Abstract
Introduction
Amphiphilic copolymer nanoparticle-encapsulated multi-target chemotherapeutic drugs have attracted considerable attention due to their favorable drug efficiency and potential application prospect. Studies have shown that an amphiphilic copolymer, methoxypoly(ethylene glycol)-poly(lactide-co-glycolide) modified with ε-polylysine, and encapsulated with hydrophilic doxorubicin, hydrophobic paclitaxel and survivin siRNA profoundly improved the therapeutic effect both in vitro and in vivo.
Objectives
To investigate how MCF-7 cells would response to the exposure of these nanoparticles over with time and assess the biological effects of these nanoparticles and their encapsulated drugs in a holistic manner.
Methods
MCF-7 cells were treated with PBS, nanocarrier and three encapsulated drugs, respectively. Metabolic alterations associated with nano-drugs exposure were investigated by performing untargeted NMR metabolomics with combination of targeted fatty acids analysis by GC-MS on cell extracts. Altered metabolic pathways were further validated by qRT-PCR approach.
Results
Copolymers showed great biocompatibility with cells as it induced transit metabolic disruptions without affecting cell survival rate. The rapid release of encapsulated doxorubicin resulted in inhibition of glycolysis and DNA synthesis, active proteolysis; these metabolic alternations were recovered after 10 h exposure. However, the combination use of multiple drugs consistently induced cell cycle arrest and apoptosis evidenced by reduction in glycolysis, active proteolysis, stimulated O-GlcNAcylation, reduced the PC:GPC ratio and fatty acids accumulation. Prolonged exposure to encapsulated-multiple-drugs also induced oxidative stress to cells.
Conclusion
These findings provide important insight into the biological effects of nanoparticles and their encapsulated drugs while demonstrate that metabolomics is a powerful approach to evaluate the biological effects of nano-drugs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
It has become common practice to use a combination of drugs to treat cancer. The different combinations of drugs are often administrated to patients sequentially rather than simultaneously, which could reduce the efficacy of the treatment (Overman et al. 2010). The rapid development of nanotechnology in the past few years has opened up an opportunity to deliver multiple cancer drugs simultaneously. There have different types of nanomaterials been designed to carry diagnostic agents, genes, and cancer targeted drugs (Portney and Ozkan 2006; Torchilin 2012). These nanomaterials include inorganic materials such as gold nanorods (Jain et al. 2007; Huang et al. 2007), quantum dots (Michalet et al. 2005) and silicon-based nanoparticles (Tang et al. 2012), and some organic nanocarriers, such as liposomes, albumin-based particles, dendrimers and smart polymers (Davis et al. 2008). Among these nanocarriers, organic nanomaterials show significant potential in cancer medicine due to their well-defined three-dimensional structures, high biocompatibility and biodegradability. The US Food and Drug Administration (FDA) have approved various nanomedicines based on organic nanocarriers. Doxil is the first liposomal drug formulation used to treat Kaposi’s sarcoma [also associated with AIDS(Northfelt et al. 1998)], whereas abraxane (a 130-nm albumin-bound paclitaxel drug) is the second-line drug used to treat patients with breast cancer (Harries et al. 2005). Benefited from enhanced permeation and retention effect (EPR effect) and controlled-release property, these nanocarriers have helped anti-tumor drugs to achieve favorable therapeutic outcomes.
Advances in the field of nanomedicine have led to the development of polymer-based systems as drug carriers, for example, poly(ethylene)glycol (PEG) and poly(DL-lactic-co-glycolic acid) (PLGA) (Lee et al. 2008; Shen et al. 2015; Han et al. 2015). PEG is hydrophilic in nature and has been used to enfold soluble drugs such as doxorubicin, whilst PLGA is lipophilic, and acts as carrier of insoluble drugs such as paclitaxel (Yoo and Park 2001; Jeong et al. 2000). Doxorubicin and paclitaxel are the most commonly used chemotherapeutic drugs. The combinational use of these compounds in cancer treatment has been effectively applied in the clinic for decades (Gehl et al. 1996). Previous study showed that the modifying of ε-polylysine (EPL) on the surface of methoxypoly(ethylene glycol)-poly(lactide-co-glycolide) (mPEG-PLGA) could improve the polydispersity and surface charge density of this amphiphilic copolymer, which facilitated the extra delivery of siRNA as well as enhanced the inhibition of cancer cell growth, highlighting its advantageous use as a drug carrier (Wang et al. 2013). This system (E-NP) is capable of simultaneously incorporating doxorubicin (DOX, D), paclitaxel (TAX, T) and negatively charged survivin siRNA (S). Survivin works as a member of the inhibitor of apoptosis protein family, is overexpressed in almost all common human cancers such as lung, prostate, pancreas, colon and breast cancers (Ambrosini et al. 1997). Survivin interacts with microtubules of the mitotic spindle to prevent apoptosis. Survivin siRNA disrupts survivin-microtubule interactions, which results in a loss of survivin’s anti-apoptotic function and an increased caspase-3 activity, a mechanism involved in cell death (Li et al. 1998). Apoptosis-based therapy using survivin siRNA to inhibit cell survival has been at the forefront of recent research in cancer medicine (Carew et al. 2015; Kar et al. 2015).
Despite the development and evaluation of a variety of similar multifunctional nanocarrier platforms for their effectiveness in cancer cell inhibition and tumor suppression (Alibolandi et al. 2015; He et al. 2015), there has been less focus on how the nanocarriers and their encapsulated drugs affect metabolism in vitro. Understanding the dynamic metabolic effects of this applicable nanocarrier and its encapsulated drugs may essentially be vital in verifying their time-dependent controlled-release property as well as providing fundamental information for their further application in biological system. Studies have highlighted that the metabolic effects as a result of drug treatment can feasibly be investigated by metabolomics (Kaddurah-Daouk et al. 2008; Bathen et al. 2010; Wang et al. 2012). Metabolomics studies of free DOX and TAX, mPEG-PLGA and its DOX/TAX encapsulated form on healthy mice have shown that nanoparticle-encapsulated drugs can relieve the energy metabolism inhibition and decrease the activation of antioxidation caused by free drug exposure (Song et al. 2015). Metabolomics technology is capable of detecting small metabolites using nuclear magnetic resonance (NMR) spectroscopy and chromatography coupled to mass spectrometry, which reflects metabolic responses of a living organism during exposure to external stimuli (Nicholson et al. 1999; Tang and Wang 2006). This technique has been successfully applied to investigate the effects of toxins (Huang et al. 2013a), host–microbe interactions (Zhao et al. 2013), cancers (Huang et al. 2013b) and obesity (An et al. 2013), as well as the underlying molecular mechanisms of nanomaterials in selective cancer cell inhibition (Zhang et al. 2013).
Herein, our nanocarrier consists of EPL, PEG and PLGA (E-NP), incorporated with different combinations of antitumor agents, doxorubicin and paclitaxel and survivin siRNA, thus fabricating a range of nano-drugs, namely E-NP-DOX, E-NP-DT, and E-NP-DTS, respectively. Both in vitro and in vivo studies have shown that E-NP-DTS is the most effective antitumor formulation and exhibits an impressive tumor inhibiting capacity among these nano-drugs (Wang et al. 2013). Instead of comparing the improved efficacy or decreased side effects of these nanoparticles as reported previously, in this study, we focused on how MCF-7 cells would response to the exposure of these nanoparticles over with time to explore their biocompatibility and dynamic biological effects. Thus, we systematically investigated the time-dependent metabolic changes of a human breast cancer cell line, MCF-7, treated with nanocarrier E-NP and three separately encapsulated drugs, E-NP-DOX, E-NP-DT and E-NP-DTS. In consideration of the simple sample preparation procedure and method stability, untargeted metabolomics based on NMR was chosen as the main technique in this work. To complete the information concerning the state of cell membrane and energy metabolism during nanomaterials exposure, targeted detection of 37 fatty acids base on GC-MS was performed. Altered metabolic pathways were identified and further validated by measuring the expression of key genes using qRT-PCR. Research findings from this study provide an insight into the dynamic biological effects of MCF-7 cells exposed to different nanomaterials, and demonstrate the feasibility of metabolomics as a useful tool to assess the biological effects of novel nanomaterials.
2 Experimental procedures
2.1 Materials
NaH2PO4·2H2O, K2HPO4·3H2O, K2CO3 and methanol (analytical grade) were obtained from Sinopharm Chemical Reagent Co. Ltd. (Shanghai, P. R. China). 3,5-Di-tertbutyl-4-hydroxytoluene (BHT), hexane, and methanol (HPLC grade) were purchased from Supelco (Bellefonte, PA) acetyl chloride, methyl heptadecanoate (99%), and methyl tricosanate (99%) were purchased from Sigma-Aldrich (P.R. China). D2O (99.9% D), and 3-(Trimethylsilyl)propionic-2,2,3,3-d4 acid sodium salt (TSP-d4) were obtained from Cambridge Isotope Laboratories (MA, USA).
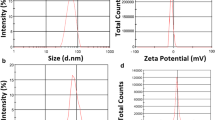
In this study, E-NP, E-NP-DOX, E-NP-DT, and E-NP-DTS were synthesized using a double-emulsion method, as described previously (Wang et al. 2011, 2013). The nanoparticles were characterized by dynamic light scattering (DLS, Malvern Instruments Ltd., Malvern, UK) and transmission electron microscopy prior to further analysis (TEM, JEM-200CX, Jeol Ltd., Japan) (Fig. S1).
2.2 Cell culture and drug exposure
MCF-7 cells were cultured in 15 cm diameter petri-dishes with 20 mL l-Dulbecco’s modified Eagle’s medium (l-DMEM) supplemented with 10% (v/v) fetal calf serum (FCS) and 1% 200 mM l-glutamine (GIBCO-Invitrogen Corporation, UK). Cells were incubated at 37 °C, with 5% CO2. All cultured MCF-7 cells were mixed and homogeneously divided into one hundred and fifty 15 cm diameter culture dishes. Before the cell confluence reached about 40%, every 30 dishes were treated with PBS, E-NP, E-NP-DOX, E-NP-DT and E-NP-DTS, respectively. Alive cells (n = 10) were collected from each group at different time intervals: 10, 24, and 36 h post treatment. Cells were washed with 10 mL 4 °C PBS and trypsinized by adding 0.25% trypsin supplemented with 0.02% ethylenediaminetetraacetic acid (EDTA) (GIBCO-Invitrogen Corporation, UK). Before cells starting to detach from culture dishes, PBS was added to remove trypsin. Cells were then transferred to a 2 mL centrifuge tube and gently washed for three times with 4 °C PBS before being stored at −80 °C (Fig. S2).
2.3 Extraction of intracellular metabolites
Cell pellets were homogenized in 600 μL methanol/H2O (2:1 v/v) solution and subjected to three freeze–thaw cycles to lyse the cells. Sonication was performed in ice for 15 min with cycles consisting of 1-min power followed by 1-min stop. Supernatant was collected after centrifugation at 12,000g for 10 min at 4 °C. The remaining cell residues were further extracted twice using the above-mentioned procedure. The supernatant from the three extractions was pooled together and lyophilized after removal of methanol using SpeedVac (ThermoFisher, America). The cell extracts were dissolved in 600 μL phosphate buffer (pH 7.4, 99.9% D2O) containing 0.001% TSP-d4 (Xiao et al. 2009). After centrifugation at 16,000g for 10 min at 4 °C, 550 μL of the supernatants was transferred into 5 mm NMR tubes for NMR analysis.
2.4 NMR measurements
1H NMR spectra of cell extracts were acquired at 298 K on a Bruker Avance III 600 MHz NMR spectrometer equipped with a cryogenic probe (Bruker, Biospin, Germany) using the first increment of NOESY sequence. Water presaturation was achieved by irradiation of water resonance during the recycle delay (2 s) and mixing time (80 ms). All 1H NMR spectra were multiplied by an exponential function with a 1 Hz line broadening factor before Fourier transformation (FT). The spectra were referenced to the TSP methyl resonance (δ 0.000). For the purposes of signal assignment, a range of two-dimensional NMR spectra including 1H-1H J-RES, COSY and TOCSY, 1H-13C HSQC and HMBC were acquired and processed as described previously (Zhang et al. 2013; Li et al. 2015b) (Fig. S3).
2.5 NMR data processing and multivariate data analysis
The spectral region δ 0.5–9.5 was integrated into regions with an equal width of 0.002 ppm (1.2 Hz) using an AMIX software package (V3.9.2, Bruker Biospin, Germany). Regions δ 4.591–5.039 and δ 3.707–3.729 were discarded to avoid the effects of water saturation and exogenous PEG, respectively. Each integral bucket was then normalized to the sum of the total integrals prior to statistical analysis.
Multivariate data analyses were performed on SIMCA-P + software (V. 11.0, Umetrics, Sweden). Principal component analysis (PCA) was firstly conducted on the mean-centered NMR data to generate an overview of the data distribution and detect potential outliers. Orthogonal projection to latent structure with discriminant analysis (OPLS-DA), a supervised multivariate data analytical tool, was subsequently applied to the unit-variance scaled data and the model was cross-validated using a sevenfold cross-validation method. The quality of the model was monitored by the parameters Q2 and R2X, indicating the model predictability and the total explained variations, respectively (Eriksson et al. 2006). The validity of all models was further ensured by permutation tests (200 permutations) and CV-ANOVA tests (p < 0.05) (Eriksson et al. 2008).
The model was interpreted by back-transformed loadings incorporated with color-coded correlation coefficients, which were achieved with an in-house developed Matlab script (MATLAB 7.1, the Mathworks Inc., Natwick, USA). The color of the correlation coefficient indicates the importance of the metabolite in contributing to class differentiation (red color more important than a blue color). In this study a correlation coefficient |r| greater than 0.602 was considered to be statistically significant (p < 0.05) (Cloarec et al. 2005).
2.6 GC-FID/MS analysis of fatty acids in cells
Fatty acids were methylated prior to GC-MS detection. A total of 5 mg cells were suspended in 0.2 mL methanol together with 4 mL methanol-hexane mixture (4:l, v/v), internal standards (C17:0 and C23:0) and butylated hydroxytoluene (BHT). The reaction was initiated by the addition of 200 mL pre-cooled acetyl chloride and halted by adding 5 mL 6% K2CO3 solution after leaving the mixture on ice for 24 h. The methylated fatty acids were extracted using 200 mL of hexane three times and the supernatants were pooled together, followed by the evaporation of hexane.
The methylated fatty acids were measured on a Shimadzu GC2010Plus GC-MS spectrometer (Shimadzu Scientific Instruments, USA) equipped with a flame ionization detector (FID) and a mass spectrometer (MS) with an electron impact (EI) ion source. A DB-225 capillary GC column with length of 10 m, inner diameter of 0.1 mm and film thickness of 0.1 mm was used. The sample injection volume was 1 mL with a splitter (1:60). Both the injection port and detector temperatures were set at 230 °C. The column temperature was set at 155 °C and kept for 1 min, then increased from 155 to 205 °C (30 °C/min), and kept at 205 °C for 3 min, and finally elevated from 205 to 230 °C (5 °C/min). Fatty acids were identified by comparing the signals with a mixture of 37 known standards, and quantified by comparing with peak integrals of internal standards C17:0 and C23:0 (Li et al. 2013, 2015a). The statistical analyses of GC-FID/MS data were performed with the student’s t test or nonparametric analysis (IBM SPSS Statistics V20.0).
2.7 Quantitative RT-PCR analysis
MCF-7 cells were harvested after 36 h of treatment, and RNA was isolated using Trizol (Transgen Biotech, Beijing, P.R. China). Reverse transcription was performed three times using a First Strand cDNA Synthesis kit (Thermo Scientific, Rockford, USA). Quantitative RT-PCR was performed using SYBR® Premix Ex Taq™ II (TaKaRa, Japan). The primer sequences are provided in the Supplementary Information (Table S1). Gene transcription data was normalized using the reference gene β-actin. The data was analyzed using SPSS (IBM SPSS Statistics V20.0) and presented as the mean ± standard deviation (SD).
3 Results
3.1 Metabolic characterization of intracellular extracts
Intracellular metabolite assignments of typical 1H NMR spectra of MCF-7 extracts (Fig. 1, Table S2) were confirmed by a set of two-dimensional NMR spectra. The metabolic profile of MCF-7 intracellular extracts consisted of a range of amino acids (AAs), membrane metabolites including choline, phosphocholine (PC), glycerophosphocholine (GPC), short chain fatty acids (SCFAs), organic acids, nucleosides, and nucleotides.
Typical 600 MHz 1H NMR spectra of aqueous methanol extracts from MCF-7 cells. The region of δ 5.2–9.5 in the spectra is vertically expanded eight times compared with the region of δ 0.5–4.7. Key: 1 isoleucine, 2 leucine, 3 valine, 4 lactate, 5 alanine, 6 lysine, 7 glutamate, 8 glutathione (reduced), 9 glutathione (oxidized), 10 succinate, 11 aspartate, 12 creatine, 13 choline, 14 phosphorylcholine, 15 glycerophosphocholine, 16 taurine, 17 methylphosphate, 18 glycine, 19 UDP-glucose, 20 UDP-glucuronate, 21 UDP-N-acetylglucosamine, 22 UDP-N-acetylgalactosamine, 23 N-acetylglucosamine, 24 tyrosine, 25 phenylalanine, 26 uracil, 27 nicotinamide, 28 hypoxanthine, 29 NAD+, 30 uridine, 31 AMP, 32 ADP, 33 histidine, 34 formate, 35 PEG, 36 fatty acid
3.2 Metabolic consequences of E-NP and its encapsulated drugs
In order to evaluate the metabolic responses of cells to the exposure of nano-carrier (E-NP) and its encapsulated drugs, NMR profiles of cells treated with E-NP were compared with those treated with PBS, E-NP-DOX, E-NP-DT or E-NP-DTS at matched time points using an O-PLS-DA strategy (Fig. 2a). The qualities for the models were confirmed by the R2 and Q2 values and further evaluated with a CV-ANOVA approach (p < 0.05) (Fig. S4, Table S3). The results of the comparison between E-NP and PBS revealed that the amphiphilic copolymer nanocarrier E-NP induced significant elevations in the levels of lactate, succinate, uridine diphosphate-glucuronate (UDP-GlcA), and adenosine diphosphate (ADP), accompanied with depletions in the levels of most amino acids at 10 h. Notably, incubation with E-NP resulted in metabolites that increased in level at 10 h dropping in level by 24 h, and vice versa. This fluctuation suggested that the recovery of metabolic disruptions occurred at 24 h and an over compensation mechanism may have contributed to the fluctuations observed.
Heat-map summarizing dynamic metabolic changes in 1H NMR spectral data of cell extracts. Metabolite changes are color-scaled with correlation coefficients where red denotes an increase of metabolite levels whereas cold color indicates a decrease in the treated MCF-7 with respect to the control group; colorless blocks meant no significant changes between two groups
To investigate the metabolic profiles of breast cancer cell MCF-7 treated with E-NP encapsulated DOX, a combination of DOX and TAX, or with additional survivin siRNA, we considered the metabolic profiles obtained from cells treated with E-NP as negative controls. The O-PLS-DA showed that elevated levels of choline, uracil, hypoxanthine and a range of amino acids, such as glutamate, leucine, and tyrosine, were associated with MCF-7 cells treated with encapsulated DOX. These changes were companied with decreased levels of lactate, ADP, and UDP-N-acetylglucosamine (GlcNAc) at 10 h. Metabolic recovery of cells treated with encapsulated DOX was noted at 24 h post treatment, where either fluctuations of metabolites, or complete recovery of metabolites was observed 36 h (Fig. 2b). This observation was in contrast to the metabolic profiles of cells treated with encapsulated DT and DTS. Encapsulated DT and DTS treated cells displayed similar metabolic profiles and were characterized by up-regulated levels of a range of amino acids, GlcNAc, and UDP-N-acetylgalactosamine (GalNAc), and down-regulated levels of lactate, succinate, creatine, choline, and PC. Furthermore, results demonstrated that encapsulated DT and DTS treatment did not result in a recovery of metabolic profiles even at 36 h post treatment. Other altered metabolites, such as taurine, hypoxanthine, NAD and ADP showed a certain degree of recovery at 24 h post treatment (Fig. S5, S6 and S7; Table S3). Notably, the PC:GPC ratio decreased significantly at 36 h exposure of encapsulated DT and DTS, suggesting that an up-regulation of GPC accompanied with down-regulation of PC was associated with exposure of both E-NP-DT and E-NP-DTS (Fig. 3).
3.3 GC-FID/MS analysis of fatty acid composition
Lipid analysis was performed on MCF-7 cells treated with PBS, E-NP, E-NP-DOX, E-NP-DT and E-NP-DTS for 10, 24 and 36 h. A total of 17 fatty acids, including C16:0, C18:0, C20:0, C22:0 and their relevant unsaturated forms were detected and quantified in MCF-7 cells. Compared to cells treated with PBS, the nanocarrier E-NP induced significant elevations in the levels of saturated fatty acids (C14:0, C20:0) and total monounsaturated fatty acid (MUFA) at 10 h post treatment (Table S4). Similar to the NMR profiles obtained from cells treated with E-NP, these changes of fatty acids were diminished when incubating with E-NP for 24 h and complete recovery was achieved at 36 h post treatment. However, for cells treated with encapsulated drugs, a high level of disruption in the levels of fatty acids was observed (Table S5). Encapsulated DOX resulted in down-regulated levels of C14:0, C20:0, C16:1n7 and C18:1n9 at 10 h post treatment, while a full recovery was made at 24 h and up-regulated levels of C18:2n6 and C20:5n3 was observed at 36 h post exposure. Both encapsulated DT and DTS induced in a lower level of series of fatty acids at 10 h and these alterations were reversed at 24 h. After 36 h exposure, the increase of a number of polyunsaturated fatty acids (PUFA) in encapsulated DT and DTS treated cells was observed (Table S5).
3.4 The transcriptional levels of related genes
The levels of UDP-GlcNAc and UDP-GalNAc increased markedly in both encapsulated DT and DTS treatment groups. UDP-GlcNAc and UDP-GalNAc are end-products of the hexosamine pathway. In order to confirm changes in these pathways, mRNA expression of enzymes involved in the hexosamine pathway were measured (Fig. 4). The transcriptional levels of GFPT1, which encodes glutamine-fructose-6-phosphate amidotransferase (GFPT1), a rate-limited enzyme in hexosamine biosynthesis pathway (HBP), was significantly reduced in cells treated with encapsulated DOX and DT, (as compared with cells treated with E-NP) and significantly increased in cells treated with encapsulated DTS. We subsequently investigated the expression levels of O-GlcNAc transferases (OGT) and O-GlcNAcases (OGA) since these two enzymes catalyze O-GlcNAcylation, where UDP-GlcNAc donates a GlcNAc (Hart et al. 2007; Zeidan and Hart 2010). Our findings showed that encapsulated DT and DTS resulted in a significant up-regulation of OGA, whereas OGT was not significantly responsive to these encapsulated drug treatments (Fig. 4).
The PC:GPC ratio significantly reduced in both encapsulated DT and DTS treatment groups (Fig. 3). Fatty acids, choline and phosphocholine (PC) are the precursors for the biosynthesis of phosphatidylcholine, which is the most abundant phospholipid within biological membranes (Esko et al. 1982). Choline kinase A (CHKA) catalyzes choline into PC, and the reverse reaction is catalyzed by phosphocholine phosphatase (PHOSPHO1) (Roberts et al. 2004). Our results demonstrated a higher level of PHOSPHO1 expression in E-NP treated group as compared with the PBS negative control (Fig. 4). The expression of PHOSPHO1 was higher in cells treated with encapsulated DTS, whereas the expression of CHKA was higher in cells treated with encapsulated DOX. The expressions of both choline/ethanolamine phosphotransferase 1 (CEPT1) and choline phosphotransferase 1 (CHPT1) were downgraded in encapsulated DT and DTS treated groups while the transcriptional level of PCYT1A was elevated in encapsulated DTS treated cells (Fig. 4). In CDP-Choline cycle, phosphocholine cytidylyltransferase A (PCYT1A) catalyzes the production of CDP-choline while CDP-choline is transferred into phosphatidylcholine (PTC) by both CEPT1 and CHPT1.
4 Discussion
Doxorubicin and paclitaxel are anti-tumor drugs and have been used in combination to treat malignant neoplasm for decades (Gehl et al. 1996). In order to overcome the side effects as well as improve efficacy of treatments, these drugs, combined with survivin siRNA were incorporated into an amphiphilic nanocarrier, E-NP. The encapsulated drug has shown to improve antitumor outcomes in both in vitro and in vivo studies (Wang et al. 2011, 2013). To investigate the metabolic effects of an amphiphilic nanocarrier and its encapsulated drugs, we employed untargeted NMR-based metabolomics combining free fatty acids targeted metabolomics based on GC-MS approach and analyzed the metabolome of MCF-7 breast cancer cells exposed to these encapsulated drugs at different time intervals, i.e. at 10, 24 and 36 h. In addition, the relative expression levels of genes concerning several metabolic pathways were validated by qRT-PCR.
The exposure of E-NP posed no impact on the survival of MCF-7 cells (Fig. 5), which suggested potential biocompatibility of this material for clinical use. Nonetheless, E-NP exposure resulted in some metabolic alterations, suggesting that this copolymer vehicle was not completely inert in vitro. However, a fluctuation in the levels of metabolites in MCF-7 cells after 10 h of E-NP treatment suggested a potential partial recovery of metabolic perturbation. Fluctuations in the metabolisms has been observed before (Zhao et al. 2013; Wan et al. 2015), and is likely due to an over compensation mechanism. The initial depletion of amino acids, choline and some nucleotides, as well as elevated UDP-GlcA, lactate, succinate and ADP observed after E-NP exposure could be explained by endocytosis. The uptake of E-NP through endocytosis via clathrin- or caveolae-mediated pathway is known to be energy-dependent (Rejman et al. 2004; Frohlich 2012; Kim et al. 2006). The active endocytic trafficking at the start of E-NP exposure may have promoted the TCA activity and up-regulated levels of succinate to provide sufficient levels of ATP. The down-regulation of amino acids observed in cells exposed to E-NP could be a result of cells supplying additional energy or synthesizing certain functional proteins that may have become insufficient due to non-specific binding between E-NPs and intracellular proteins (Lynch and Dawson 2008).
Amphiphilic nanocarrier-encapsulated drug, doxorubicin, significantly inhibit the proliferation of MCF-7, whilst encapsulated DT and DTS show similar and improved capability in inhibiting cancer cell proliferation compared with single drug incorporation (Fig. 5). The metabolic profiles perturbed by the three nano-drugs mirrored the cell proliferation profiles: both encapsulated DT and DTS induced similar metabolic disruptions with prolonged effects (Fig. 2b) whereas cells exposed to encapsulated DOX displayed metabolic recovery after 24 h post treatment (Fig. 2b). The metabolic recovery in encapsulated DOX treated cells may due to the rapid release of limited DOX in 10 h.
The most noticeable metabolic perturbation of the three nano-drugs was their steady increase in the levels of a range of amino acids in exposed cells (Fig. 6). This observation can be explained by the activation of apoptosis induced by these nano-drugs. DOX intercalates itself into DNA strands and results in the activation of molecular signals from AMPK to alter the Bcl-2/Bax ratio, subsequently activating different caspases and apoptosis (Reed 1994; Leung and Wang 1999; Tacar et al. 2013). Apoptosis ultimately results in hydrolysis of some proteins. In this current study, protein concentration was significantly reduced in cells treated with nano-drugs (Fig. S8A), which provides supportive evidence for drug-induced protein hydrolysis and subsequent cell death.
The metabolic changes as a result of exposure to nano-drugs were manifested in the down-regulated levels of lactate, suggesting an inhibition of glycolysis within the cancer cells (Fig. 6). Cancer cells undergo anaerobic glycolysis regardless of whether oxygen is sufficient, thereby producing high levels of lactate, which is known as the “Warburg effect” (Gatenby and Gillies 2004). The decrease in the levels of lactate suggested that these nano-drugs might be capable of modifying the microenvironment of cancer cells by reducing lactate levels through inhibition of glycolysis, which impairs the nutrient supply to cancer cells and eventually results in cell death. The down-regulated levels of glycolysis are in agreement with concurrent down-regulated TCA activity, in which reduced levels of succinate are evident in encapsulated DT and DTS treated cells (Fig. 2b). The persistent inhibition of lactate production and TCA activity observed in encapsulated DT and DTS treated cells could be attributed to the drug synergy effect. The decreased levels of a series of fatty acids in encapsulated drugs treated cells at 10 h could be a consequence of energy metabolism adjustment while the glycolysis in MCF-7 cells was suppressed. After being exposed to encapsulated drugs for 36 h, the alteration of fatty acids in encapsulated drugs treated cells was reversed to some extent (Table S5). A decreased GSH to GSSG ratio related to ROS stress was observed at 36 h (Fig. S8B), which could lead to mitochondrial dysfunctions and divert fatty acids away from β-oxidation. Besides, lipid accumulation due to increased de novo synthesis was reported as a character of apoptosis (Boren and Brindle 2012). The breakdown of biological membranes followed by apoptosis could also result in an elevated level of fatty acids (Table S5).
The synergy effect of encapsulated DT and DTS was manifested in the disruption the hexosamine biosynthetic pathway (HBP), evident by an increase in levels of the end products of HBP, UDP-GlcNAc and UDP-GalNAc (Fig. 2b and 6). The net levels of both nucleotide sugars depend on the rate of their production and consumption. GFPT1 is the rate-limiting enzyme controlling the production of UDP-GlcNAc and UDP-GalNAc, and its expression levels were significantly up-regulated in encapsulated DTS treated cells (Fig. 4b). However, similar trend was not observed in encapsulated DT treated cells. The expression of GFPT1 was down-regulated. An alternative pathway, such as, deglycosylation, could contribute to the accumulated levels of UDP-GlcNAc and UDP-GalNAc for cells treated with encapsulated DT. The deglycosylation of proteins is catalyzed by O-GlcNAcase (OGA), whereas glycosylation is mediated by OGT (Hart et al. 2007; Zeidan and Hart 2010). Herein, the expression levels of OGA were significantly up-regulated in both encapsulated DT and DTS treated cells (Fig. 4a). Therefore, the increased levels of nucleotide sugars in the encapsulated DT treated cells could be a consequence of the accelerated deglycosylation process. Glycosylation is a critical post-translational modification for many functional proteins (Zachara and Hart 2006), and O-glycosylation in particular, attenuates nuclear factor kappa B (NF-κB) and has shown to be involved in increased tolerance to stresses such as cardiac myopathy, diabetes, and severe injury from trauma (Zachara and Hart 2004; Chatham et al. 2008).
The metabolic disruptions resulted from the synergy effect of multiple drugs, also exhibited by the up-regulated concentration of GPC accompanied with down-regulated concentrations of PC, consequently reduced PC:GPC ratio in encapsulated DT and DTS treated cells (Fig. 3). The depleted PC levels are consistent with the enhanced expressions of PHOSPHO1 and PCYT1A (Fig. 4). It was also observed that the expressions of CEPT1 and CHPT1, that catalyzed CDP-choline to generate PTC, were down-regulated. PC, GPC, and PTC are important membrane constituents, and play an integral role in cancer proliferation and cancer treatment. Higher levels of PC and the total choline-containing phospholipids are features of many malignant cells. The levels of PC and tCho increase as cells transform from normal to immortalized cells, and then to oncogenic cells (Iorio et al. 2005; Glunde et al. 2004; Aboagye and Bhujwalla 1999). A decrease in PC:GPC ratio and reduced PTC biosynthesis has been observed during cell growth arrest, followed by apoptosis (Mirbahai et al. 2011; Lamego et al. 2014). Our current observation of reduced PC:GPC ratio after treatment with E-NP-DT and E-NP-DTS implies a significant reduction in malignant proliferation, highlighting the effectiveness of these nano-drugs. As a consequence of cell death, disruptions of purine and pyridine metabolisms were observed in treated cells.
5 Conclusion
Here we have shown a time-dependent response of human breast cancer cells exposed to different formulations of anti-tumor agents carried by a copolymer nanocarrier. First of all, E-NP exposure only induced transit metabolic disruptions without affecting cell survival rate, indicating high biocompatibility of this material. Secondly, encapsulated doxorubicin could suppress the proliferation of MCF-7 cells while perturb several metabolic pathways, including glycolysis, purine and pyridine metabolism and amino acids metabolism. The metabolic alterations were diminished after 10 h. Furthermore, the nanoparticles incorporating multiple drugs consistently resulted in significant cell cycle arrest and apoptosis, evidenced by marked proteolysis, reduced levels of PC:GPC, anaerobic glycolysis inhibition, and increased O-glycosylation. These findings provide important insights into the dynamic biological effects of polymer-based nano-materials and their encapsulated chemotherapeutic drugs in vitro. Our work has strongly demonstrated the combination of NMR and GC-FID/MS-based metabonomics and qRT-PCR as a powerful tool for understanding the influence of nanomaterials at a molecular level.
References
Aboagye, E. O., & Bhujwalla, Z. M. (1999). Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Research, 59(1), 80–84.
Alibolandi, M., Ramezani, M., Sadeghi, F., Abnous, K., & Hadizadeh, F. (2015). Epithelial cell adhesion molecule aptamer conjugated PEG-PLGA nanopolymersomes for targeted delivery of doxorubicin to human breast adenocarcinoma cell line in vitro. International Journal of Pharmaceutics, 479, 241–251. doi:10.1016/j.ijpharm.2014.12.035.
Ambrosini, G., Adida, C., & Altieri, D. C. (1997). A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nature Medicine, 3(8), 917–921. doi:10.1038/Nm0897-917.
An, Y. P., Xu, W. X., Li, H. H., Lei, H. H., Zhang, L. M., Hao, F. H., et al. (2013). High-fat diet induces dynamic metabolic alterations in multiple biological matrices of rats. Journal of Proteome Research, 12(8), 3755–3768. doi:10.1021/pr400398b.
Bathen, T. F., Sitter, B., Sjobakk, T. E., Tessem, M. B., & Gribbestad, I. S. (2010). Magnetic resonance metabolomics of intact tissue: a biotechnological tool in cancer diagnostics and treatment evaluation. Cancer Research, 70(17), 6692–6696. doi:10.1158/0008-5472.CAN-10-0437.
Boren, J., & Brindle, K. M. (2012). Apoptosis-induced mitochondrial dysfunction causes cytoplasmic lipid droplet formation. Cell Death and Differentiation, 19(9), 1561–1570. doi:10.1038/cdd.2012.34.
Carew, J. S., Espitia, C. M., Zhao, W. G., Mita, M. M., Mita, A. C., & Nawrocki, S. T. (2015). Targeting survivin inhibits renal cell carcinoma progression and enhances the activity of temsirolimus. Molecular Cancer Therapeutics, 14(6), 1404–1413. doi:10.1158/1535-7163.MCT-14-1036.
Chatham, J. C., Not, L. G., Fulop, N., & Marchase, R. B. (2008). Hexosamine biosynthesis and protein O-glycosylation: The first line of defense against stress, ischemia, and trauma. Shock (Augusta, Ga.), 29(4), 431–440. doi:10.1097/SHK.0b013e3181598bad.
Cloarec, O., Dumas, M. E., Trygg, J., Craig, A., Barton, R. H., Lindon, J. C., et al. (2005). Evaluation of the orthogonal projection on latent structure model limitations caused by chemical shift variability and improved visualization of biomarker changes in H-1 NMR spectroscopic metabonomic studies. Analytical Chemistry, 77(2), 517–526. doi:10.1021/ac048803i.
Davis, M. E., Chen, Z. G., & Shin, D. M. (2008). Nanoparticle therapeutics: An emerging treatment modality for cancer. Nature Reviews Drug Discovery, 7(9), 771–782. doi:10.1038/nrd2614.
Eriksson, L., Andersson, P. L., Johansson, E., & Tysklind, M. (2006). Megavariate analysis of environmental QSAR data. Part I: A basic framework founded on principal component analysis (PCA), partial least squares (PLS), and statistical molecular design (SMD). Molecular Diversity, 10(2), 169–186. doi:10.1007/s11030-006-9024-6.
Eriksson, L., Trygg, J., & Wold, S. (2008). CV-ANOVA for significance testing of PLS and OPLS (R) models. Journal of Chemometrics, 22(11–12), 594–600. doi:10.1002/cem.1187.
Esko, J. D., Nishijima, M., & Raetz, C. R. H. (1982). Animal-cells dependent on exogenous phosphatidylcholine for membrane biogenesis. Proceedings of the National Academy of Sciences of the United States of America-Biological Sciences, 79(6), 1698–1702. doi:10.1073/pnas.79.6.1698.
Frohlich, E. (2012). The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. International Journal of Nanomedicine, 7, 5577–5591. doi:10.2147/Ijn.S36111.
Gatenby, R. A., & Gillies, R. J. (2004). Why do cancers have high aerobic glycolysis? Nature Reviews Cancer, 4(11), 891–899. doi:10.1038/nrc1478.
Gehl, J., Boesgaard, M., Paaske, T., Jensen, B. V., & Dombernowsky, P. (1996). Combined doxorubicin and paclitaxel in advanced breast cancer: Effective and cardiotoxic. Annals of Oncology, 7(7), 687–693.
Glunde, K., Jie, C., & Bhujwalla, Z. M. (2004). Molecular causes of the aberrant choline phospholipid metabolism in breast cancer. Cancer Research, 64(12), 4270–4276. doi:10.1158/0008-5472.Can-03-3829.
Han, L., Tang, C., & Yin, C. (2015). Dual-targeting and pH/redox-responsive multi-layered nanocomplexes for smart co-delivery of doxorubicin and siRNA. Biomaterials, 60, 42–52. doi:10.1016/j.biomaterials.2015.05.001.
Harries, M., Ellis, P., & Harper, P. (2005). Nanoparticle albumin-bound paclitaxel for metastatic breast cancer. Journal of Clinical Oncology, 23(31), 7768–7771. doi:10.1200/Jco.2005.08.002.
Hart, G. W., Housley, M. P., & Slawson, C. (2007). Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature, 446(7139), 1017–1022. doi:10.1038/nature05815.
He, C. L., Ma, H. C., Cheng, Y. L., Li, D. S., Gong, Y. B., Liu, J. G., et al. (2015). PLK1shRNA and doxorubicin co-loaded thermosensitive PLGA-PEG-PLGA hydrogels for localized and combined treatment of human osteosarcoma. Journal of Controlled Release, 213, E18–E18. doi:10.1016/j.jconrel.2015.05.026.
Huang, C. Y., Lei, H. H., Zhao, X. J., Tang, H. R., & Wang, Y. L. (2013a). Metabolic Influence of Acute Cyadox Exposure on Kunming Mice. Journal of Proteome Research, 12(1), 537–545. doi:10.1021/pr301033a .
Huang, Q., Tan, Y. X., Yin, P. Y., Ye, G. Z., Gao, P., Lu, X., et al. (2013b). Metabolic characterization of hepatocellular carcinoma using nontargeted tissue metabolomics. Cancer Research, 73(16), 4992–5002. doi:10.1158/0008-5472.CAN-13-0308.
Huang, X. H., Jain, P. K., El-Sayed, I. H., & El-Sayed, M. A. (2007). Gold nanoparticles: Interesting optical properties and recent applications in cancer diagnostic and therapy. Nanomedicine: The Official Journal of the American Academy of Nanomedicine, 2(5), 681–693. doi:10.2217/17435889.2.5.681.
Iorio, E., Mezzanzanica, D., Alberti, P., Spadaro, F., Ramoni, C., D’Ascenzo, S., et al. (2005). Alterations of choline phospholipid metabolism in ovarian tumor progression. Cancer Research, 65(20), 9369–9376. doi:10.1158/0008-5472.CAN-05-1146.
Jain, P. K., El-Sayed, I. H., & El-Sayed, M. A. (2007). Au nanoparticles target cancer. Nano Today, 2(1), 18–29. doi:10.1016/S1748-0132(07)70016-6.
Jeong, B., Bae, Y. H., & Kim, S. W. (2000). Drug release from biodegradable injectable thermosensitive hydrogel of PEG-PLGA-PEG triblock copolymers. Journal of Controlled Release, 63(1–2), 155–163. doi:10.1016/S0168-3659(99)00194-7.
Kaddurah-Daouk, R., Kristal, B. S., & Weinshilboum, R. M. (2008). Metabolomics: A global biochemical approach to drug response and disease. Annual Review of Pharmacology and Toxicology, 48, 653–683. doi:10.1146/annurev.pharmtox.48.113006.094715.
Kar, R., Palanichamy, J. K., Banerjee, A., Chattopadhyay, P., Jain, S. K., & Singh, N. (2015). Survivin siRNA increases sensitivity of primary cultures of ovarian cancer cells to paclitaxel. Clinical & Translational Oncology, 17(9), 737–742. doi:10.1007/s12094-015-1302-2.
Kim, J. S., Yoon, T. J., Yu, K. N., Noh, M. S., Woo, M., Kim, B. G., et al. (2006). Cellular uptake of magnetic nanoparticle is mediated through energy-dependent endocytosis in A549 cells. Journal of Veterinary Science, 7(4), 321–326.
Lamego, I., Duarte, I. F., Marques, M. P., & Gil, A. M. (2014). Metabolic markers of MG-63 osteosarcoma cell line response to doxorubicin and methotrexate treatment: Comparison to cisplatin. Journal of Proteome Research, 13(12), 6033–6045. doi:10.1021/pr500907d.
Lee, K. S., Chung, H. C., Im, S. A., Park, Y. H., Kim, C. S., Kim, S. B., et al. (2008). Multicenter phase II trial of Genexol-PM, a Cremophor-free, polymeric micelle formulation of paclitaxel, in patients with metastatic breast cancer. Breast Cancer Research and Treatment, 108(2), 241–250. doi:10.1007/s10549-007-9591-y.
Leung, L. K., & Wang, T. T. Y. (1999). Differential effects of chemotherapeutic agents on the Bcl-2/Bax apoptosis pathway in human breast cancer cell line MCF-7. Breast Cancer Research and Treatment, 55(1), 73–83. doi:10.1023/A:1006190802590.
Li, D., Zhang, L. L., Dong, F. C., Liu, Y., Li, N., Li, H. H., et al. (2015a). Metabonomic changes associated with atherosclerosis progression for LDLR−/− mice. Journal of Proteome Research, 14(5), 2237–2254. doi:10.1021/acs.jproteome.5b00032.
Li, F. Z., Ambrosini, G., Chu, E. Y., Plescia, J., Tognin, S., Marchisio, P. C., et al. (1998). Control of apoptosis and mitotic spindle checkpoint by survivin. Nature, 396(6711), 580–584.
Li, H. D., Zhu, W. D., Zhang, L. K., Lei, H. H., Wu, X. Y., Guo, L., et al. (2015b). The metabolic responses to hepatitis B virus infection shed new light on pathogenesis and targets for treatment. Scientific Reports, 5, 8421. doi:10.1038/srep08421.
Li, H. H., An, Y. P., Zhang, L. L., Lei, H. H., Zhang, L. M., Wang, Y. L., et al. (2013). Combined NMR and GC-MS analyses revealed dynamic metabolic changes associated with the carrageenan-induced rat pleurisy. Journal of Proteome Research, 12(12), 5520–5534. doi:10.1021/pr400440d.
Lynch, I., & Dawson, K. A. (2008). Protein-nanoparticle interactions. Nano Today, 3(1–2), 40–47. doi:10.1016/S1748-0132(08)70014-8.
Michalet, X., Pinaud, F. F., Bentolila, L. A., Tsay, J. M., Doose, S., Li, J. J., et al. (2005). Quantum dots for live cells, in vivo imaging, and diagnostics. Science, 307(5709), 538–544. doi:10.1126/science.1104274.
Mirbahai, L., Wilson, M., Shaw, C. S., McConville, C., Malcomson, R. D. G., Griffin, J. L., et al. (2011). (1)H magnetic resonance spectroscopy metabolites as biomarkers for cell cycle arrest and cell death in rat glioma cells. International Journal of Biochemistry & Cell Biology, 43(7), 990–1001. doi:10.1016/j.biocel.2010.07.002.
Nicholson, J. K., Lindon, J. C., & Holmes, E. (1999). ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica; The Fate of Foreign Compounds in Biological Systems, 29(11), 1181–1189.
Northfelt, D. W., Dezube, B. J., Thommes, J. A., Miller, B. J., Fischl, M. A., Friedman-Kien, A., et al. (1998). Pegylated-liposomal doxorubicin versus doxorubicin, bleomycin, and vincristine in the treatment of AIDS-related Kaposi’s sarcoma: Results of a randomized phase III clinical trial. Journal of Clinical Oncology, 16(7), 2445–2451.
Overman, M. J., Kazmi, S. M., Jhamb, J., Lin, E., Yao, J. C., Abbruzzese, J. L., et al. (2010). Weekly docetaxel, cisplatin, and 5-fluorouracil as initial therapy for patients with advanced gastric and esophageal cancer. Cancer, 116(6), 1446–1453. doi:10.1002/cncr.24925.
Portney, N. G., & Ozkan, M. (2006). Nano-oncology: Drug delivery, imaging, and sensing. Analytical and Bioanalytical Chemistry, 384(3), 620–630. doi:10.1007/s00216-005-0247-7.
Reed, J. C. (1994). Bcl-2 and the regulation of programmed cell-death. Journal of Cell Biology, 124(1–2), 1–6. doi:10.1083/Jcb.124.1.1.
Rejman, J., Oberle, V., Zuhorn, I. S., & Hoekstra, D. (2004). Size-dependent internalization of particles via the pathways of clathrin-and caveolae-mediated endocytosis. Biochemical Journal, 377, 159–169. doi:10.1042/Bj20031253.
Roberts, C. J., Stewart, A. J., Sadler, P. J., & Farquharson, C. (2004). Human PHOSPHO1 displays high specific phosphoethanolamine and phosphocholine phosphatase activities: A means of generating inorganic phosphate in mineralising cells. Journal of Bone and Mineral Research, 19(6), 1041–1041.
Shen, S., Du, X. J., Liu, J., Sun, R., Zhu, Y. H., & Wang, J. (2015). Delivery of bortezomib with nanoparticles for basal-like triple-negative breast cancer therapy. Journal of Controlled Release: Official Journal of the Controlled Release Society, 208, 14–24. doi:10.1016/j.jconrel.2014.12.043.
Song, Y. P., Zhao, R. F., Hu, Y. L., Hao, F. H., Li, N., Nie, G. J., et al. (2015). Assessment of the biological effects of a multifunctional nano-drug-carrier and its encapsulated drugs. Journal of Proteome Research, 14(12), 5193–5201. doi:10.1021/acs.jproteome.5b00513.
Tacar, O., Sriamornsak, P., & Dass, C. R. (2013). Doxorubicin: An update on anticancer molecular action, toxicity and novel drug delivery systems. Journal of Pharmacy and Pharmacology, 65(2), 157–170. doi:10.1111/j.2042-7158.2012.01567.x.
Tang, F., Li, L., & Chen, D. (2012). Mesoporous silica nanoparticles: Synthesis, biocompatibility and drug delivery. Advanced Materials, 24(12), 1504–1534. doi:10.1002/adma.201104763.
Tang, H. R., & Wang, Y. L. (2006). Metabonomics: A revolution in progress. Progress in Biochemistry and Biophysics, 33(5), 401–417.
Torchilin, V. P. (2012). Multifunctional nanocarriers. Advanced Drug Delivery Reviews, 64, 302–315. doi:10.1016/j.addr.2012.09.031.
Wan, Q., Wu, G., He, Q., Tang, H., & Wang, Y. (2015). The toxicity of acute exposure to T-2 toxin evaluated by the metabonomics technique. Molecular BioSystem, 11(3), 882–891. doi:10.1039/c4mb00622d.
Wang, H., Wu, Y., Zhao, R., & Nie, G. (2013). Engineering the assemblies of biomaterial nanocarriers for delivery of multiple theranostic agents with enhanced antitumor efficacy. Advanced Materials, 25(11), 1616–1622. doi:10.1002/adma.201204750.
Wang, H., Zhao, Y., Wu, Y., Hu, Y. L., Nan, K., Nie, G., et al. (2011). Enhanced anti-tumor efficacy by co-delivery of doxorubicin and paclitaxel with amphiphilic methoxy PEG-PLGA copolymer nanoparticles. Biomaterials, 32(32), 8281–8290. doi:10.1016/j.biomaterials.2011.07.032.
Wang, Z., Chen, Z., Yang, S., Wang, Y., Yu, L., Zhang, B., et al. (2012). (1)H NMR-based metabolomic analysis for identifying serum biomarkers to evaluate methotrexate treatment in patients with early rheumatoid arthritis. Experimental and Therapeutic Medicine, 4(1), 165–171. doi:10.3892/etm.2012.567.
Xiao, C. N., Hao, F. H., Qin, X. R., Wang, Y. L., & Tang, H. R. (2009). An optimized buffer system for NMR-based urinary metabonomics with effective pH control, chemical shift consistency and dilution minimization. The Analyst, 134(5), 916–925. doi:10.1039/b818802e.
Yoo, H. S., & Park, T. G. (2001). Biodegradable polymeric micelles composed of doxorubicin conjugated PLGA-PEG block copolymer. Journal of Controlled Release, 70(1–2), 63–70. doi:10.1016/S0168-3659(00)00340-0.
Zachara, N. E., & Hart, G. W. (2004). O-GlcNAc a sensor of cellular state: The role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochimica Et Biophysica Acta-General Subjects, 1673(1–2), 13–28. doi:10.1016/j.bbagen.2004.03.016.
Zachara, N. E., & Hart, G. W. (2006). Cell signaling, the essential role of O-GlcNAc! Biochimica Et Biophysica Acta-Molecular and Cell Biology of Lipids, 1761(5–6), 599–617. doi:10.1016/j.bbalip.2006.04.007.
Zeidan, Q., & Hart, G. W. (2010). The intersections between O-GlcNAcylation and phosphorylation: Implications for multiple signaling pathways. Journal of Cell Science, 123(1), 13–22. doi:10.1242/jcs.053678.
Zhang, L., Wang, L., Hu, Y., Liu, Z., Tian, Y., Wu, X., et al. (2013). Selective metabolic effects of gold nanorods on normal and cancer cells and their application in anticancer drug screening. Biomaterials, 34(29), 7117–7126. doi:10.1016/j.biomaterials.2013.05.043.
Zhao, Y., Wu, J. F., Li, J. V., Zhou, N. Y., Tang, H. R., & Wang, Y. L. (2013). Gut microbiota composition modifies fecal metabolic profiles in mice. Journal of Proteome Research, 12(6), 2987–2999. doi:10.1021/pr400263n.
Acknowledgements
This work was supported by the grants from the National Natural Science Foundation of China (21675169, 21175149, 91439102 and 21375144) and National Distinguished Young Scientists program (31325010), and the Ministry of Science and Technology 973 (2012CB934004).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hu, Y., Zhang, L., Wang, H. et al. Biological effects of amphiphilic copolymer nanoparticle-encapsulated multi-target chemotherapeutic drugs on MCF-7 human breast cancer cells. Metabolomics 13, 49 (2017). https://doi.org/10.1007/s11306-017-1187-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-017-1187-x