we carried out the identification procedure based on their MS/MS spectra.
3.3 Identification of potential radical scavenger compounds based on in silico fragmentation
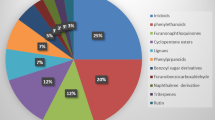
Using the OPLS regression analysis results, the top eight compounds were tentatively identified by interrogating local natural product databases integrated in MS-FINDER: PlantCyc, UNPD, KNApSAcK and ChEBI (Table 1, Hit NP databases). For each compound, the results afforded several candidates and ranked them according to their similarity score, which was based on comparison between experimental MS/MS fragments and in silico spectra of candidates. Interestingly, for compounds 1, 2, 3, 6 and 8, the top three candidates ranked by MS-FINDER each contained a coumarin core, and for compounds 4, 5 and 7, each contained a flavonoid derivative. UV spectra of each peak (Table 1; UV λmax) supported our assumptions. The coumarin nucleus displays two absorption bands near 270 and 310 nm. Substitutions on this nucleus tend to produce a bathochromic shift (Masrani et al. 1974). For the flavonoids, two absorption bands of interest are around 250 to 295 and 310 to 370 nm, depending on the flavonoid class and the substitution pattern (Olsen et al. 2009). Therefore, the higher wavelength band gives differentiation between coumarins and flavonoids. Still, the candidates were numerous and not always relevant, e.g., compound 5 had hundreds of candidates and feruloyl derivatives were proposed for compound 1. To refine the results, we carried out a second identification step by creating a user-defined database restricting results of UNPD to compounds found in Viola genus (Table 1, Hit UNPD-Viola). Thus, fewer and more pertinent candidates could be proposed. In our case, this led to the identification of compounds 4, 5 and 7 as flavonoids, indicating that all the other significant features have never been described in Viola genus. Moreover, compounds 3 and 8 remained totally unknown as no hit matched with any database.
3.4 Extending the dereplication process via a mass spectral similarity network
The mass spectral similarity network displayed two clusters of radical scavenger compounds, i.e., antioxidants (Fig. 2). Each cluster was closely related to one SPE fraction: according to our previous identification results, flavonoids were mainly found in F3 while coumarins were mainly found in F2.
The flavonoid cluster displayed three compounds with related structures active in reducing DPPH· (Fig. 3). Compounds 4 and 5 presented a statistically significant radical scavenging potential according to their coefficient score (Table 1; OPLS coefficient). Interestingly, the molecular formulae of the compounds from this cluster matched structures already identified in Violaceae. Experimental MS/MS patterns mirrored to in silico fragmentation allowed the preliminary identification of schaftoside (4), isovitexin 6″-O-β-d-glucopyranoside (5) and 6,8-di-C-α-l-arabinopyranosylapigenin (7) (see Supplementary Data S4). These structures were based on a user-defined Viola database, as hits were more pertinent and the final candidate had the highest total score, generally above 7.
Expansion on the redox-active compounds clusters. Putative coumarin and flavonoids scaffold correspond to top ranked hits from MS-FINDER. Node size was emphased based on OPLS coefficient value. Compound number is indicated in brackets in each node in correspondence to Table 1
The other compounds of interest belonged to the coumarin cluster (Fig. 3). Compounds 3 and 8 were connected with the identified glycosylated coumarins C16H18O11 (compound 2), confirmed by 1D and 2D NMR (see Supplementary Data S5). Thus, we applied a de novo dereplication based on the interpretation of mass loss to propose potential chemical structures. We interpreted a difference of 162 Da between 3 and 2 as a glucose substituent. We suggest a new 8-O-diglycosylated coumarin structure for 3, based on the MS/MS spectrum and the loss of a 324.12 Da, correlated to two linked glucose units (see Supplementary Data S4). Regarding 8, a difference of 14 Da was indicative of a di-methoxy-8-O-glycosylated coumarin.
Compound 6 was tentatively identified as an 8-C-glycolsylated coumarin from comparison with in silico fragmentation data using MS-FINDER local databases.
3.5 Validation of the model
To validate our OPLS model, we used a TLC–DPPH–MS assay for fractions F2 (Fig. 4a) and F3 (Fig. 4b). Overall, the four coumarins (1, 2, 3 and 8) and the three flavonoids (4, 5 and 7) were identified and matched with the OPLS ranking.
We then tested purified compounds 1 and 2 for their respective scavenging activities against DPPH (plain bar plots) and superoxide radicals (striped bar plots) (Fig. 5). Trolox and rutoside were used as references for both assays. The difference in radical scavenging capacity of Trolox against DPPH and superoxide radicals can be easily explained by considering the rate constants of the scavenging reactions: kTrolox/DPPH = 1.1 × 104 M−1 s−1 (Friaa and Brault 2006) and \({{k}_{{Trolox}/{O}_{2}^{{ \cdot - }}}}=0.1\;{{M}^{ - 1}}\;{{s}^{ - 1}}\) (Cabelli and Bielski 1986).
The radical scavenging properties of compound 1 (97% pure) were confirmed by comparison with the active controls, Trolox (DPPH IC50: 1 9 ± 2 mg/L vs. Trolox 7 mg/L), and rutoside (ESR double integration: 1 0.88 ± 0.01 vs. rutoside 1.78 ± 0.10).
For compound 2 (90% pure), the DPPH IC50 of 49 ± 1 mg/L and ESR double integration of 15 ± 1, suggests this compound is a less active radical scavenger than compound 1, meeting the statistical ranking.
4 Discussion
The objective of this work was to characterise redox active metabolites from the leaves of Viola alba subsp. dehnhardtii together with de novo dereplication of active compounds and the establishment of a non-volatile metabolite fingerprint.
In order to decipher the redox potential in few steps, we correlated DPPH results to UHPLC–HRMS fingerprints by OPLS regression. This method allowed the ranking of detected features according to their potential radical scavenging properties. In addition, a mass spectral similarity network allowed de novo dereplication of top ranked features, based on the acquisition of UHPLC–HRMS profiles in data dependent analysis mode. This provided accurate mass-to-charge ratio for molecular formula determination along with MS/MS fragments used for peak assignments. NI mode MS and MS/MS spectra were mainly used as they were of better quality than PI spectra, thus improving in silico identification using MS-FINDER. Processing with MS-DIAL allowed the acquisition of a clean peak list with deconvoluted MS/MS data for each feature. This meant we could easily remove adducts and other ionisation artefacts from files uploaded to MetamapR. This resulted in a cleaner molecular network compared with uploading raw data files. The mass spectral similarity network allowed the organisation of features in clusters and dereplication results highlighted mainly coumarins in SPE fraction F2 and mainly flavonoids in fraction F3. These scaffolds are known for having several biological activities, including redox activity (Procházková et al. 2011; Gacche and Jadhav 2012). We identified a mixture of C-glycosylated flavonoids along with O-glycosylated coumarins. The enrichment of these molecules in the two fractions explains the improved antioxidant properties of the fractions over the crude extracts. Although some of the compounds, e.g., compounds 5 and 7, are well-known in Viola genus (Vukics et al. 2008; Xie et al. 2003), the other compounds we identified have not previously been described in this genus.
The protective effect of phenolic coumarins against oxidative damage depends on the hydrogen-donating capacity of hydroxyl groups (Kostova 2006; Borges et al. 2005) and then on their oxidisability. Previous structure–activity relationship studies have identified the importance of the number and location of the phenolic hydroxyl groups (Kancheva et al. 2010). As compounds 1 and 2 have the same substituents, energy minimisation calculations were carried out using the MM2 forcefield (Chemdraw 3D, Cambridge Soft, USA) to understand the difference between their radical reducing capacity. These calculations demonstrate that the hydroxyl group in position 6 in compound 1 was the most readily oxidisable into OH·+ and thus responsible for the strong antioxidant capacity measured for 1 compared with 2. Purification of coumarins C16H18O11 1 and 2 tested in DPPH and ESR redox assays confirmed the workflow used in this study: as predicted by the model, compound 1 appeared as a reducer that is a good antioxidant compound in vitro.
Our statistical analysis was validated by TLC–DPPH–MS assays, with detection of seven DPPH-active compounds ranked in top positions of the analysis. A big advantage of our approach is the limited manipulation steps (crude extraction followed by a single SPE fractionation) and low sample demand. For instance, 1 g of dried plant material should be sufficient to conduct the entire workflow. The workflow could easily be implemented to any kind of extract with other biological activities. However, ranking results of variables may not accurately reflect the biological activity. Ranked positions may vary depending on scaling and normalisation processes. The first 20 ranked variables should be considered for thorough analysis. Another drawback of this method concerns the correlation between biological assay results (Y) and the detection method used to generate variables (X). In our case, we only took into account ionisable compounds in electrospray ionisation (ESI) mode for OPLS modelling. The ionisation process is compound dependent and does not reflect a quantitative measure of each detected feature. We could reach more realistic models by using universal-type detectors such as Evaporating Light Scattering Detector (ELSD) or Charged Aerosol Detector (CAD). While 1H- or 13C-NMR could be an interesting alternative, the lack of sensitivity of this method is not compatible with complex samples like crude natural extracts.
Finally, the framework of our dereplication strategy capitalised on UHPLC–HRMS–MS/MS profiles, using agreement between modelled and experimental MS/MS fragmentation patterns and mass spectral similarity network to propagate assignments of clustered features. As illustrated with the coumarin cluster, using the molecular network and propagating from a known compound allows identification of others belonging to the same class by interpreting the neutral mass loss.
5 Conclusion
A UHPLC–HRMS based metabolomic study combined with molecular networking tools allowed the dereplication of redox-active metabolites present in the violet of Toulouse. These compounds were mainly flavonoid and coumarin derivatives. Our approach highlighted two main metabolite clusters displaying functional groups with redox-active properties. We identified seven compounds of interest, of which five were found through MS/MS fragmentation and comparison with references found in databases. Two unknown coumarins were de novo dereplicated through a molecular networking approach. Overall, the workflow proposed here allowed early identification of redox active compounds within a complex mixture with limited effort and crude materials.
In this work, it was chosen to correlate redox-active properties with LC–MS profiles to identify compounds of interest but more generally, provided variability is brought to the data set, this method can be implemented in any study of other chemical and biological properties employing different types of assays.
References
Allard, P.-M., Péresse, T., Bisson, J., Gindro, K., Marcourt, L., Pham, V. C., et al. (2016). Integration of molecular networking and in-silico MS/MS fragmentation for natural products dereplication. Analytical Chemistry, 88(6), 3317–3323. doi:10.1021/acs.analchem.5b04804.
Ayouni, K., Berboucha-Rahmani, M., Kim, H. K., Atmani, D., Verpoorte, R., & Choi, Y. H. (2016). Metabolomic tool to identify antioxidant compounds of Fraxinus angustifolia leaf and stem bark extracts. Industrial Crops and Products, 88, 65–77. doi:10.1016/j.indcrop.2016.01.001.
Borges, F., Roleira, F., Milhazes, N., Santana, L., & Uriarte, E. (2005). Simple coumarins and analogues in medicinal chemistry: Occurrence, synthesis and biological activity. Current Medicinal Chemistry, 12(8), 887–916.
Bylesjö, M., Rantalainen, M., Cloarec, O., Nicholson, J. K., Holmes, E., & Trygg, J. (2006). OPLS discriminant analysis: Combining the strengths of PLS-DA and SIMCA classification. Journal of Chemometrics, 20(8-10), 341–351.
Cabelli, D. E., & Bielski, B. H. (1986). Studies of the reactivity of trolox with Mn3+/Fe3+ complexes by pulse radiolysis. Journal of Free Radicals in Biology & Medicine, 2(1), 71–75.
Cox, D. G., Oh, J., Keasling, A., Colson, K. L., & Hamann, M. T. (2014). The utility of metabolomics in natural product and biomarker characterization. Biochimica et Biophysica Acta (BBA)-General Subjects, 1840(12), 3460–3474.
Forman, H. J., Davies, K. J., & Ursini, F. (2014). How do nutritional antioxidants really work: Nucleophilic tone and para-hormesis versus free radical scavenging in vivo. Free Radical Biology and Medicine, 66, 24–35.
Friaa, O., & Brault, D. (2006). Kinetics of the reaction between the antioxidant Trolox® and the free radical DPPH in semi-aqueous solution. Organic & Biomolecular Chemistry, 4(12), 2417–2423.
Gacche, R. N., & Jadhav, S. G. (2012). Antioxidant activities and cytotoxicity of selected coumarin derivatives: Preliminary results of a structure–activity relationship study using computational tools. Journal of Experimental & Clinical Medicine, 4(3), 165–169.
Grapov, D., Wanichthanarak, K., & Fiehn, O. (2015). MetaMapR: Pathway independent metabolomic network analysis incorporating unknowns. Bioinformatics. doi:10.1093/bioinformatics/btv194.
Hubert, J., Berger, M., Nepveu, F., Paul, F., & Daydé, J. (2008). Effects of fermentation on the phytochemical composition and antioxidant properties of soy germ. Food Chemistry, 109(4), 709–721.
Jung, M., Geiger, H., & Zinsmeister, H. D. (1995). Tri-and tetrahydroxycoumarin derivatives from Tetraphis pellucida. Phytochemistry, 39(2), 379–381.
Kancheva, V. D., Saso, L., Boranova, P. V., Khan, A., Saroj, M. K., Pandey, M. K., et al. (2010). Structure–activity relationship of dihydroxy-4-methylcoumarins as powerful antioxidants: Correlation between experimental & theoretical data and synergistic effect. Biochimie, 92(9), 1089–1100.
Kind, T., & Fiehn, O. (2016). Strategies for dereplication of natural compounds using high-resolution tandem mass spectrometry. Phytochemistry Letters. doi:10.1016/j.phytol.2016.11.006.
Kostova, I. (2006). Synthetic and natural coumarins as antioxidants. Mini Reviews in Medicinal Chemistry, 6(4), 365–374.
López-Alarcón, C., & Denicola, A. (2013). Evaluating the antioxidant capacity of natural products: A review on chemical and cellular-based assays. Analytica Chimica Acta, 763, 1–10.
Marzouk, M. M., Elkhateeb, A., Ibrahim, L. F., Hussein, S. R., & Kawashty, S. A. (2012). Two Cytotoxic Coumarin Glycosides from the aerial parts of Diceratella elliptica (DC.) Jonsell growing in Egypt. Records of Natural Products, 6(3), 237.
Masrani, K., Rama, H., & Bafna, S. (1974). Ultraviolet absorption spectra: Some substituted coumarins. Journal of Chemical Technology and Biotechnology, 24(6), 331–341.
Muhammad, N., Saeed, M., Aleem, A., & Khan, H. (2012). Ethnomedicinal, phytochemical and pharmacological profile of genus Viola. Phytopharmacol, 3(1), 214–226.
Nguyen, H. Y. T., Vo, B. H. T., Nguyen, L. T. H., Bernad, J., Alaeddine, M., Coste, A., et al. (2013). Extracts of Crinum latifolium inhibit the cell viability of mouse lymph oma cell line EL4 and induce activation of anti-tumour activity of macrophages in vitro. Journal of Ethnopharmacology, 149(1), 75–83.
Olsen, H., Aaby, K., & Borge, G. I. A. (2009). Characterization and quantification of flavonoids and hydroxycinnamic acids in curly kale (Brassica oleracea L. convar. acephala var. sabellica) by HPLC DADESI-MS. Journal of Agricultural and Food Chemistry, 57(7), 2816–2825.
Procházková, D., Boušová, I., & Wilhelmová, N. (2011). Antioxidant and prooxidant properties of flavonoids. Fitoterapia, 82(4), 513–523.
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research, 13(11), 2498–2504.
Tsugawa, H., Cajka, T., Kind, T., Ma, Y., Higgins, B., Ikeda, K., et al. (2015). MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nature Methods. doi:10.1038/nmeth.3393.
Tsugawa, H., Kind, T., Nakabayashi, R., Yukihira, D., Tanaka, W., Cajka, T., et al. (2016). Hydrogen rearrangement rules: Computational MS/MS fragmentation and structure elucidation using MS-FINDER software. Analytical Chemistry. doi:10.1021/acs.analchem.6b00770.
Vukics, V., Kery, A., Bonn, G. K., & Guttman, A. (2008). Major flavonoid components of heartsease (Viola tricolor L.) and their antioxidant activities. Analytical and Bioanalytical Chemistry, 390(7), 1917–1925.
Vukics, V., Ringer, T., Kery, A., Bonn, G. K., & Guttman, A. (2008). Analysis of heartsease (Viola tricolor L.) flavonoid glycosides by micro-liquid chromatography coupled to multistage mass spectrometry. Journal of Chromatography A, 1206(1), 11–20.
Witkowska-Banaszczak, E., Bylka, W., Matławska, I., Goślińska, O., & Muszyński, Z. (2005). Antimicrobial activity of Viola tricolor herb. Fitoterapia, 76(5), 458–461.
Wolfender, J. L., Marti, G., & Ferreira Queiroz, E. (2010). Advances in techniques for profiling crude extracts and for the rapid identificationof natural products: Dereplication, quality control and metabolomics. Current Organic Chemistry, 14, 1808–1832.
Wolfender, J.-L., Marti, G., Thomas, A., & Bertrand, S. (2015). Current approaches and challenges for the metabolite profiling of complex natural extracts. Journal of Chromatography A, 1382, 136–164. doi:10.1016/j.chroma.2014.10.091.
Worley, B., & Powers, R. (2013). Multivariate analysis in metabolomics. Current Metabolomics, 1(1), 92–107.
Xiao, J. F., Zhou, B., & Ressom, H. W. (2012). Metabolite identification and quantitation in LC-MS/MS-based metabolomics. TrAC Trends in Analytical Chemistry, 32, 1–14.
Xie, C., Veitch, N. C., Houghton, P. J., & Simmonds, M. S. (2003). Flavone C-glycosides from Viola yedoensis Makino. Chemical and Pharmaceutical Bulletin, 51(10), 1204–1207.
Yang, J. Y., Sanchez, L. M., Rath, C. M., Liu, X., Boudreau, P. D., Bruns, N., et al. (2013). Molecular networking as a dereplication strategy. Journal of Natural Products, 76(9), 1686–1699.
Zhu, H., Qin, S., Zhang, N., Yang, D., Han, H., Wei, K., & Li, M. (2015). Chemical constituents and biological activities of plants from the genus Viola. Chemistry & Biodiversity, 12(12), 1777–1808.
Acknowledgements
The authors would like to thank the Regional Council Midi-Pyrénées for sponsoring this project (Project CLE 13053062) as well as the Toulouse Municipal Greenhouses for making the violet collection available to its collaborators.
Funding
This study was funded by the Regional Council Midi-Pyrénées (Project CLE 13053062).
Author information
Authors and Affiliations
Contributions
GM proposed the study and designed the experiments. JC performed the extraction of crude material, the radical scavenging assays under the supervision of PP, processed the LC-HRMS profiles acquired by NMF and performed MVA and molecular networking analysis under the supervision of GM. JC analysed the results and wrote the manuscript which was reviewed by all the collaborators of the work.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
No human participants or animals were involved in this study.
Informed consent
No human participants were involved in this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chervin, J., Perio, P., Martins-Froment, N. et al. Dereplication of natural products from complex extracts by regression analysis and molecular networking: case study of redox-active compounds from Viola alba subsp. dehnhardtii . Metabolomics 13, 96 (2017). https://doi.org/10.1007/s11306-017-1227-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-017-1227-6