Abstract
Objectives
Meta-analysis was used to determine the association between rs3751143 polymorphism of P2RX7 gene and the risk of chronic lymphocytic leukemia (CLL).
Methods
Search for published articles about the association between the rs3751143 and CLL in PubMed, MEDINE, Web of Science, and Embase databases, with a calculated odds ratio of (OR) and 95% confidence interval (95%CI).
Results
A total of 1184 cases and 1725 controls in 8 studies were pooled together for evaluation of the overall association between rs3751143 and risk of CLL. Allele model (A vs C, p = 0.16, OR = 0.85, 95%CI = 0.71–1.17), homozygous model (AA vs CC, p = 0.07; OR = 0.78, 95%CI = 0.84–1.08), and heterozygous model (AC vs CC, p = 0.76; OR = 0.85; 95%CI = 0.68–0.79) did not show decreased risk of developing CLL. Similarly, dominant model (AA + AC vs. CC: p = 0.58; OR = 1.10, 95%CI = 0.69–1.75), and recessive model (AA vs AC + CC, p = 0.21, OR = 1.18, 95%CI = 0.70–1.99) failed to show decreased risk of developing CLL. However, in familial, heterozygous model (AC vs. CC: p = 0.0006, OR = 0.64, 95%CI = 0.67–1.50) and recessive model (AA vs. AC + CC: p = 0.0017; OR = 1.02, 95%CI = 0.73–2.35) indicated the association between the inheritance of rs3751143 and the risk of developing CLL. In the overall survival prognosis, no significant association between rs3751143 and CLL was detected with relatively high heterogeneity.
Conclusions
Our pooled data indicates that there is a correlation between the inheritance of rs3751143 and the risk of CLL in familial.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chronic lymphocytic leukemia (CLL) is a malignant tumor occurring in the blood system, which is difficult to treat clinically. The diagnosed patients are mainly treated by chemotherapy but also by radiation therapy, immunotherapy (such as interferon, anti-CD20 monoclonal antibodies, and anti-CD52 antibodies) and hematopoietic stem cell transplantation, but it is easy to relapse [1, 2]. Therefore, it is necessary to explore the relevant molecular factors in the pathogenesis of CLL, and better targeted and individualized treatment. P2RX7 is dependent on ATP ion channel receptor, and ATP is a natural activator of P2RX7. When the body is subjected to noxious stimulation, high concentration of extracellular ATP can activate P2RX7, opens the ion channels on the cell membrane (mainly calcium ion influx), changes the permeability of the cell membrane, and affects the molecular metabolism in the cell [3]. P2RX7 is widely expressed in blood-derived cells (such as lymphocytes, macrophages, bone marrow-derived cells). Activation of P2RX7 can activate different intracellular signaling pathways (such as ERK and NF-kB), induce the release of inflammatory factors (such as TNF-a and IL-1β), and play an important regulatory role in cell proliferation, apoptosis and death [4,5,6]. P2RX7 activation participates in the pathogenesis of inflammatory and immune diseases by activating macrophages and lymphocytes. In lymphocytic leukemia, P2RX7 plays an important regulatory role in hematological tumors [7]. Overexpression of P2RX7 promotes the proliferation of lymphocytes and increases the levels of leukemia stem cell and accelerates the progression of leukemia [8]. Studies have shown that P2RX7 is highly expressed in patients with CLL, which can promote the progression of leukemia via the P2RX7/NLRP3 axis [9]. These data indicate that over-expression of P2RX7 promotes the progression of lymphocytic leukemia. With the study of the function of P2RX7, it is determined that the polymorphism of P2RX7 gene is closely related to the development of most diseases [10, 11]. It has been found that the allele frequency of rs3751143 may be related to age [12]. Gu et al. found that glu-496-ala polymorphism leaded to the loss of P2RX7 function and suggested that the function of P2RX7 required glutamate at position 496 [13]. Studies have shown that the A > C substitution in exon 13 at position 1513 can eliminate P2RX7-mediated apoptosis and death, and promote cell growth and proliferation [14].
However, the role of rs3751143 in the risk of developing CLL has not been clearly confirmed, and the results obtained in different studies are different. Related studies have shown that there are a certain correlation between rs3751143 and the risk of developing CLL [15], while other studies have shown no correlation between rs3751143 and CLL [16]. In addition, the results of previous meta analyses showed that P2RX7 gene rs3751143 polymorphism had no significant correlation with the risk of CLL [16, 17]. Therefore, it is necessary to conduct a complete meta-analysis to evaluate the association between the rs3751143 and the risk of developing CLL. This meta analysis differs from the previous meta-analyses in that we searched all case-control studies on between rs3751143 and CLL and evaluated the association between rs3751143 in different models and overall survival prognosis of patients with CLL. Moreover, we further analyzed the association between rs3751143 and familial/sporadic CLL through subgroups.
Methods
Publication search
A computer was used to retrieve all documents conforming to standards from PubMed, MEDLINE, Web-Science and Embase databases. The period for document retrieval is limited to January 1, 2018. Keyword searches include P2X7, P2RX7, P2X7 receptor, 1513A/C polymorphism, genotype, rs3751143 AND chronic lymphocytic leukemia, lymphocytic leukemia, and leukemia. Relevant documents were obtained (including original papers, reviews, and conference articles) from these databases, and then further screened the articles for meta-analysis by reading the title and abstract, and only include published papers.
Inclusion and exclusion criteria
All case-control studies on P2RX7-rs3751143 polymorphism and CLL were included in this meta-analysis, and the inclusion criteria were as follows: (1). case–control studies on human P2RX7-rs3751143 polymorphism and CLL, (2) containing available genotype data in cases and controls for estimating an OR and 95%CI, and (3) the genotype distribution of the control population conforms to Hardy–Weinberg equilibrium (HWE). The exclusion criteria were (1) reviews, letters, editorial articles, and case reports and (2) study on CLL and other genotypes of P2RX7. This study was performed in accordance with the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) guidelines for meta-analysis of randomized clinical trials (http://prisma-statement.org/statement.htm).
Data extraction and quality evaluation
Two researchers examined and evaluated the selected literature respectively, read the full text of each study that met the criteria after screening, and extracted the data from these articles, including author information, follow-up time, family characteristics, evaluation methods, number of cases and controls, results, and possible bias. If the outcome was assessed multiple times during a study, the last measurements were included. If data were not presented in the article, the authors were contacted, and in cases of no response, two reminders were sent with 1-week intervals.
Statistical analysis
Chi-square test was used to check whether the genotypes in the control were consistent with Hardy–Weinberg equilibrium. (HWE), and STATA10.0 software and Revman were used for meta analysis of all data. The relationship between P2RX7-rs3751143 polymorphism and CLL was obtained by calculating OR and 95%CI. X2 test was used to evaluate heterogeneity between studies; when the test level is set to P < 0.05, there is heterogeneity between studies; when P > 0.05, there is no heterogeneity between studies. At the same time, I2 was used to quantitatively analyze the heterogeneity between studies, and the significance level was set to 50%. That is, I2 > 50% indicates significant heterogeneity; less than 50% means there is no significant heterogeneity. No heterogeneity, fixed effect model was used for data analysis, otherwise used random effect model for data analysis. An estimate of potential publication bias was performed using the funnel plot.
Results
Study characteristics
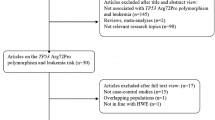
According to the keyword entries, through a complete database search, a total of 32 papers were retrieved, repeating 18, 6 conferences, and reviews. After reading the title and abstract, a total of 8 articles met the meta analysis inclusion criteria (Fig. 1). These eight papers are all about case–control studies between P2RX7-rs3751143 polymorphism and CLL, from different countries, including 1184 patients with CLL and 1725 healthy control with rs3751143 polymorphism [15, 16, 18,19,20,21,22,23]. Three studies have investigated the association between P2RX7-rs3751143 polymorphism and the risk of familial or sporadic CLL [15, 16, 18]. Four studies investigated the correlation between P2RX7-rs3751143 polymorphism and the overall survival prognosis of patients with CLL [19,20,21,22]. For the case group, the frequency of rs3751143 polymorphism among AA-homozygous individuals was 63.85%. However, 27.02% of AC-heterozygous individuals and 0.29% of CC-homozygous individuals displayed the rs3751143 polymorphism. In control groups, the frequencies of rs3751143 polymorphism among AA-homozygous individuals, AC heterozygous individuals, and CC-homozygous individuals were 69.33%, 26.70%, and 0.28%, respectively. The A allelic frequencies in the case and control groups were 80.27% and 79.53%, respectively (Tables 1 and 2).
Meta-analysis results
A total of 1184 cases and 1725 controls in 8 studies were pooled together for evaluation of the overall association between P2RX7-rs3751143 polymorphism and risk of CLL. The pooled OR indicated no significant association between the P2RX7-rs3751143 polymorphism and the risk of CLL. Allele model (A vs C, p = 0.16; OR = 0.85; 95%CI = 0.71–1.17), homozygous model (AA vs CC, p = 0.07; OR = 0.78, 95%CI = 0.84–1.08), and heterozygous model (AC vs CC, p = 0.76; OR = 0.85; 95%CI = 0.68–0.79) did not show decreased risk of developing CLL. Similarly, dominant model (AA + AC vs. CC: p = 0.58; OR = 1.10, 95%CI = 0.69–1.75) and recessive model ((AA vs AC + CC, p = 0.21, OR = 1.18; 95%CI = 0.70–1.99) failed to show decreased risk of developing CLL. Large heterogeneity (I2 = 85%, P = 0.002) was detected among 8 studies. In order to eliminate heterogeneity, we further conducted subgroup analysis according to familial and overall prognostic survival rate. A total of 3 studies involved familial and 4 involved survival prognosis analysis. In familial, heterozygous model (AC vs. CC: p = 0.0006, OR = 0.64, 95%CI = 0.67–1.50) and recessive model (AA vs. AC + CC: p = 0.0017; OR = 1.02, 95%CI = 0.73–2.35) indicated the association between the inheritance of P2RX7-rs3751143 polymorphism and the risk of developing CLL. Heterogeneity disappeared in subgroup of familial subjects, which revealed that most of the studies could not be grouped based on familial. In the overall survival prognosis, no significant association between P2RX7-rs3751143 polymorphism and CLL were detected with relatively high heterogeneity (Table 3).
Publication bias
Begg–Mazumdar test and Egger test were performed to assess the publication bias in the literature. All of the studies investigating the 1513 A allele vs the C allele yielded a Begg’s test score of p = 0.242 and an Egger’s test score of p = 0.233. These results do not indicate a potential for publication bias.
Discussion
The understanding of the correlation between the risk of CLL and genetic variation has attracted researchers to explore the correlation between various genetic polymorphisms and CLL. This has lead to the gradual mining of a large number of candidate genes to find out the possible relationship between them and the risk of CLL. Different studies have evaluated the association between P2RX7-rs3751143 gene polymorphism and the risk of CLL. However, studies related to the P2RX7-rs3751143 polymorphism and CLL have yielded conflicting results. Some studies have reported that P2RX7-rs3751143 polymorphism may increase the risk of CLL [15, 22, 24]. However, some studies have failed to show the intrinsic association between P2RX7-rs3751143 gene polymorphism and the risk of CLL [16, 19, 20]. Therefore, it is necessary to conduct a meta-analysis to more accurately assess the relationship between the two. In this meta analysis, we collected all the data on the relationship between rs3751143 polymorphism and CLL and comprehensively evaluated and analyzed these data to explore whether there is an association between P2RX7-rs3751143 polymorphism and the risk of CLL. Our results indicate that there is no significant association between the P2RX7-rs3751143 polymorphism and the risk of CLL. Similarly, no association between P2RX7-rs3751143 polymorphism and survival prognosis of patients with CLL was observed. However, through subgroup analysis, we found that the inheritance of P2RX7-rs3751143 polymorphism had a certain association in the family CLL. The inheritance of P2RX7-rs3751143 polymorphism can increase the risk of CLL in familial. Through this meta analysis, our comprehensive data show that there is a certain correlation between the inheritance of P2RX7-rs3751143 polymorphism and the risk of CLL in familial.
Heterogeneity is a potential problem that may affect the interpretation of the results. We selected 8 studies that meet the HWE standard to ensure the reliability of association analysis. Among them, the allele distribution in the P2RX7 gene polymorphism in the two studies [15, 21] is different from other studies. The possible reason is that the 1513C allele may be in linkage disequilibrium with a nearby susceptibility gene. Although there are differences in study design, sample size, sample selection, and population, there are no statistically significant heterogeneity among the eight studies included in the meta-analysis. This indicates that the overall estimation of the relationship between the P2RX7-rs3751143 polymorphism and the risk of CLL is appropriate. Compared with previous meta-analyses, our research has been significantly improved. We not only evaluated different genetic models and overall survival prognosis of CLL but also comprehensively evaluated familial and sporadic characteristics, confirming the results of the previous studies [15].
CLL is known to be caused by polygenic mutations [25]. Therefore, mutations in a single gene cannot be considered sufficient to bear the risk of this terrible disease [26]. It has been found that rs3751143 mutations exist in 3 of 17 potential haplotypes [27]. A recent study revealed that rs3751143 was associated with a novel regulatory polymorphism (rs11615992). The area around rs11615992 can interact with the P2RX7 promoter and act as an enhancer, which indicates that the single-nucleotide polymorphism in linkage disequilibrium can regulate P2RX7 expression [28]. Studies have shown that P2RX7-rs3751143 polymorphism plays a role in familial CLL susceptibility, which may be related to linkage disequilibrium of nearby susceptibility genes [15]. The genomic detection of 18 families with CLL by Goldin et al. also supports this possibility [24]. Our results indicate that P2RX7-rs3751143 polymorphism has a certain correlation with the susceptibility of familial CLL, which is consistent with previous studies. Together raising the possibility that a certain P2RX7 haplotype rather than rs3751143 per se is responsible for the possible association with familial CLL. Based on this information, this may provide some biological explanation for the difference of P2RX7 between familial and sporadic CLL.
Considering the limitations of this meta analysis, our results should be interpreted carefully. First of all, our results are based on unadjusted estimates, personal data should be used for more accurate analysis, and researchers need to adjust for covariates, including age, gender, immune status, race, and environment. Second, we only use published data for analysis, although we did not find potential publication bias. However, some unimportant or negative research results may not have been published and are not included in this meta-analysis. Third, only three familial related studies may not be sufficient to produce decisive results, nor to assess heterogeneity or publication bias. Therefore, the results we obtained are for reference only, and more relevant studies are needed to determine and judge them.
Data availability
All data generated or analyzed during this study are included in this article. And we have not used other data that has already been published. All the data presented in this article are original results derived from this study.
References
Takizawa J (2017) Chronic lymphocytic leukemia: pathophysiology and current therapy. Rinsho Ketsueki 58:471–479
Francesc B, Dalla-Favera R (2019) Chronic lymphocytic leukaemia: from genetics to treatment. Nat Rev Clin Oncol 16:684–701
Zhang WJ, Zhu ZM, Liu ZX (2020) The role and pharmacological properties of the P2X7 receptor in neuropathic pain. Brain Res Bull 155:19–28
Alves LA, Bezerra RJ, Faria RX, Ferreira L, da Silva Frutuoso V (2013) Physiological roles and potential therapeutic applications of the P2X7 receptor in inflammation and pain. Molecules. 18:10953–10972
Arandjelovic S, McKenney KR, Leming SS et al (2012) ATP induces protein arginine deiminase 2-dependent citrullination in mast cells through the P2X7 purinergic receptor. J Immunol 189:4112–4122
Collo G, Neidhart S, Kawashima E, Kosco-Vilbois M, North RA, Buell G (1997) Tissue distribution of the P2X7 receptor. Neuropharmacology. 36:1277–1283
Zhang XJ, Zheng GG, Ma XT, Yang YH, Li G, Rao Q, Nie K, Wu KF (2004) Expression of P2X7 in human hematopoietic cell lines and leukemia patients. Leuk Res 28:1313–1322
Feng W, Yang X, Wang L et al (2019) P2X7 promotes the progression of MLL-AF9 induced acute myeloid leukemia by upregulation of Pbx3. Haematologica. 2020:243360
Salaro E, Rambaldi A, Falzoni S, Amoroso FS, Franceschini A, Sarti AC, Bonora M, Cavazzini F, Rigolin GM, Ciccone M, Audrito V, Deaglio S, Pelegrin P, Pinton P, Cuneo A, di Virgilio F (2016) Involvement of the P2X7-NLRP3 axis in leukemic cell proliferation and death. Sci Rep 6:26280
Ozdemir FA, Deniz Erol VK, Hüseyin Y et al (2014) Lack of association of 1513 a/C polymorphism in P2X7 gene with susceptibility to pulmonary and extrapulmonary tuberculosis. Tuberk Toraks 62:7–11
Dardano A, Falzoni S, Caraccio N, Polini A, Tognini S, Solini A, Berti P, Virgilio FD, Monzani F (2009) 1513A>C polymorphism in the P2X7 receptor gene in patients with papillary thyroid cancer: correlation with histological variants and clinical parameters. J Clin Endocrinol Metab 94:695–698
Sanz JM, Falzoni S, Morieri ML, Passaro A, Zuliani G, Di Virgilio F (2020) Association of hypomorphic P2X7 receptor genotype with age. Front Mol Neurosci 13:8
Gu BJ, Zhang W, Worthington RA, Sluyter R, Dao-Ung P, Petrou S, Barden JA, Wiley JS (2001) A Glu-496 to Ala polymorphism leads to loss of function of the human P2X7 receptor. J Biol Chem 276:11135–11142
Gong M, Dong W, Shi Z, Qiu S, Yuan R (2017) Vascular endothelial growth factor gene polymorphisms and the risk of renal cell carcinoma: evidence from eight case-control studies. Oncotarget. 8:8447–8458
Dao-Ung LP, Fuller SJ, Sluyter R, SkarRatt KK, Thunberg U, Tobin G, Byth K, Ban M, Rosenquist R, Stewart GJ, Wiley JS (2004) Association of the 1513C polymorphism in the P2X7 gene with familial forms of chronic lymphocytic leukaemia. Br J Haematol 125:815–817
Sellick GS, Rudd M, Eve P et al (2004) The P2X7 receptor gene A1513C polymorphism does not contribute to risk of familial or sporadic chronic lymphocytic leukemia. Cancer Epidemiol Biomark Prev 13:1065–1067
Zintzaras E, Kitsios GD (2009) Synopsis and synthesis of candidate-gene association studies in chronic lymphocytic leukemia: the CUMAGAS-CLL information system. Am J Epidemiol 170:671–678
Wiley JS, Dao-Ung LP, Gu BJ, Sluyter R, Shemon AN, Li C, Taper J, Gallo J, Manoharan A (2002) A loss-of-function polymorphic mutation in the cytolytic P2X7 receptor gene and chronic lymphocytic leukaemia: a molecular study. Lancet. 359:1114–1119
Nückel H, Frey UH, Dürig J, Dührsen U, Siffert W (2004) 1513A/C polymorphism in the P2X7 receptor gene in chronic lymphocytic leukemia: absence of correlation with clinical outcome. Eur J Haematol 72:259–263
Starczynski J, Pepper C, Pratt G, Hooper L, Thomas A, Hoy T, Milligan D, Bentley P, Fegan C (2003) The P2X7 receptor gene polymorphism 1513 A-->C has no effect on clinical prognostic markers, in vitro sensitivity to fludarabine, Bcl-2 family protein expression or survival in B-cell chronic lymphocytic leukaemia. Br J Haematol 123:66–71
Zhang LY, Ibbotson RE, Orchard JA, Gardiner AC, Seear RV, Chase AJ, Oscier DG, Cross NCP (2003) P2X7 polymorphism and chronic lymphocytic leukaemia: lack of correlation with incidence, survival and abnormalities of chromosome 12. Leukemia. 17:2097–2100
Thunberg U, Tobin G, Johnson A, Söderberg O, Padyukov L, Hultdin M, Klareskog L, Enblad G, Sundström C, Roos G, Rosenquist R (2002) Polymorphism in the P2X7 receptor gene and survival in chronic lymphocytic leukaemia. Lancet. 360:1935–1939
Cabrini G, Falzoni S, Forchap SL et al (2005) A His-155 to Tyr polymorphism confers gain-of-function to the human P2X7 receptor of human leukemic lymphocytes. J Immunol 17:82–89
Goldin LR, Ishibe N, Sgambati M, Marti GE, Fontaine L, Lee MP, Kelley JM, Scherpbier T, Buetow KH, Caporaso NE (2003) A genome scan of 18 families with chronic lymphocytic leukaemia. Br J Haematol 121:866–873
Crowther-Swanepoel D, Mansouri M, Enjuanes A, Vega A, Smedby KE, Ruiz-Ponte C, Jurlander J, Juliusson G, Montserrat E, Catovsky D, Campo E, Carracedo A, Rosenquist R, Houlston RS (2010) Verification that common variation at 2q37.1, 6p25.3, 11q24.1, 15q23, and 19q13.32 influences chronic lymphocytic leukaemia risk. Br J Haematol 150:473–479
Xiao W, Gong C, Liu X, Liu Y, Peng S, Luo D, Wang R, Li T, Zhao J, Xiong C, Liang S, Xu H (2019) Association of P2X7R gene with serum lipid profiles in Chinese postmenopausal women with osteoporosis. Climacteric. 22:498–506
Jørgensen NR, Husted LB, Skarratt KK, Stokes L, Tofteng CL, Kvist T, Jensen JEB, Eiken P, Brixen K, Fuller S, Clifton-Bligh R, Gartland A, Schwarz P, Langdahl BL, Wiley JS (2012) Single-nucleotide polymorphisms in the P2X7 receptor gene are associated with post-menopausal bone loss and vertebral fractures. Eur J Hum Genet 20:675–681
Peng T, Zhong L, Gao J, Wan Z, Fu WP, Sun C (2020) Identification of rs11615992 as a novel regulatory SNP for human P2RX7 by allele-specific expression. Mol Gen Genomics 295:23–30
Acknowledgments
We thank Wen-jun Zhang for writing this article and Zheng-ming Zhu for his guidance and support.
Author information
Authors and Affiliations
Contributions
Wen-jun Zhang carried out the whole study and drafted the manuscript. Zheng-ming Zhu carried out the design of the study and helped revise this paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing of interest
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, WJ., Zhu, ZM. Association between the rs3751143 polymorphism of P2RX7 gene and chronic lymphocytic leukemia: A meta-analysis. Purinergic Signalling 16, 479–484 (2020). https://doi.org/10.1007/s11302-020-09737-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11302-020-09737-8