Abstract
The use of single nucleotide polymorphism (SNP) molecular markers has provided advances in selection methodologies used in breeding programs of different crops, reducing cost and time of cultivar release. Despite the great economic and social importance of Coffea arabica, studies with SNP markers are scarce and a small number of SNP are available for this species, when compared with other crops of agronomic importance. Thus, the objective of this study was to identify and validate SNP molecular markers for the species Coffea arabica and to introduce these markers to genetic breeding by means of an accurate analysis of the diversity and genetic structure of breeding populations of this species. After quality filtering, 11,187 SNP markers were selected from the coffee population obtained from crosses between the genotypes Catuaí and Híbrido de Timor. A great number of markers were distributed in the 11 chromosomes, within transcribed regions, and were used to estimate the genetic dissimilarity among the individuals of the breeding population. Dendrogram analysis and a Bayesian approach demonstrated the formation of two groups and the discrimination of all genotypes evaluated. The expressive number of SNP molecular markers distributed throughout C. arabica genome was efficient to discriminate all the accessions evaluated in the experiment, clustering them according to their genealogies. This work identified mixtures within the progenies. The genotyping data also provided detailed information about the parental genotypes and led to the identification of new candidate parents to be introduced to the breeding program. The study discussed population structure and its consequence in obtaining improved varieties of C. arabica.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rapid population growth associated with several climate changes and with restricted expansion of agricultural frontiers are challenges for modern agriculture in the continuous food supply. To overcome these limitations, genetic breeding programs must have precise and accurate selective methods to maximize selection gains and maintain the genetic variability of the population (Hallauer 2011).
However, the progress of breeding programs has culminated in the reduction of genetic variability of improved populations (Rodgers et al. 1983; Ortiz et al. 2003; Smith et al. 2004). This problem may be worse in species with narrow genetic base, such as Coffea arabica. The Coffea genus comprises 104 species (Davis et al. 2006; Davis 2010; Davis 2011), and C. arabica and C. canephora are the most important agronomic species, corresponding to 63 and 37% of the world’s coffee production in 2016, respectively (ICO 2017).
C. arabica is allotetraploid, originated from the natural cross between the species C. eugenioides and C. canephora (Lashermes et al. 1999), and is autogamous with about 10% cross fertilization (Carvalho and Krug 1949). C. arabica is also known to have a narrow genetic basis, which is explained by the autogamous reproduction system and the low number of plants initially distributed throughout the world (Setotaw et al. 2013). This low genetic variability hinders the identification and selection of superior genotypes using traditional breeding methods. To overcome this problem, molecular markers have been used as an auxiliary tool to accurately discriminate genotypes (Ferrão et al. 2015; Sousa et al. 2017) in order to accelerate and enable breeding programs (Ceccarelli 2015).
The use of markers is particularly desirable for coffee, a perennial species with a narrow genetic base, due to the time and resources required to release a new cultivar (Ferrão et al. 2015). The molecular information allows the knowledge of the genetic structure and similarity between the individuals, which when combined with phenotypic measures allows the selection of superior genotypes, maximizing selection gains (Sousa et al. 2017). Several studies using molecular markers have demonstrated the lower genetic diversity of C. arabica in relation to C. canephora (Lashermes et al. 1999; Cubry et al. 2008; Lashermes et al. 2011; Ferrão et al. 2015). In Brazil, three research institutions (EPAMIG, Universidade Federal de Viçosa—UFV, and EMBRAPA-Café) have partnered to breed arabica coffee. In order to deal with the low genetic diversity, the program has aimed at the introgression of genes of interest, present in C. canephora, into C. arabica. This introgression is facilitated by the use of Hibrido de Timor (HdT), a natural hybrid between the species C. arabica and C. canephora (Lashermes et al. 1999). HdT’s germplasm carries genes that confer durable resistance to Hemileia vastatrix (Diola et al. 2011; Romero et al. 2014; Alkimim et al. 2017), Colletotrichum kahawae (Gichimu et al. 2013; Gichimu et al. 2014; Alkimim et al. 2017), and Meloidogyne exigua (Bertrand et al. 2001; Pereira et al. 2016). All cultivars released by this breeding program are derived from HdT and have demonstrated superior cup quality when compared with traditional arabica coffee cultivars (Pereira et al. 2010b; Sobreira et al. 2015).
Different molecular markers have been identified and made available for coffee (Rovelli et al. 2000; Combes et al. 2000; Moncada and McCouch 2004; Cubry et al. 2008; Missio et al. 2009a; Vieira et al. 2010; Ferrão et al. 2015). These markers have been used for different genetic studies in C. arabica, such as analyses of genetic diversity and molecular characterization of germplasm (Maluf et al. 2005; Oliveira et al. 2007; Missio et al. 2009b; Missio et al. 2011; Aerts et al. 2013), linkage maps (Pearl et al. 2004; Diola et al. 2011; Pestana et al. 2015; Moncada et al. 2016), marker-assisted selection (Alkimim et al. 2017), and cultivar fingerprinting (Sousa et al. 2017). However, one limitation of the marker platforms currently available is the difficulty in identifying polymorphic markers well-distributed throughout the genome of the species. Previous estimates identified that only 10% of the markers screened were polymorphic in C. arabica, which increases the genotyping cost and hinders molecular breeding applications (Sousa et al. 2017).
Single nucleotide polymorphism (SNP) markers are the most common marker type used in breeding programs. The platforms, initially designed to characterize the SNPs on a chip, have been developed for several plant and animal species. These markers can be associated with the genes that control the main traits of agronomic interest (Heffner et al. 2009), and have low cost per data point (Elsik et al. 2009; Matukumalli et al. 2009). These facts, combined with the development of genomic selection algorithms, have increased the accuracy of the selection methods used in breeding programs for several plant species (Crossa et al. 2010). However, in order for a chip to be developed for a novel species, it is necessary that the SNPs be known a priori. Sequencing-based technologies, on the other hand, allow the genotyping process to be carried in parallel to the discovery, enabling the application of SNPs for species with little genomic resources.
To introduce new molecular markers and increase the efficiency of their use in coffee genetic breeding, the present work aimed at developing sequencing-based genotyping in arabica coffee, based on targeted capture coupled with next-generation sequencing. The SNPs identified in this work were subsequently used to genotype a breeding population aiming at the study of genetic diversity and genetic structure.
Material and methods
Genetic material
A population of C. arabica from EPAMIG/UFV/EMBRAPA’s breeding program was selected for the identification of the SNP markers. The genotypes have been maintained in the experimental area of the Department of Plant Pathology of the UFV.
The population consisted of 72 coffee genotypes generated from parents with contrasting rust resistance phenotypes (Fig. 1). The colors dark green, dark blue, dark yellow, light green, light blue, and light yellow correspond, respectively, to rust-susceptible parents, rust-resistant parents, F1 generation, genotypes obtained from the backcross between the F1 hybrid with the recurrent susceptible parent (BCs), genotypes obtained from the backcross between the F1 hybrid with recurrent rust-resistant parent (BCr), and the F2 generation.
Pedigree of the parents, hybrids and of the 13 progenies that compose the Coffea arabica breeding program analyzed in this study; Tables 1, 2, and 3 present the relation of the respective genotypes corresponding to each letter and number; ⊗ = selfing generation; The colors dark green, dark blue, dark yellow, light green, light blue, and light yellow correspond, respectively, to rust-susceptible parents, rust-resistant parents, F1 generation, genotypes obtained from the backcross between the F1 hybrid with the recurrent susceptible parent (BCs), genotypes obtained from the backcross between the F1 hybrid with recurrent rust-resistant parent (BCr), and the F2 generation
Crosses were carried out between three rust-resistant HdT accessions with three rust-susceptible Catuaí varieties (Table 1). Five coffee rust-resistant F1 hybrids (Table 2) were selected, which were selfed and backcrossed, resulting in 13 arabica coffee progenies (Table 3). In each progeny, four to five individuals were selected. In addition to these genotypes, four accessions of arabica coffee belonging to the germplasm bank of these institutions were selected to compose the study population, in order to increase the genetic variability of the population under analysis (Table 4). Thus, the population was composed of 72 genotypes of coffee plants of different generations.
Genomic DNA extraction
Young and fully expanded leaves from 72 genotypes were collected, and the genomic DNA was extracted using the methodology described by Diniz et al. (2005). DNA concentration was measured using the NanoDrop 2000, and DNA quality was evaluated in a 1% agarose gel. DNA concentration of the samples was standardized and sent to Rapid Genomics, Florida, USA, for the construction of probes, sequencing, and identification of SNP molecular markers.
Probes design and SNP identification
The sequencing-based genotyping was performed using targeted enrichment followed by next-generation sequencing, as previously described (Gnirke et al. 2009). We designed a total of 40,000 120-bp probes in order to reduce the complexity of the genome and enrich the libraries for the targeted regions. These probes were designed using a combination of genomic resources available for C. arabica and C. canephora. The genomic resources utilized were C. arabica specific expressed sequence tags (ESTs) and C. canephora specific ESTs obtained from a database generated by the Brazilian Coffee Genome Project, which contains more than 200,000 ESTs, and corresponds to about 33,000 transcripts (Vieira et al. 2006). In addition, probes were also developed from the reference genome of C. canephora (Denoeud et al. 2014), which has an estimated size of 710 Mb. The EST database was also mapped against the reference genome to identify candidate SNPs that represented good candidate regions to become a polymorphic probe. Furthermore, all the probes were designed in regions that were not repetitive in the C. canephora genome and not present in the plastid genome.
We started the process targeting ESTs that were unique to C. arabica and did not match the canephora reference genome. Based on these unigenes, we designed 1879 probes derived exclusively from C. arabica contigs. Secondly, we identified 8236 probes mapping to 7347 annotated genes that contained at least one SNP between the EST database and the reference genome. Next, we used the reference genome to design an additional set of 11,879 probes that mapped uniquely in the genome and represented 11,879 genes in addition to the 7347 previously described. Finally, to add to 40,000 probes, a total of 18,006 probes were selected in non-genic regions of the reference genome by breaking the genome in 10 kb intervals and selecting proves that are well-distributed and covered the entire genome.
The 72 coffee samples were prepared for next-generation sequencing and hybridized against the probes synthesized in-solution. Protocols were based on Neves et al. (2013). After capturing 40,000 target regions in the coffee genome, C. canephora and C. arabica genotypes were sequenced using the Illumina Hi-Seq sequencing platform. The sequencing product was separated in each individual barcodes representing each genotyped sample. Low quality bases with less than 20 quality score in the 3′ end were trimmed out followed by a low quality filter that removed reads with more than 10% of the read with less than 20 quality score. Filtered reads were aligned against the C. canephora reference genome using Mosaik (Lee et al. 2014). The identification of SNPs was performed using diploid settings on Freebayes (Garrison and Gabor 2012), and resulted in the identification of 162,026 SNPs (SNPs specific to C. arabica, specific to C. canephora, and common to both species) in 27,651 polymorphic probes, with a mean of five SNPs per probe.
Quality analysis of SNP molecular markers
In order to evaluate the effect of SNP quality on the subsequent analysis, we created three datasets with different filtering criteria. Quality analyses were performed in the VCFtools software (Danecek et al. 2011) and Rbio (Bhering 2017). The quality parameters used in the first filter were MinDP3, DPrange15-750, Miss0.4, MAF0.01, and MinQ10. For the second filter, the quality parameters were MinDP5, DPrange20-350, Miss0,2, MAF0,03 and MinQ20. And finally, the quality parameters used in the third filter were MAF0,1 and CR0,95. In addition, SNP sets that presented identical genotyping results in all individuals evaluated were identified in filter 3 by analysis of variance, and removed from the data set to avoid redundancy and multicollinearity problems.
MinDP is defined as the minimum sequencing depth per SNP and per individual. Values below the critical level determined by this parameter were defined as missing data; DPrange is the mean sequencing depth range of the population for which an SNP is maintained. SNP markers with mean coverage in the population outside the range determined in each filter were removed. Miss refers to the percentage of missing data accepted in the population after filtering by the MinDP parameter. SNP markers with Miss values above that set in the parameter were removed. MAF represents the minor allele frequency that refers to the frequency at which the alternative allele occurs in a given population. MinQ is the minimum quality accepted for SNP, and SNP with quality values lower than those established by this criterion were removed. CR is the minimum proportion of genotypes per marker with non-missing data.
Genetic-statistical analyses
For the genetic-statistical analyses of population data, since the marker was bi-allelic and codominant, the genotypes A1A1, A1A2, or A2A2 were coded as 11, 12, and 22, respectively.
Allele and genotype frequencies of the selected SNP markers were estimated. In addition, polymorphism information content (PIC) (Botstein et al. 1980) of each SNP obtained from the following equation was calculated as follows:
Where p i is the frequency of the ith allele of the studied locus.
Population structure was also studied. This analysis was performed by the Bayesian clustering algorithm of the InStruct software (Gao et al. 2007). The InStruct software makes inference of optimal number of subpopulations underlying a sample via the Deviance Information Criteria. InStruct is an extended Bayesian clustering approach of STRUCTURE (Pritchard et al. 2000) that absorbs inbreeding or selfing rate for population inference. It quantifies the contribution of two forms of non-random mating: inbreeding (mating among relatives) and population substructure (limited dispersal of gametes) when determining the pattern of existing genetic variation (Gao et al. 2007). K values ranged from 1 to 6, in mode 2 for joint inference of population selfing rate and population substructure for five independent chains. Each run was implemented with a period of 50,000 burn-in, followed by 200,000 iteration steps, and a thinning interval of 10 steps, assuming different starting points.
The genetic dissimilarity matrix was obtained by the arithmetic complement of the weighted index implemented in the GENES software (Cruz 2013). Genetic distance was obtained from the equation:
where
Dii′=genetic distance between the pairs of accessions i and i′
\( {p}_j=\frac{a_j}{A} \)weigh associated with locus j, determined by:
ajtotal number of alleles of locus j;
Atotal number of alleles studied;
c j number of common alleles between the pairs of accessions i and i′
The dendrogram was constructed using the UPGMA methodology (Unweighted Pair Group Method using Arithmetic averages) implemented in the MEGA 7.0 software (Kumar et al. 2016).
The principal coordinate analysis (PCoA) was performed by the Genes software (Cruz 2013) using the vegan package implemented in the R software. PCoA is the multivariate statistical analysis where eigenvalues were extracted from the genetic dissimilarity matrix. This analysis is advantageous since it can be applied when the relations between the variables are not linear (Inácio et al. 2002).
Results
Identification and analysis of SNP markers
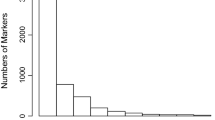
A total of 91,517 SNP markers were identified with the probes distributed throughout the coffee genome. After analyzing the quality of the SNP markers using the first, the second, and the third filter, 34,892, 7058, and 38,380 SNP were eliminated, respectively. Filter 1, filter 2, and filter 3 reduced the original set of SNP markers in 38.13, 45.84, and 87.78%, respectively. Therefore, after quality analyses, 11,187 SNP markers were selected (Fig. 2). We made available a file with 11,187 SPNs used in the genetic analyzes and their respective positions in the genome (Online Resource 1).
SNP molecular markers distributed throughout the UNIGENES from the EST sequences of Coffea arabica and of the 11 Coffea canephora chromosomes and chromosome “0” of Coffea canephora. Chromosome “0” is just a pool of non-ordered sequence scaffolds (Denoeud et al. 2014)
By the analysis of the four datasets (data without a filter, data after filter 1, data after filter 2, and data after filter 3) in the C. canephora reference genome and in the database of ESTs of C. arabica (UNIGENE), markers were confirmed to be distributed throughout the genome. The greatest number of SNP markers was identified on chromosomes 0 and 1 of C. canephora. The “0” chromosome is not a true chromosome; in fact, it is just a set of unsorted sequence scaffolds. The lowest number of SNP markers was observed in the UNIGENE (Fig. 2).
Genetic-statistical analyses
Allelic frequency, genotypic frequency, and PIC
All remaining SNP markers were listed per chromosome and UNIGENE probes, and had the means of allele and genotype frequencies per chromosome estimated (Table 5). The mean allelic frequency was 0.5725 and 0.4275 for alleles A1 and A2, respectively. The mean genotypic frequency was 0.1852, 0.7745, and 0.0402 for genotypes A1A1, A1A2, and A2A2, respectively. The mean PIC per chromosome was also estimated (Table 5). The SNP of chromosomes 1 and 9 had the highest mean PIC values, 0.3526 and 0.3546, respectively. The mean PIC value was 0.3503.
Study of population structure
To analyze the population structure of the genotypes of interest for breeding in studies, 11,187 SNP markers were used. Two groups (K = 2) would be the optimal number (Fig. 3). The Deviance Information Criterion of this model was 534,926.84. The log-likelihood were posterior mean equal − 267,463.421 and posterior variance equal 72,846,714.79. The Gelman-Rubin statistics for the convergence of log-likelihood was 0.85. The significance level for Posterior Credible Interval was 0.90.
Bar graphs of the InStruct software used to determine the population structure of the 72 Coffea arabica genotypes, showing the formation of two groups (k = 2); Tables 1 and 2 list the genotypes corresponding to each letter; SUS = coffee rust-susceptible genotype; RES = coffee rust resistant genotype; BCr = backcross of F1 hybrid with resistant recurrent parent; BCs = backcross of F1 hybrid with recurrent susceptible parent; F2 = generation obtained by the selfing of F1 hybrids
In the first group, individuals were represented by bars, and the blue color was predominant. In the second group, individuals were represented by bars, and the red color was predominant. Parents of the Catuaí group, the progenies obtained by susceptible backcrosses, and most of the plants of the F2 progenies were clustered in the first group. The second group clustered the HdT parents (except the parent UFV 530 [F]), all individuals obtained from resistant backcrosses (except one individual of progeny 3 [3-2]), and plants of progenies 10, 11, and 12 (F2). The genotype UFV 530 (F) was genetically intermediate between the commercial cultivars of the Catuaí group and HdT UFV 445,46 (B) and UFV 440-10 (D), accounting for approximately 54% of the blue color bar. Results of this analysis showed the greater similarity of the genotypes obtained by susceptible and resistant backcrosses with the parents of the Catuaí group and HdT, respectively. In addition, all F1 hybrids (except for H513-5 [K]) had similarity of about 50% with the two groups. Hybrid H513-5 (K) presented intermediate similarity between its parents, UFV 2148-57 (E) and UFV 530 (F). All individuals of progeny 9, obtained from the backcrossing between hybrid H513-5 (K) and UFV 2148-57 (E) also showed similarity between their parents, demonstrating the high discriminatory ability of the SNP markers selected in this study.
Genetic distance between pairs of genotypes
A genetic distance matrix between the pairs of coffee genotypes was constructed using the 11,187 SNP markers. The greatest genetic distance (0.2542) was observed between genotypes CIFC 134/4 and UFV 440-10 (D). The smallest genetic distance (0.0673) was observed between genotypes H419-10 (H) and H514-7 (I) (Table 6).
Genetic distances between the pairs of parents used to obtain the study population were evaluated (Table 7). The greatest genetic distance (0.2336) was observed between the parents UFV 2148-57 (E) and UFV 440-10 (D). The smallest genetic distance (0.0836) was observed between the parents UFV 2143-236 (A) and UFV 2154-344 (C). The mean genetic distances between pairs of parents Catuaí and the Híbrido de Timor were 0.0910 and 0.1442, respectively.
The coffee plants population was divided into four groups, based on their generation: (1) F1 hybrids; (2) F2 generation; (3) BCr; and (4) BCs. The greatest genetic distances were observed between genotypes belonging to the F2 progenies. The smallest genetic distance was observed between the F1 hybrids (Table 8).
Clustering analysis
A dendrogram was constructed using the UPGMA clustering technique from the values of the distance matrix generated by the arithmetic complement of the weighted index (Fig. 4). All genotypes were accurately discriminated. Results of the clustering analysis using the UPGMA methodology were in accordance with those obtained by the Bayesian analysis, forming two large clusters.
Dendrogram obtained by the UPGMA technique based on the dissimilarity matrix of the arithmetic complement of the weighted index from 72 Coffea arabica genotypes; The list of the genotypes corresponding to each letter, from A to K, are presented in Tables 1 and 2; SUS = coffee rust-susceptible genotype; RES = coffee rust resistant genotype; BCr = backcross of F1 hybrid with recurrent resistant parent; BCs = backcross of F1 hybrid with recurrent susceptible parent; F2 = generation obtained by the selfing of F1 hybrids
The first group clustered the rust-susceptible parents and the rust-resistant parent UFV 530 (K), all individuals from the backcross with the recurrent susceptible parent, all hybrid F1, and most individuals of progeny in F2 generation.
Two large subgroups (1.a and 1.b) were formed in this group. Subgroup 1.a clustered all the rust-susceptible parents, the parent HdT UFV 530 (F), the hybrid F1 H513-5 (K), plants of progenies 2, 6, 8, and 9 in BCs, progeny 13 in F2 generation, and three of the four clones of differential coffee plant hosts for Hemileia vastatrix Berk. et Br CIFC 128/2, CIFC 635/3, and CIFC 33/1. Subgroup 1.b clustered four of the five F1 hybrids, individuals of progenies 10, 11, and 12 in F2 generation, individuals of progenies 2, 4, and 8 in BCs generation, and two plants of progeny 3 in BCr generation.
The second group clustered two resistant parents, UFV 445-46 (B) and UFV 440-10 (D), four plants in F2 generation (progenies 10, 11, and 12), and all progenies resistant backcross, except for two plants of progeny 3 (3-2 and 3-3).
One plant of progeny 1 (1-5) and the accessions of the germplasm that correspond to clones of differential coffee plant hosts for Hemileia vastatrix Berk. et Br CIFC 134/4 were not clustered in any of the major groups.
Principal coordinate analysis
Figure 5 shows the graphical dispersion of the scores obtained from the PCoA from the genetic dissimilarity matrix of coffee plants. As in the Bayesian analysis, PCoA showed the genetic distance between the parents of the Catuaí group and the HdT parents. In addition, this analysis demonstrated the genetic dissimilarity between the parent HdT UFV 530 (F) and the other HdT evaluated, and also the genetic dissimilarity between this parent and the F1 hybrids evaluated in this study. Progenies 9 and 13 and hybrid H513-5 (K) were clustered in the graphic region comprised between their parents, UFV 2148-57 (E) and UFV 530 (F). The same occurred with the other F1 hybrids and progenies obtained from the other crosses. As demonstrated in the InStruct software, the graphic dispersion showed the highest similarity of the progenies obtained by susceptible and resistant backcrosses with their respective parents, Catuaí and HdT. Moreover, the greatest graphic dispersion was observed among F2 individuals.
Principal coordinates analysis (PCoA) of the 72 Coffea arabica genotypes; a groups formed according to type of generation; b groups formed according to analysis in InStruct software; c groups formed according to dendrogram analysis; P-SUS = parents of the Catuaí group; P-RES = parents of Híbrido de Timor; F1 = hybrid obtained by the cross between rust-resistant and rust-susceptible genotypes; F2 = generation obtained by the selfing of F1 hybrids; BCr = backcross of F1 hybrid with recurrent resistant parent; BCs = backcross of F1 hybrid with recurrent susceptible parent; DCr = resistant clones of differential coffee plants hosts for Hemileia vastatrix Berk. et Br; DCs = susceptible clones of differential coffee plants hosts for Hemileia vastatrix Berk. et Br; the list of the genotypes corresponding to each letter, from A to K, are presented in Tables 1 and 2
Results of PCoA were in agreement with those of population structure. The region where the four of the five F1 hybrids were clustered divides the population into two groups, and similar result was observed in the other analyses.
Discussion
Identification and quality analysis of SNP markers
The data set composed of the SNP markers was subjected to quality analysis, which eliminated 80,330 SNP. To ensure that only informative SNP markers were applied to C. arabica, the 11,187 SNP markers obtained with more stringent parameters (filter 3) were used in the other analyses. With this strategy, more accurate and safer markers were developed with the potential to generate a lower missing percentage in the populations to be analyzed and greater probability of success in determining the nitrogen bases of the SNP markers (Laurie et al. 2010; Yang et al. 2011).
The SNP markers obtained in this species are distributed on the 11 chromosomes and on chromosome “0,” which is a pool of non-ordered sequence scaffolds of C. canephora. The identification of SNP in these chromosomes is the result of the construction of probes from the reference genome of C. canephora (Denoeud et al. 2014), since the genome of C. arabica is not available. The expressive number and distribution of SNP markers throughout the genome of C. canephora evidenced the efficiency and precision of the technique used in the construction of polymorphic probes (Resende et al. 2016). The greatest number of SNP markers was identified on chromosomes 0 and 1. This fact may be related to their length, since, for this species, a variation of 2.06 to 5.30 μM was observed (Clarindo and Carvalho 2008).
Because C. arabica originates from the cross between C. canephora and C. eugenioides (Lashermes et al. 1999), some arabica chromosome that are exclusively originated from C. eugenioides might not be sampled with the probes constructed from the reference genome of C. canephora. To solve this problem, polymorphic probes were constructed from the ESTs sequences (Expressed Sequence Tag), originated from the Brazilian Coffee Genome Project. This database has more than 200,000 ESTs, of which 130,792 are originated from C. arabica, which identified approximately 33,000 transcripts (UNIGENE) of the Coffee Genome Project (Vieira et al. 2006). The SNP markers identified from the UNIGENE probes are crucial for C. arabica breeding programs for the EST sequences are functional gene sequences, which increases the probability of finding markers associated with genes of interest.
With this large number of informative SNP markers distributed throughout the genome of the species, it is evident the possibility of application of accurate studies on diversity and population structure, as well as selection and broad genomic association in C. arabica. In vegetables, this type of analysis has provided selection gains in polygenic traits, with low heritability, which has stimulated the adoption of such procedures (Crossa et al. 2010; Garcia et al. 2011; Heslot et al. 2012; Resende et al. 2012a; Resende et al. 2012b; Azevedo Peixoto et al. 2017; Ventorim Ferrão et al. 2017).
Genetic-statistical analyses
Allele frequency, genotype frequency, and PIC
PIC, genotype, and allele frequencies for the distinct loci were estimated. The mean PIC value of the 11,187 SNP markers in this population was of 0.3503. The low PIC value evidences the narrow genetic base of C. arabica. Similar results were observed in other studies in which PIC values were obtained from SSR and RFLP molecular markers, being multi-allelic and very informative (Lashermes et al. 1999; Poncet et al. 2006; Hendre et al. 2008; Ferrão et al. 2015). Thus, it can be inferred that despite being bi-allelic, the SNP were very informative.
The mean frequency of heterozygote genotypes (A1A2) was higher than the mean frequencies of homozygote genotypes (A1A1 and A2A2). Despite being autogamous, this result was expected for C. arabica, since most of the individuals that compose the population evaluated in this study were derived from contrasting crosses.
The C. arabica species is allotetraploid, but the genotypes were determined as diploid. However, in the quality analysis, we used variance criterion in order to eliminate the false SNPs (Vidal et al. 2010).
Study of population structure
Similar to the dendrogram analysis, population structure formed two groups. The first group clustered all the accessions of the germplasm bank, which evidenced the greater dissimilarity of these accessions with the HdT parents in relation to the Catuaí parents. All BCs progenies were clustered in the first group. These results can be explained by the fact that BCs progenies present greater genetic similarity with the parents of the Catuaí group. Of the three HdT parents and of the five F1 hybrids evaluated, only the parent UFV 530 (F) and the F1 hybrid H513-5 (K) were clustered in the first group. This information is of great relevance to breeding programs, since the parent HdT UFV 530 carries the SH5, SH6, SH7, SH8, SH9, and SH? genes, which confer durable resistance to rust (Bettencourt and Rodrigues-Júnior 1988), and is genetically closer to parents of the Catuaí group than the other HdT parents. Accordingly, in a study on genetic variability and kinships of Coffea species using molecular markers, high similarity was observed between CIFC 832/1 (derived from the same plant as HdT UFV 530 [F]) and C. arabica, suggesting that, after initial interspecific hybridization, several spontaneous backcrosses occurred with C. arabica to generate Híbrido de Timor (Lashermes et al. 1993). H513-5 (K) was the only F1 hybrid clustered in group I, and this was expected, since this genotype comes from the cross between the parents UFV 2148-57 (E) and UFV 530 (F). All individuals of progeny 9 were also clustered in the first group, and were genetically clustered between the parent UFV 2148-57 (E) and the hybrid H513-5 (K). This can be explained by the genealogy of this progeny, obtained from backcross between the hybrid H513-5 (K) and the parent of the Catuaí group UFV 2148-57 (E).
The other HdT parents, the other F1 hybrids, all BCr plants, except BCr 3-2, and about 44% of the plants in F2 generation were clustered in group II. These results can be explained by the introgression of C. canephora in C. arabica and prove the genetic diversity between these two germplasm, Catuaí and HdT. For being genetically divergent, these results confirm the capacity of the HdT germplasm to amplify the genetic base of C. arabica (Setotaw et al. 2010; Setotaw et al. 2013).
As observed in this work, a significant genetic divergence between the parents HdT UFV 530 (F) and UFV 445-46 (B) was reported in a study with RAPD, SSR, and AFLP molecular markers (Setotaw et al. 2010). These authors also verified greater genetic similarity between HdT UFV 445-46 (B) and UFV 440-22 in relation to HdT UFV 530 (F). These results corroborate those observed in the present study, since HdT UFV 440-22 and UFV 440-10 (D) are derived from HdT ERU 209-6.
This result evidences the capacity of the molecular markers and of this type of genetic-statistical analysis to identify genotypes with greater recovery of the recurrent parent’s genome in backcross programs. In addition, molecular markers provide more accurate and less subjective genetic information than that generated from phenotypic data, which is useful in breeding programs.
Genetic distance between pairs of genotypes
Genetic diversity analysis showed that genotype CIFC 134/4 is present in the greatest genetic distances detected in this study. This genotype is an accession that belongs to a germplasm bank that has not yet been genetically improved, but is important genotype for the breeding program, since it carries genes of interest that can be introgressed in commercial cultivars. The genotype 1-5, belonging to the generation of resistant backcross (progeny 1), was involved in 4 of the 10 greatest genetic distances in relation to the evaluated genotypes. By the analysis of all genotypes of this progeny, the other individuals presented small genetic distance between them, whereas the mean genetic distance between genotype 1-5 and the other genotypes was high. This indicates possible mixture of pollen, during the crosses, or mixture of seedlings, during planting. By the analyses of the phenotypic characteristics of the individuals of this progeny, all the individuals presented red fruits, except for the genotype 1-5, which presented yellow fruits.
The smallest genetic distances between the pairs of genotypes under study were observed between F1 hybrids H419-1 (G), H419-10 (H), H514-7 (I), and H514-8-1 (J). F1 hybrids H419-1 (G) and H419-10 (H) were originated from the cross between UFV 2143-236 (A) and UFV 445-46 (B), and the hybrids H514-7 (I) and H514-8-1 (J) were originated from the cross between the parents UFV 2154-344 (C) and UFV 440-10 (D). Genetic similarity between hybrids H419-1 (G) and H419-10 (H), and between hybrids H514-7 (I) and H514-8 (J) was expected for they are full-sibs. However, similarity was observed between the four hybrids, including non-siblings. This result suggests that the parents of the crosses analyzed in this study, both the parents Catuaí (A and C) and the parents Híbrido de Timor (B and D), are genetically close. HdT UFV 445-46 (B) and UFV 440-10 (D) correspond to accessions introduced by seeds from genotypes ERU 209/15 and ERU 209/6, respectively, which originated from genotype CIFC 2570 (Pereira et al. 2008). These data demonstrate the narrow genetic base of the breeding populations.
By the analysis of the genetic distances between the parents of all the studied populations, parents of the Catuaí group presented smaller distances in relation to HdT parents. In addition, among the Híbrido de Timor genotypes, UFV 530 (F) is the most divergent. These results demonstrate the importance of using HdT parents, especially the most divergent, in the development of new cultivars, in order to increase the genetic distance between them. In addition, accessions of HdT coffee plants carry genes of resistance to different diseases and pests, being the main source of resistance used in breeding programs around the world. HdTs have been used in the development of rust-resistant varieties, such as Catimor and Sarchimor, which present small size, high yield, and good cup quality. Despite being genetically similar, parents of the Catuaí group stand out for the high mean yield for they have great vegetative vigor and small size (Pereira et al. 2010a). Varieties of this coffee group are flexible in relation to climatic variations, and are well-adapted to several producing regions.
The parent UFV 530 (F) was the most genetically divergent among the HdT parents; however, it was the most genetically similar to the parents of the Catuaí group. These results demonstrate the importance of using this genotype in crosses in the development of new arabica cultivars, since it carries resistance genes and is more related to the genetically improved cultivars.
By the analysis of the genetic diversity among the individuals of the same generation (F1, F2, BCr, and BCs), the greatest genetic distances among genotypes were observed in the F2 generation. The smallest genetic distances were observed between genotypes in F1 generation. However, the values of the mean genetic distances between the four generations (F1, F2, BCr, and BCs) are very close and much higher than the mean genetic distance of the parents of the Catuaí group.
An increase in the genetic distance between the individuals originated from the crosses was observed when using parents that carry resistance genes. These results confirm the importance of the introduction of HdT genotypes (which carry resistance genes) to the crosses for the development of new cultivars. Setotaw et al. (2013) also observed an increase in genetic diversity after the introduction of HdT to C. arabica breeding. The authors demonstrated that rust-resistant coffee cultivars, which are basically derived from HdT, are more genetically distant than rust-susceptible coffee cultivars, and contributed to increase the genetic base of the improved Brazilian varieties.
Clustering analysis
The high efficiency of SNP markers in studies on genetic diversity was observed by the analysis of the dendrogram. In spite of being highly endogamic, all the genotypes of the study population were discriminated. This indicates that these markers are powerful tools to discriminate accessions in germplasm banks, avoiding redundancy in the collections; and to discriminate cultivars, supporting the morphological descriptors (Sousa et al. 2017).
Two main groups were formed in the dendrogram. In general, rust-susceptible parents were clustered in the first group, and rust-resistant parents were clustered in the second group. The susceptible backcross progenies were also clustered in the first group. The formation of these two large groups can be explained by the fact that the rust-susceptible parents was Catuaí Amarelo, which is the most commercially planted cultivar in Brazil, and corresponds to related genotypes. The resistant parents are accessions of Híbridos de Timor; therefore, they contain introgression of C. canephora genome (Lashermes et al. 2000; Bertrand et al. 2003). Híbrido de Timor is a natural hybrid between C. arabica and C. canephora, in which probably an unreduced gamete of C. canephora was combined with another normal gamete of C. arabica (Bettencourt 1973). HdT presents a phenotype similar to that of C. arabica, it is autogamous and tetraploid with 2n = 44 chromosomes, being easily crossed with C. arabica plants. The use of HdTs in the crosses results in increased genetic diversity (Setotaw et al. 2013) for they carry several genes of C. canephora.
The F1 hybrids were closely clustered in group I.b, except for the hybrid H513-5 (K).The H513-5 and its progenies, 9 (BCs) and 13 (F2), were separated into a subgroup (I.a). This greater diversity of H513-5 in relation to other hybrids obtained from crosses between arabica coffee and HdT was also observed by Fontes et al. (2002). The authors analyzed 29 hybrids of arabica coffee originated from crosses with different accessions of HdT, and observed that the hybrid H513-5 (K) did not cluster with any of the other evaluated genotypes. In addition, the hybrid H513-5 was resistant to rust and presented high yield capacity (Fontes et al. 2002). This F1 hybrid is the result of the cross between Catuaí Amarelo IAC 64 (UFV 2148-57 [E]) and HdT UFV 530 (F). The dendrogram, in agreement with the Bayesian analysis, demonstrated that parent UFV 530 (F) is the genetically closest HdT to the parent of the Catuaí group. UFV 530 (F) has been introduced from the Coffee Rust Research Center, Portugal, to the UFV/Epamig germplasm bank by cloning the plant CIFC 832/2. This genotype is considered as one of the main sources of resistance to rust, since it contains at least six resistance genes (Eskes 1989), and its resistance has not been supplanted yet. The other HdT parents of the studied progenies were introduced to the germplasm bank by using seeds of the same plant, CIFC 2570, justifying the genetic proximity of these HdTs and their F1 hybrids in the dendrogram.
Three accessions belonging to the germplasm bank, CIFC 128/2, CIFC 635/3, and CIFC 33/1, were also clustered in group I.a. The accessions CIFC 128/2 and CIFC 635/3 correspond to coffee plants introduced from Ethiopia and do not contain introgression of C. canephora into the genome, which explains why they were clustered in the same group of rust-susceptible parents. The last accession, CIFC 33/1, carries a resistance gene from another coffee species, C. liberica (Bettencourt and Rodrigues-Júnior 1988). The introgression of the genome of another species justifies the distance between these genotypes and the others. The greater genetic divergence and the presence of rust resistance gene different from the other coffee plants demonstrate the importance of including CIFC 33/1 in future crosses in breeding programs. The other accession of the germplasm bank, CIFC 134/4, was not clustered. This accession is not related to the parents used in this work; thus, greater genetic divergence between CIFC 134/4 and the population evaluated is expected.
Principal coordinate analysis
By the graphic dispersion of the scores obtained by PCoA, it was possible to observe the structuring of progenies based on the genetic distance between them. Similar to the cluster analysis, PCoA demonstrated the genetic distinction between the parent HdT UFV 530 and the F1 hybrids evaluated in this study. The parents of the Catuaí group were clustered on the opposite side to that where HdT parents were clustered; F1 hybrids were plotted between their respective parents; the progenies BCs and BCr were clustered close to their respective recurrent parents. These results are in agreement with other studies and can be explained by the kinship degree between the evaluated progenies.
Conclusion
In this research, the methodology of detection of the SNP molecular markers by constructing polymorphic probes was efficient to identity expressive number of SNP distributed throughout the C. arabica genome. The use of these SNP markers allowed the accurate discrimination of all the genotypes analyzed in the study, and even detected genetic mixtures among full-sib individuals, which are highly inbred, and therefore recommended for backcross. Analyses of the diversity and genetic structure of the population allowed evaluating the contribution of the HdT parents in the amplification of the genetic base of coffee plants. The analyses were precise in the division of the individuals into subpopulations according to the degree of introgression of C. canephora and its consequence in the obtainment of improved varieties was discussed. In addition, they enabled the identification of genetically different coffee plants to be introduced to breeding programs, as well as the molecular analysis of the parents that are already used.
For being numerous, polymorphic and widely distributed throughout the genome of the species, this set of markers has proved to be a useful auxiliary tool for breeding, and it can also be used for fingerprinting of accessions of germplasm banks and cultivars, for and genomic selection and association.
References
Aerts R, Berecha G, Gijbels P et al (2013) Genetic variation and risks of introgression in the wild Coffea arabica gene pool in south-western Ethiopian montane rainforests. Evol Appl 6:243–252. https://doi.org/10.1111/j.1752-4571.2012.00285.x
Alkimim ER, Caixeta ET, Sousa TV et al (2017) Marker-assisted selection provides arabica coffee with genes from other Coffea species targeting on multiple resistance to rust and coffee berry disease. Mol Breed 37:6. https://doi.org/10.1007/s11032-016-0609-1
Bertrand B, Anthony F, Lashermes P (2001) Breeding for resistance to Meloidogyne exigua in Coffea arabica by introgression of resistance genes of Coffea canephora. Plant Pathol 50:637–643. https://doi.org/10.1046/j.1365-3059.2001.00597.x
Bertrand B, Guyot B, Anthony F, Lashermes P (2003) Impact of the Coffea canephora gene introgression on beverage quality of C. arabica. TAG Theor Appl Genet 107:387–394. https://doi.org/10.1007/s00122-003-1203-6
Bettencourt A (1973) Considerações gerais sobre o híbrido de Timor: Origem e possibilidades de cultivo, 23rd edn. Instituto Agronômico, Campinas
Bettencourt A, Rodrigues-Júnior C (1988) Principles and practice of coffee breeding for resistance to rust and other diseases. In: Clarke RJ, Macrae R (eds) Coffee agronomy. Elsevier Applied Science Publishers LTD, London, pp 199–234
Bhering LL (2017) Rbio: a tool for biometric and statistical analysis using the R platform. Crop Breed Appl Biotechnol 17:187–190. https://doi.org/10.1590/1984-70332017v17n2s29
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Carvalho A, Krug CA (1949) Agentes de polinização da flor do cafeeiro (Coffea arabica L.) Bragantia. https://doi.org/10.1590/S0006-87051949000100002
Ceccarelli S (2015) Efficiency of plant breeding. Crop Sci 55:87. https://doi.org/10.2135/cropsci2014.02.0158
Clarindo WR, Carvalho CR (2008) First Coffea arabica karyogram showing that this species is a true allotetraploid. Plant Syst Evol 274:237–241. https://doi.org/10.1007/s00606-008-0050-y
Combes MC, Andrzejewski S, Anthony F et al (2000) Characterization of microsatellite loci in Coffea arabica and related coffee species. Mol Ecol 9:1178–1180. https://doi.org/10.1046/j.1365-294x.2000.00954-5.x
Crossa J, de los Campos G, Perez P et al (2010) Prediction of genetic values of quantitative traits in plant breeding using pedigree and molecular markers. Genetics 186:713–724. https://doi.org/10.1534/genetics.110.118521
Cruz CD (2013) GENES—a software package for analysis in experimental statistics and quantitative genetics. Acta Scientiarum Agronomy 35:271–276. https://doi.org/10.4025/actasciagron.v35i3.21251
Cubry P, Musoli P, Legnate H et al (2008) Diversity in coffee assessed with SSR markers: structure of the genus Coffea and perspectives for breeding. Genome 51:50–63. https://doi.org/10.1139/G07-096
Danecek P, Auton A, Abecasis G et al (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158. https://doi.org/10.1093/bioinformatics/btr330
Davis AP (2010) Six species of Psilanthus transferred to Coffea (Coffeeae, Rubiaceae). Phytotaxa 10:41–45
Davis AP (2011) Psilanthus mannii, the type species of Psilanthus, transferred to Coffea. Nord J Bot 29:471–472. https://doi.org/10.1111/j.1756-1051.2011.01113.x
Davis AP, Govaerts R, Bridson DM, Stoffelen P (2006) An annotated taxonomic conspectus of the genus Coffea (Rubiaceae). Bot J Linn Soc 152:465–512. https://doi.org/10.1111/j.1095-8339.2006.00584.x
de Azevedo Peixoto L, Laviola BG, Alves AA et al (2017) Breeding Jatropha curcas by genomic selection: a pilot assessment of the accuracy of predictive models. PLoS One 12:e0173368. https://doi.org/10.1371/journal.pone.0173368
Denoeud F, Carretero-Paulet L, Dereeper A et al (2014) The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science (New York, NY) 345:1181–1184. https://doi.org/10.1126/science.1255274
Diniz LEC, Sakiyama NS, Lashermes P et al (2005) Analysis of AFLP markers associated to the Mex-1 resistance locus in Icatu progenies. Cropp Breed Appl Biotechnol 5:387–393. 10.12702/1984-7033.v05n04a03
Diola V, de Brito GG, Caixeta ET et al (2011) High-density genetic mapping for coffee leaf rust resistance. Tree Genet Genomes 7:1199–1208. https://doi.org/10.1007/s11295-011-0406-2
Elsik CG, Tellam RL, Worley KC (2009) The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science (New York, NY) 324:522–528. https://doi.org/10.1126/science.1169588
Eskes AB (1989) Resistance. In: Kushalapa AC, Eskes AB (eds) Coffee rust: epidemiology, resistance and management. CRC Press, Boca Raton, FL, pp 171–293
Ferrão LFV, Caixeta ET, Pena G et al (2015) New EST–SSR markers of Coffea arabica: transferability and application to studies of molecular characterization and genetic mapping. Mol Breed 35:31. https://doi.org/10.1007/s11032-015-0247-z
Fontes JRM, Sakiyama NS, Cardoso AA et al (2002) Avaliação de híbridos F1 de café (coffee arabica L.) e respectivos progenitores com marcadores RAPD/Evaluation of F1 coffe hybrids (Coffee arabica L.) and their respective progenitors with RAPD markers. Ceres 49:283–294
Gao H, Williamson S, Bustamante CD (2007) A Markov chain Monte Carlo approach for joint inference of population structure and inbreeding rates from multilocus genotype data. Genetics 176:1635–1651. https://doi.org/10.1534/genetics.107.072371
Garcia C, Lima B, Almeida A et al (2011) Genome wide selection for Eucalyptus improvement at international paper in Brazil. BMC Proc 5:44. https://doi.org/10.1186/1753-6561-5-S7-P44
Garrison E, Gabor M (2012) Haplotype-based variant detection from short-read sequencing
Gichimu B, Gicheru E, Mamati G, Nyende A (2013) Variation and association of cup quality attributes and resistance to Coffee Berry Disease in Coffea arabica L. composite cultivar, Ruiru 11. African Journal of Hortic Sci 7:22–35
Gichimu BM, Gichuru EK, Mamati GE, Nyende AB (2014) Occurrence of Ck-1 gene conferring resistance to Coffee Berry Disease in Coffea arabica cv. Ruiru 11 and its parental genotypes. J Agric Crop Res 2:51–61
Gnirke A, Melnikov A, Maguire J et al (2009) Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol 27:182–189. https://doi.org/10.1038/nbt.1523
Hallauer AR (2011) Evolution of plant breeding. Crop Breed Appl Biotechnol 11:197–206. https://doi.org/10.1590/S1984-70332011000300001
Heffner EL, Sorrells ME, Jannink J-L (2009) Genomic selection for crop improvement. Crop Sci 49(1). https://doi.org/10.2135/cropsci2008.08.0512
Hendre P, Phanindranath R, Annapurna V et al (2008) Development of new genomic microsatellite markers from robusta coffee (Coffea canephora Pierre ex A. Froehner) showing broad cross-species transferability and utility in genetic studies. BMC Plant Biol 8:51. https://doi.org/10.1186/1471-2229-8-51
Heslot N, Yang H-P, Sorrells ME, Jannink J-L (2012) Genomic selection in plant breeding: a comparison of models. Crop Sci 52:146. https://doi.org/10.2135/cropsci2011.06.0297
ICO (2017) International coffee organization. In: Note regarding the review of statistical data published by the ICO. http://www.ico.org/prices/po-production.pdf. Accessed 6 Apr 2017
Inácio P, Lewinsohn T, do Carmo RL, Hogan DJ (2002) Ordenação multivariada na ecologia e seu uso em ciências ambientais. Ambiente & Sociedade 69–83. doi: https://doi.org/10.1590/S1414-753X2002000100005
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Lashermes P, Andrzejewski S, Bertrand B et al (2000) Molecular analysis of introgressive breeding in coffee (Coffea arabica L.) TAG Theor Appl Genet 100:139–146. https://doi.org/10.1007/s001220050019
Lashermes P, Combes M-C, Robert J et al (1999) Molecular characterisation and origin of the Coffea arabica L. genome. Mol Gen Genet MGG 261:259–266. https://doi.org/10.1007/s004380050965
Lashermes P, Combes MC, Ansaldi C et al (2011) Analysis of alien introgression in coffee tree (Coffea arabica L.) Mol Breed 27:223–232. https://doi.org/10.1007/s11032-010-9424-2
Lashermes P, Cros J, Marmey P, Charrier A (1993) Use of random amplified DNA markers to analyse genetic variability and relationships of Coffea species. Genet Resour Crop Evol 40:91–99. https://doi.org/10.1007/BF00052639
Laurie CC, Doheny KF, Mirel DB et al (2010) Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol 34:591–602. https://doi.org/10.1002/gepi.20516
Lee W-P, Stromberg MP, Ward A et al (2014) MOSAIK: a hash-based algorithm for accurate next-generation sequencing short-read mapping. PLoS One 9:e90581. https://doi.org/10.1371/journal.pone.0090581
Maluf MP, Silvestrini M, Ruggiero LM de C et al (2005) Genetic diversity of cultivated Coffea arabica inbred lines assessed by RAPD, AFLP and SSR marker systems. Sci Agric 62:366–373. https://doi.org/10.1590/S0103-90162005000400010
Matukumalli LK, Lawley CT, Schnabel RD et al (2009) Development and characterization of a high density SNP genotyping assay for cattle. PLoS One 4:e5350. https://doi.org/10.1371/journal.pone.0005350
Missio RF, Caixeta ET, Zambolim EM et al (2009a) Development and validation of SSR markers for Coffea arabica L. Cropp Breed Appl Biotechnol 9:361–371. 10.12702/1984-7033.v09n04a11
Missio RF, Caixeta ET, Zambolim EM et al (2011) Genetic characterization of an elite coffee germplasm assessed by gSSR and EST-SSR markers. Genet Mol Res 10:2366–2381. https://doi.org/10.4238/2011.October.6.2
Missio RF, Caixeta ET, Zambolim EM et al (2009b) Assessment of EST-SSR markers for genetic analysis on coffee. Bragantia 68:573–581. https://doi.org/10.1590/S0006-87052009000300003
Moncada MDP, Tovar E, Montoya JC et al (2016) A genetic linkage map of coffee (Coffea arabica L.) and QTL for yield, plant height, and bean size. Tree Genet Genomes 12:5. https://doi.org/10.1007/s11295-015-0927-1
Moncada P, McCouch S (2004) Simple sequence repeat diversity in diploid and tetraploid Coffea species. Genome 47:501–509. https://doi.org/10.1139/g03-129
Neves LG, Davis JM, Barbazuk WB, Kirst M (2013) Whole-exome targeted sequencing of the uncharacterized pine genome. Plant J 75:146–156. https://doi.org/10.1111/tpj.12193
Oliveira ACB, Sakiyama NS, Caixeta ET et al (2007) Partial map of Coffea arabica L. and recovery of the recurrent parent in backcross progenies. Crop Breed Appl Biotechnol 7:196–203
Ortiz R, Lund B, Andersen SB (2003) Breeding gains and changes in morphotype of Nordic spring wheat (1901-1993) under contrasting environments. Genet Resour Crop Evol 50:455–459. https://doi.org/10.1023/A:1023902110224
Pearl HM, Nagai C, Moore PH et al (2004) Construction of a genetic map for arabica coffee. TAG Theor Appl Genet 108:829–835. https://doi.org/10.1007/s00122-003-1498-3
Pereira AA, Carvalho GR, Moura WM et al (2010a) Cultivares: Origem e suas Características. In: Reis PR, Cunha RL (eds) Café arábica do plantio à colheita. EPAMIG, Lavras, pp 167–221
Pereira AA, Oliveira ACB, Sakiyama NS (2008) Híbrido de Timor como fonte de resistência a doenças e de qualidade da bebida do cafeeiro. In: Fernandes LH (ed) Manejo Fitossanitário da Cultura do Cafeeiro. Sociedade Brasileira de Fitopatologia, Brasília-DF, pp 13–24
Pereira MC, Chalfoun SM, de Carvalho GR, Savian TV (2010b) Multivariate analysis of sensory characteristics of coffee grains (Coffea arabica L.) in the region of upper Paranaíba. Acta Sci Agron. https://doi.org/10.4025/actasciagron.v32i4.4283
Pereira TB, Setotaw TA, Santos DN et al (2016) Identification of microsatellite markers in coffee associated with resistance to Meloidogyne exigua. Genet Mol Res. https://doi.org/10.4238/gmr.15038054
Pestana KN, Capucho AS, Caixeta ET et al (2015) Inheritance study and linkage mapping of resistance loci to Hemileia vastatrix in Híbrido de Timor UFV 443-03. Tree Genet Genomes 11:72. https://doi.org/10.1007/s11295-015-0903-9
Poncet V, Rondeau M, Tranchant C et al (2006) SSR mining in coffee tree EST databases: potential use of EST–SSRs as markers for the Coffea genus. Mol Gen Genomics 276:436–449. https://doi.org/10.1007/s00438-006-0153-5
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959. https://doi.org/10.1111/j.1471-8286.2007.01758.x
Resende M, Caixeta E, Alkimim ER, et al (2016) High-throughput targeted genotyping of Coffea arabica and Coffea canephora using next generation sequencing. San Diego, CA, p 1
Resende MFRJ, Muñoz P, Acosta JJ et al (2012a) Accelerating the domestication of trees using genomic selection: accuracy of prediction models across ages and environments. New Phytol 193:617–624. https://doi.org/10.1111/j.1469-8137.2011.03895.x
Resende MFRJ, Munoz P, Resende MDV et al (2012b) Accuracy of genomic selection methods in a standard data set of loblolly pine (Pinus taeda L.) Genetics 190:1503–1510. https://doi.org/10.1534/genetics.111.137026
Rodgers DM, Murphy JP, Frey KJ (1983) Impact of plant breeding on the grain yield and genetic diversity of spring oats. Crop Sci 23:737. https://doi.org/10.2135/cropsci1983.0011183X002300040032x
Romero G, Vásquez LM, Lashermes P, Herrera JC (2014) Identification of a major QTL for adult plant resistance to coffee leaf rust (Hemileia vastatrix) in the natural Timor hybrid (Coffea arabica x C. canephora). Plant Breed 133:121–129. https://doi.org/10.1111/pbr.12127
Rovelli P, Mettulio R, Anthony F et al (2000) Microsatellites in Coffea arabica L. In: Sera T, Soccol C, Pandey A, Roussos S (eds) Coffee biotechnology and quality. Springer Netherlands, Dordrecht, pp 123–133
Setotaw TA, Caixeta ET, Pena GF et al (2010) Breeding potential and genetic diversity of “Híbrido do Timor” coffee evaluated by molecular markers. Crop Breed Appl Biotechnol 10:298–304. https://doi.org/10.1590/S1984-70332010000400003
Setotaw TA, Caixeta ET, Pereira AA et al (2013) Coefficient of parentage in Coffea arabica L. cultivars grown in Brazil. Crop Sci 53:1237–1247. https://doi.org/10.2135/cropsci2012.09.0541
Smith JSC, Duvick DN, Smith OS et al (2004) Changes in pedigree backgrounds of Pioneer Brand maize hybrids widely grown from 1930 to 1999. Crop Sci 44:1935. https://doi.org/10.2135/cropsci2004.1935
Sobreira FM, de Oliveira ACB, Pereira AA et al (2015) Sensory quality of arabica coffee (Coffea arabica) genealogic groups using the sensogram and content analysis. Aust J Crop Sci 9:486–493
Sousa TVTV, Caixeta ETET, Alkimim ERER et al (2017) Molecular markers useful to discriminate Coffea arabica cultivars with high genetic similarity. Euphytica 213:75. https://doi.org/10.1007/s10681-017-1865-9
Ventorim Ferrão LF, Gava Ferrão R, Ferrão MAG et al (2017) A mixed model to multiple harvest-location trials applied to genomic prediction in Coffea canephora. Tree Genet Genomes 13:95. https://doi.org/10.1007/s11295-017-1171-7
Vidal RO, Mondego JMC, Pot D et al (2010) A high-throughput data mining of single nucleotide polymorphisms in Coffea species expressed sequence tags suggests differential homeologous gene expression in the allotetraploid Coffea arabica. Plant Physiol 154:1053–1066. https://doi.org/10.1104/pp.110.162438
Vieira ESN, Von Pinho ÉV d R, Carvalho MGG et al (2010) Development of microsatellite markers for identifying Brazilian Coffea arabica varieties. Genet Mol Biol 33:507–514. https://doi.org/10.1590/S1415-47572010005000055
Vieira LGE, Andrade AC, Colombo CA et al (2006) Brazilian coffee genome project: an EST-based genomic resource. Braz J Plant Physiol 18:95–108. https://doi.org/10.1590/S1677-04202006000100008
Yang H-C, Lin H-C, Kang M et al (2011) SAQC: SNP array quality control. BMC Bioinf 12:100. https://doi.org/10.1186/1471-2105-12-100
Zambolim L (2016) Current status and management of coffee leaf rust in Brazil. Trop Plant Pathol 41:1–8. https://doi.org/10.1007/s40858-016-0065-9
Funding
This work was financially supported by the Brazilian Coffee Research and Development Consortium (Consórcio Brasileiro de Pesquisa e Desenvolvimento do Café—CBP&D/Café), by the Foundation for Research Support of the state of Minas Gerais (FAPEMIG), by the National Council of Scientific and Technological Development (CNPq), and by the National Institutes of Science and Technology of Coffee (INCT/Café).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Data archiving statement
The authors have not submitted biological data to any of the public databases.
Additional information
Communicated by P. Ingvarsson
Electronic supplementary material
ESM 1
(XLSX 249 kb).
Rights and permissions
About this article
Cite this article
Sousa, T.V., Caixeta, E.T., Alkimim, E.R. et al. Population structure and genetic diversity of coffee progenies derived from Catuaí and Híbrido de Timor revealed by genome-wide SNP marker. Tree Genetics & Genomes 13, 124 (2017). https://doi.org/10.1007/s11295-017-1208-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-017-1208-y