Abstract
The sucrose non-fermenting 1-related protein kinase 2 (SnRK2) gene family belongs to a group of plant-specific serine/threonine kinase family involved in abscisic acid (ABA) signaling and biotic and abiotic stress response. Although genome-wide analyses of the SnRK2 gene family have been conducted in some species, little is known about the SnRK2 gene family in rubber tree (Hevea brasiliensis). In this study, we identified 10 SnRK2s designated as HbSnRK2.1 to HbSnRK2.10 in the rubber tree genome. The subsequently constructed phylogenetic tree demonstrated that HbSnRK2s have three subfamilies that correlate well with those of Arabidopsis sp. and rice subfamilies. All SnRK2 genes contained nine exons and eight introns. Although the C-terminus was divergent, eight conserved motifs were found. Motifs 1–6 were common to all HbSnRK2s. Expression analysis results showed that 7 of the 10 HbSnRK2s were highly expressed in latex. HbSnRK2.7 was predominantly expressed and simultaneously regulated by abscisic acid, jasmonic acid, and ethylene treatment in laticifers. HbSnRK identification and characterization provided further understanding on the role of ABA signal in the rubber tree.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Abscisic acid (ABA) is a classical plant hormone involved in abiotic stress response. ABA is important to plant growth and development, particularly to cell division and elongation, embryo maturation, seed dormancy, germination, root growth, floral induction, and biotic and abiotic stress response to osmotic stress, chilling, high salinity, drought, pathogen attack, and UV radiation (Finkelstein 2013; Miyakawa et al. 2013; Sah et al. 2016; Finkelstein et al. 2002, 2008; Yoshida et al. 2014; Lopez-Molina et al. 2001). As a stress hormone, ABA acts through regulatory pathways that control stomatal closure and gene expression (Zhu 2002; Wasilewska et al. 2008; Lee and Luan 2012). The ABA signaling pathway consists of three major protein classes, namely, ABA receptors (PYR/PYL/RCARs), type 2C protein phosphatases (PP2Cs), and sucrose non-fermenting 1-related protein kinase 2 (SnRK2; Wasilewska et al. 2008; Zhang et al. 2015; Cutler et al. 2010). Signaling starts with molecular recognition of ABA by the ABA receptor protein family. ABA binds to a PYR/PYL/RCAR protein, thereby inhibiting the phosphatase activity of PP2Cs and relieving inhibition of SnRKs required to activate downstream gene expression that convert ABA signals into appropriate cellular responses (Vilela et al. 2015; Park et al. 2009; Raghavendra et al. 2010; Ma et al. 2009).
SnRK2s belong to a group of plant-specific serine/threonine kinase family. As major contributors in ABA signaling and osmotic stress response, SnRK2s are well studied (Coello et al. 2011; Fujii and Zhu 2009; Kobayashi et al. 2004; Nakashima et al. 2009; Shao et al. 2014; Wang et al. 2015). SnRK2 families had been identified in some plants, such as Arabidopsis (Hrabak et al. 2003; Saha et al. 2014), rice (Kobayashi et al. 2004), maize (Huai et al. 2008), Malus prunifolia (Shao et al. 2014), Brachypodium distachyon (Wang et al. 2015), and Brassica rapa (Huang et al. 2015). All SnRK2 members have a conserved N-terminal catalytic domain that is similar to SNF1/AMP kinases and a short C-terminal regulatory domain that is not highly conserved. These families are classified into three subgroups according to amino acid sequences. The C-terminal domain contains stretches of acidic amino acids, either glutamic acid (group I) or aspartic acid (groups II and III; Kulik et al. 2011). The C-terminal consists of two subdomains. Domain I is present in all SnRK2 family members and is located 20 amino acids away from the catalytic domain. Domain II is essential for ABA response and specific to group III (Kobayashi et al. 2004; Belin et al. 2006; Yoshida et al. 2006). In general, ABA strongly activates SnRK2 group III members, weakly induces group II members, and moderately triggers group I (Kobayashi et al. 2004, 2005; Huai et al. 2008; Boudsocq et al. 2004; Fujita et al. 2009).
The rubber tree (Hevea brasiliensis Muell. Arg) is a tropical tree that belongs to the Euphorbiaceae family and is wildly cultivated to produce natural rubber (cis 1,4-polyisoprene) from the tree latex. Latex is a cytoplasm of highly specialized cells, known as laticifers, in rubber tree (Hao and Wu 2000). Laticifers in rubber tree barks are vital to rubber biosynthesis and defense against pathogen attacks (Chow et al. 2007). Furthermore, plant hormones are crucial to natural rubber biosynthesis. Laticifers are responsive to jasmonic acid (JA) because exogenous jasmonate specifically induces their differentiation (Hao and Wu 2000). JA signaling may also regulate natural rubber biosynthesis in laticifers (Peng et al. 2009; Tian et al. 2010; Pirrello et al. 2014). Ethylene (ET) is widely used for the stimulation of latex production (Zhu and Zhang 2009; Tungngoen et al. 2009). ABA-treated rubber trees exhibit early significant response, leading to significant increases in latex yield (Tungngoen et al. 2011) and suggesting that ABA signaling may also regulate latex production. In addition, ABA controls H. brasiliensis small rubber particle protein (HbSRPP; Guo et al. 2014), and Taraxacum brevicorniculatum small rubber particle protein (TbSRPP) gene (Fricke et al. 2013), indicating the possible regulation of natural rubber biosynthesis by ABA. HbSRPP is a major component of H. brasiliensis latex and apparently participates in natural rubber biosynthesis (Chow et al. 2007; Oh et al. 1999). However, knowledge on ABA signaling pathway in rubber trees is currently limited.
Despite the elucidation of several SnRK2 gene functions in Arabidopsis and other model species, less information on this gene family in rubber tree is available. Sequencing the rubber tree genome (Rahman et al. 2013; Tang et al. 2016) allows the identification and description of ABA signaling pathway genes. In this study, a total of 10 SnRK2 genes (designated as HbSnRK2.1–10) were identified in the genome of rubber tree. We also analyzed their phylogenetic relationships, gene structures, protein motifs, and expression patterns in five different tissues. Our results indicated that HbsnRK2s are highly expressed in latex, and ABA, JA, or ET regulated these genes. Our study provided a basis for further investigation of various HbSnRK2 gene functions in rubber trees.
Materials and methods
Plant materials and treatments
Rubber trees (H. brasiliensis cultivar RY 7-33-97) were planted in the experimental farm of the Chinese Academy of Tropical Agricultural Sciences, Hainan, China. Rubber tree shoots were treated with 0.5 w/v% ethrel, 0.1 v/v% methyl jasmonate, or 100 μM abscisic acid, according to a previous method (Hao and Wu 2000). Latex samples were collected at 1, 3, 6, 9, 12, 24, and 48 h after treatments from 12 shoots for each interval, and stored at −80 °C for RNA extraction. For latex RNA extraction, the latex was dropped directly into liquid nitrogen. Rubber tree roots, flowers, leaves, and barks were washed with double-distilled H2O and immediately frozen in liquid nitrogen.
Genome-wide identification of SnRK2 gene family in rubber tree
Multiple database searches were conducted to identify the SnRK2s in the rubber trees. Annotated (predicted) genes and proteins of the rubber trees were obtained from the tree genome data (DDBJ/EMBL/GenBank under the accession nos. AJJZ01000000 and LVXX01000000; Rahman et al. 2013; Tang et al. 2016). The SnRK2 family genes of Arabidopsis and rice were acquired from Phytozome v10.1 (https://phytozome.jgi.doe.gov/pz/portal.html#). SnRK2 complementary DNA (cDNA) sequences from Arabidopsis and rice served as queries for the search against rubber tree genome databases. Default Blast settings were used, but the low complexity filter and redundant sequences were removed manually. All search hits of the candidate sequences were analyzed using the NCBI Conserved Domain Search database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) to confirm that each gene belonged to the SnRK2 family. The specific primers (Table 1) were designed according to the annotated (predicted) HbSnRK2 genes, and the cDNA sequences of the HbSnRK2 genes were amplified and sequenced. Basing on the BlastP and BlastN search results from the rubber tree genome database, we obtained information on cDNA and genomic sequences. The molecular weight (MW) and isoelectric point (pI) of each protein sequence were calculated using ExPASy (http://web.expasy.org/compute_pi/).
Multiple sequence alignment and phylogenetic analysis
The multiple sequence alignments for the HbSnRK2 proteins were performed using ClustalW (Larkin et al. 2007). Further processing of the alignment data was carried out using ESPript 3.0 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi; Robert and Gouet 2014) with default parameter settings. An unrooted phylogenetic tree of SnRK2 protein sequences was constructed using MEGA 6.0 (Tamura et al. 2013) through the neighbor joining (NJ) method, and bootstrap analysis was conducted using 1000 replicates.
Gene structure analysis and motif detection of HbSnRK2 family genes
The exon-intron structures of the HbSnRK2 family genes were determined on the basis of the alignments of their coding sequences and corresponding genomic sequences, and a diagram was obtained with Gene Structure Display Server (GSDS 2.0; http://gsds.cbi.pku.edu.cn/) (Hu et al. 2015). Motif detection was performed with the MEME program (version 4.11.2, http://meme-suite.org/tools/meme; Bailey et al. 2015). The parameters were as follows: number of repetitions, any; maximum number of motifs, 20; and optimum motif widths, between 9 and 30 residues. Other options used default values.
RNA extraction and gene expression assay by qRT-PCR
Total RNA was extracted according to a previous method by Tang (Tang et al. 2007). Table S1 lists the quality assessment of RNAs preferably RIN values. cDNA strand was synthesized using RevertAid™ First Strand cDNA Synthesis Kit (Fermentas, Lithuania). RT-PCR was conducted with primers (Table 2) and rubber tree actin gene (GenBank HQ260674), which served as an internal control. RT-PCR was performed using the fluorescent dye SYBR-Green (TaKaRa, China), and the melt curve analysis of amplification products was conducted in Stratagene Mx3005P Real Time Thermal Cycler (Agilent, USA). The RT-PCR conditions were as follows: 5 min at 95 °C for denaturation, 45 cycles for 8 s at 95 °C, 30 s at 58 °C, and 20 s at 72 °C for amplification. Three biological replicates were used per treatment or control.
Statistical analysis
Relative RNA expression levels were determined through the 2−ΔΔCT method (Livak and Schmittgen 2001). Values are expressed as means ± SE of three different experiments with three replicate measurements. ANOVA was used to compare the statistical difference based on Fisher LSD test, at a significance level of P < 0.05 and P < 0.01.
Results
Identification of SnRK2 gene in rubber tree
To extensively identify rubber tree SnRK2 genes, we used BLAST and hidden Markov model to search the rubber tree genome database with SnRK2 sequences from Arabidopsis and rice as queries. After removing redundant sequences, we identified 10 SnRK2 genes that have complete serine/threonine protein kinase catalytic domains (Table 3) and designated them as HbSnRK2.1–10. The identified HbSnRK2 genes in rubber tree encode proteins with amino acid residues ranging from 338 (HbSnRK2.1, HbSnRK2.2, and HbSnRK2.3) to 364 (HbSnRK2.7). Table 1 lists the other characteristics of the HbSnRk2 genes, including gene and open reading frame length, pI, MW, and exons. The gene length ranged from 2096 bp (HbSnRK2.1) to 5193 bp (HbSnRK2.8). The MWs were from 38.18 (HbSnRK2.2) to 41.38 kDa (HbSnRK2.7), and all proteins have low pI values (pI <7.0) which ranged from 4.47 (HbSnRK2.7) to 6.19 (HbSnRK2.3).
The conserved residues of HbSnRK2s
To confirm the identity of putative full-length coding sequences, we performed RT-PCR amplifications on cDNAs from rubber tree latex. The 10 complete sequences were obtained and validated through sequencing. Amino acid sequence alignment indicated that 10 HbSnRK2s had at least 65.79% amino acid identity, and the maximum percentage of amino acid sequence identities was between HbSnRK2.7 and HbSnRK2.8 at 96.41% (Table 4).
Figure 1 shows the multiple sequence alignment of the HbSnRK2 proteins. As stated previously, the HbSnRK2s have highly conserved N-terminal kinase domains and divergent C-terminal domains that contain acidic amino acid-rich regions (Halford and Hardie 1998). All the members of the HbSnRK2 family have two conserved signatures in the kinase domains of their N-terminal regions. The first conserved signature is an ATP-binding region signature with a lysine residue as ATP-binding site, and the second is a serine/threonine protein kinase active-site signature with an aspartic acid residue as active site. These two signatures possibly belong to the protein kinase domain (Fig. 1). The C-terminal also contained two distinct domains. Domain I, which is necessary for activation by osmotic stress, was conserved within all SnRK2s, whereas domain II was only present in strongly ABA-responsive kinases. The domain II in the HbSnRK2 family was observed in HbSnRK2.7, HbSnRK2.8, HbSnRK2.9, and HbSnRK2.10 (Fig. 1).
Sequence and secondary structure alignment of HbSnRK2 proteins. The predicted secondary structure of the HbSnRK2 proteins was indicated, and the crystallographic structure of SnRK2.3 was used as a model (Protein Data Bank Code 3UC3). Espript interface (http://espript.ibcp.fr/) was used. Underlined stretches represent a conserved ATP-binding region and Ser/Thr protein kinase active site. Black boxes indicate the domain I and domain II at C-terminus
Phylogenetic analysis
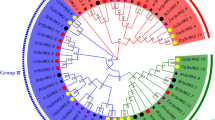
To characterize phylogenetic relationships between HbSnRK2s and SnRK2 family members from Arabidopsis and rice, phylogenetic analysis was conducted using MEGA version 6 by comparing the full-length protein sequences of 10 HbSnRK2s with SnRK2s from Arabidopsis and rice. All SnRK2 proteins were clustered into three groups, denoted as groups I, II, and III, which contain HbSnRK2.4 to HbSnRK2.6, HbSnRK2.1 to HbSnRK2.3, and HbSnRK2.7 to HbSnRK2.10, respectively (Fig. 2). Four HbSnRK2s (HbSnRK2.7 through HbSnRK2.10) that contain the domain II of the C-terminal were present in group III, suggesting that they are necessary for ABA response (Kulik et al. 2011).
Phylogenetic analysis of SnRK2 proteins from rubber tree, Arabidopsis, and rice. A total of 30 SnRK2s from rubber tree, Arabidopsis, and rice were used to create the NJ tree with 1000 bootstrap. The SnRK2 proteins are classified into three groups. GenBank accession numbers of selected SnRK2 proteins from Arabidopsis (blue) and rice (green) used for drawing phylogenetic tree: SnRK2.1–2.10 NP_196476, NP_190619, NP_001318893, NP_001031021, NP_201170, NP_567945, NP_195711, NP_974170, NP_179885, and NP_176290 and SAPK1–10 BAD17997, BAD17998, BAD17999, BAD18000, BAD18001, BAD18002, BAD18003, BAD18004, BAD18005, and BAD18006
Gene structure and conserved motifs in the C-terminus
Intron/exon structures were detected according to their evolutionary relationships to understand the structural features of the HbSnRK2 genes. Each gene structure was obtained by comparing its coding sequences to its genomic sequences. As shown in Fig. 3, all HbSnRk2 genes have eight introns and nine exons, and the exons have strictly conserved exon lengths. The lengths of the second until the eighth exons (except the second exon of HbSnRK2.4) were 75, 102, 54, 93, 93, 105, and 99 bp, respectively. In addition, the first and ninth exons of the HbSnRK2 genes in each subfamily have the same length. This finding supports the close evolutionary relationships among these genes and the classification of subfamilies.
Exon-intron structure of HbSnRK2s based on evolutionary relationship. The NJ evolutionary tree was created with 1000 bootstraps based on the full-length sequences of HbSnRK2s. Exon-intron analyses of HbSnRK2 genes were carried out with GSDS. Black lines and green boxes represent the introns and exons, respectively. The length of each exon is indicated
Although the C-terminal regions of SnRK2s were divergent in comparison with the highly conserved N-terminus, 10 conserved motifs were identified (Fig. 4). Motifs 1, 2, and 3 were present in all HbSnRK2 members. Motifs 5, 6, and 4 were unique to groups I, II, and III HbSnRK2s, respectively. Motifs 7 and 8 existed in group I TaSnRK2s except HbSnRK2.4, and motifs 9 and 10 were unique to HbSnRK2.4.
Expression profiles of HbSnRK2s in different tissues
The qRT-PCR of HbSnRK2s were performed in the roots, barks, leaves, flowers, and latex to investigate the expression patterns of HbSnRK2s in rubber tree. Figure 5 reveals that tissue expression profiles have differential patterns. The 10 HbSnRK2s were expressed in at least one of the five tissues, but their expression levels were relatively weak in the roots. The expression levels of four genes (HbSnRK2.2, 7, 8, and 9) were significantly higher in latex than in other tissues, but the content of the three genes (HbSnRK2.1, 3, and 4) were either extremely low or not expressed in the latex. The amounts of HbSnRK2.1 and HbSnRK2.3 were significantly higher in flowers than in other tissues. HbSnRK2.5, 6, and 10 showed high expression levels in leafs, flowers, and latex. With the exception of latex, HbSnRK2.7 had the highest expression levels in barks among other tissues.
Expression patterns of HbSnRK2s in different tissues. Relative transcript abundances of HbSnRK2s were examined by qRT-PCR. The Y axis is the scale of the relative transcript abundance level. X axis represents the tissues of rubber tree. R root, B bark, Le leaf, F flower, La latex. Total RNA was isolated from roots, barks, leaves, flowers, and latex. The rubber tree actin gene (GenBank HQ260674.1) served as an internal control
Expression patterns of HbSnRK2s in the latex respond to JA, ET, and ABA treatment
Regular application of ET on the rubber tree trunks stimulates latex yield. JA is also a key factor related to the production of rubber trees. ABA is an ubiquitous hormone that regulates plant growth, development, and environmental stress responses. Given that SnRK2s are important in abiotic stress responses, we examined the expression levels of HbSnRK2s in latex by treating the rubber tree shoots with JA, ET, or ABA. In this study, 7 of 10 HbSnRK2s (except for HbSnRK2.1, 3, and 4) had relative high levels of transcript abundance in latex under JA, ET, and ABA treatment. As shown in Fig. 6, the expression level of eight HbSnRK2s was differentially regulated in each of the three different treatments.
Expression patterns of the seven HbSnRK2s responding to ET, JA, and ABA treatment. Relative transcript abundances of HbSnRK2s were examined by qRT-PCR. Y axis is the scale of the relative transcript abundance level, and X axis is the time course of ET (a), JA (b), and ABA (c) treatment. Rubber tree actin gene (GenBank HQ260674.1) served as internal control. The significant difference was assessed by ANOVA (single or double asterisks corresponding to P < 0.05 and P < 0.01)
Analysis of HbsnRK2 expression levels in latex treated by ET showed that HbSnRK2.5, 7, 8, and 10 were upregulated to different degrees upon treatment, whereas HbSnRK2.6 was downregulated. No obvious change was observed in the expression level of the other two HbSnRK2s (HbSnRK2.2 and 9) analyzed (Fig. 6a). After the JA treatment, HbSnRK2.5, 6–8, and 10 exhibited elevated expression, whereas HbSnRK2.2 was downregulated. The expression level of HbSnRK2.9 had no considerable change (Fig. 6b). For ABA treatment, seven HbSnRK2s also showed altered expression levels in latex. In particular, HbSnRK2.5, 7–9, and 10 were markedly upregulated, whereas HbSnRK2.2 was downregulated. Meanwhile, HbSnRK2.6 was not responsive to ABA treatment (Fig. 6c).
Discussion
In this study, genome-wide analysis revealed 10 SnRK2 genes in H. brasiliensis. Rubber tree, Arabidopsis (Hrabak et al. 2003; Saha et al. 2014), rice (Kobayashi et al. 2004), and Brassica napus (Yoo et al. 2016) all have 10 SnRK2 genes and maize has 11 (Huai et al. 2008). In contrast, the grape genome contains only six SnRK2s (Boneh et al. 2012), suggesting the possible expansion of genes in the SnRK2 family along with whole-genome duplication events after the separation of plant lineages (Yoo et al. 2016).
According to the information obtained from the in silico survey of H. brasiliensis genome (Rahman et al. 2013; Tang et al. 2016), we identified the SnRK2 genes expressed in laticifer cells of rubber trees. Laticifer is a specific tissue essential to natural rubber biosynthesis and storage in rubber trees (Hao and Wu 2000). The transcriptome profiles of laticifers are relatively simple and have high prevalence of rubber biosynthesis-related genes and high level of defense-related genes, which are involved in rubber biosynthesis and defense (Chow et al. 2007). ABA may regulate natural rubber biosynthesis in laticifers (Fricke et al. 2013; Guo et al. 2014). Therefore, ABA signaling can be potential targets for genetic manipulation for the improvement of rubber productivity. In this study, our survey resulted in the identification of 10 HbSnRK2 genes from H. brasiliensis. All HbSnRK2 genes were mainly expressed in latex except HbSnRK2.1, 3, and 4, which were barely expressed in laticifers. The expression patterns found in various tissues can provide hints on the functional relevance and significance of HbSnRK2. For example, HbSnRK2.2, 7, 8, and 9 were preferentially expressed in laticifers, and thus, their potential involvement in rubber biosynthesis and defense appears logical. HbSnRK2.2, 5, 6, and 10 exhibited strong expression in laticifers and had high expression levels in leaf and flower tissues. Thus, we can infer that HbSnRK2.2, 5, 6, and 10 might be important in rubber biosynthesis and in other leaf and flower functions. HbSnRK2.1 and HbSnRK2.3 expression levels were high in flower, and HbSnRK2.4 expression was high in leaf, implying their potential functions in flower and leaf development. Nevertheless, further studies are necessary to determine whether HbSnRKs are crucial in rubber biosynthesis, defense, and development.
ABA is a major mediator of plant stress responses and many developmental programs. Most SnRK2 genes respond to various stresses, such as salt, drought, and cold (Yoo et al. 2016). In the present study, we applied ABA, JA, and ET treatment in latex. Among the 10 HbSnRKs that have high expression levels in latex, HbSnRK2.7 was predominant and exhibited strong responses to ABA, JA, and ET. JA and ET contribute to natural rubber biosynthesis and regulation (Hao and Wu 2000; Peng et al. 2009; Pirrello et al. 2014; Zhu and Zhang 2009; Tungngoen et al. 2009), and ABA possibly regulates natural rubber biosynthesis (Fricke et al. 2013; Guo et al. 2014). The interactions between ABA and JA/ET signaling pathways are possibly coordinated and generate combined optimal resistant responses in plants subjected to abiotic and biotic stresses (Lackman et al. 2011; Ahmad et al. 2016; Aleman et al. 2016). ABA, JA, and ET simultaneously regulated the expression of HbSnRK2.7, suggesting that HbSnRK2.7 plays a role in the interactions among ABA, JA, and ET signaling pathways. Thus, elucidating rubber biosynthesis regulated by ABA, JA, and ET as signal molecules may be of great interest in the future.
References
Ahmad P, Rasool S, Gul A, Sheikh SA, Akram NA, Ashraf M, Kazi AM, Gucel S (2016) Jasmonates: multifunctional roles in stress tolerance. Front Plant Sci 7:813. doi:10.3389/fpls.2016.00813
Aleman F, Yazaki J, Lee M, Takahashi Y, Kim AY, Li Z, Kinoshita T, Ecker JR, Schroeder JI (2016) An ABA-increased interaction of the PYL6 ABA receptor with MYC2 transcription factor: a putative link of ABA and JA signaling. Sci Rep 6:28941. doi:10.1038/srep28941
Bailey TL, Johnson J, Grant CE, Noble WS (2015) The MEME suite. Nucleic Acids Res 43:W39–W49. doi:10.1093/nar/gkv416
Belin C, de Franco PO, Bourbousse C, Chaignepain S, Schmitter JM, Vavasseur A, Giraudat J, Barbier-Brygoo H, Thomine S (2006) Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol 141:1316–1327. doi:10.1104/pp.106.079327
Boneh U, Biton I, Schwartz A, Ben-Ari G (2012) Characterization of the ABA signal transduction pathway in Vitis vinifera. Plant Sci 187:9–96. doi:10.1016/j.plantsci.2012.01.015
Boudsocq M, Barbier-Brygoo H, Lauriere C (2004) Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J Biol Chem 279:41758–41766. doi:10.1074/jbc.M405259200
Chow KS, Wan KL, Isa MN, Bahari A, Tan SH, Harikrishna K, Yeang HY (2007) Insights into rubber biosynthesis from transcriptome analysis of Hevea brasiliensis latex. J Exp Bot 58:2429–2440. doi:10.1093/jxb/erm093
Coello P, Hey SJ, Halford NG (2011) The sucrose non-fermenting-1-related (SnRK) family of protein kinases: potential for manipulation to improve stress tolerance and increase yield. J Exp Bot 62:883–893. doi:10.1093/jxb/erq331
Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61:651–679. doi:10.1146/annurev-arplant-042809-112122
Finkelstein R (2013) Abscisic acid synthesis and response. Arabidopsis Book / Am Soc Plant Biol 11:e0166. doi:10.1199/tab.0166
Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14(Suppl):S15–S45. doi:10.1105/tpc.010441
Finkelstein R, Reeves W, Ariizumi T, Steber C (2008) Molecular aspects of seed dormancy. Annu Rev Plant Biol 59:387–415. doi:10.1146/annurev.arplant.59.032607.092740
Fricke J, Hillebrand A, Twyman RM, Prüfer D, Schulze GC (2013) Abscisic acid-dependent regulation of small rubber particle protein gene expression in Taraxacum brevicorniculatum is mediated by TbbZIP1. Plant Cell Physiol 54:448–464. doi:10.1093/pcp/pcs182
Fujii H, Zhu JK (2009) Arabidopsis mutant deficient in 3 abscisic acid activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci U S A 106:8380–8385. doi:10.1073/pnas.0903144106
Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, Umezawa T, Fujita M, Maruyama K, Ishiyama K, Kobayashi M, Nakasone S, Yamada K, Ito T, Shinozaki K, Yamaguchi-Shinozaki K (2009) Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol 50:2123–2132. doi:10.1093/pcp/pcp147
Guo D, Li HL, Tang X, Peng SQ (2014) Molecular and functional characterization of the HbSRPP promoter in response to hormones and abiotic stresses. Transgenic Res 23:331–340. doi:10.1007/s11248-013-9753-0
Halford NG, Hardie DG (1998) SNF1-related protein kinases: global regulators of carbon metabolism in plants? Plant Mol Biol 37:735–748
Hao BZ, Wu JL (2000) Laticifer differentiation in Hevea brasiliensis: induction by exogenous jasmonic acid and linolenic acid. Ann Bot 85:37–43
Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, Thomas M, Walker-Simmons K, Zhu JK, Harmon AC (2003) The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132:666–680. doi:10.1104/pp.102.011999
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297. doi:10.1093/bioinformatics/btu817
Huai J, Wang M, He J, Zheng J, Dong Z, Lv H, Zhao J, Wang G (2008) Cloning and characterization of the SnRK2 gene family from Zea mays. Plant Cell Rep 27:1861–1868. doi:10.1007/s00299-008-0608-8
Huang Z, Tang J, Duan W, Wang Z, Song X, Hou X (2015) Molecular evolution, characterization, and expression analysis of SnRK2 gene family in Pak-choi (Brassica rapa ssp. chinensis). Front. Plant Sci 6:879. doi:10.3389/fpls.2015.00879
Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T (2004) Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 16:1163–1177. doi:10.1105/tpc.019943
Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T, Yamamoto A, Hattori T (2005) Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J 44:939–949. doi:10.1111/j.1365-313X.2005.02583.x
Kulik A, Wawer I, Krzywinska E, Bucholc M, Dobrowolska G (2011) SnRK2 protein kinases—key regulators of plant response to abiotic stresses. OMICS 15:859–872. doi:10.1089/omi.2011.0091
Lackman P, González-Guzmán M, Tilleman S, Carqueijeiro I, Pérez AC, Moses T, Seo M, Kanno Y, Häkkinen ST, Van Montagu MC, Thevelein JM, Maaheimo H, Oksman-Caldentey KM, Rodriguez PL, Rischer H, Goossens A (2011) Jasmonate signaling involves the abscisic acid receptor PYL4 to regulate metabolic reprogramming in Arabidopsis and tobacco. Proc Natl Acad Sci U S A 108:5891–5896. doi:10.1073/pnas.1103010108
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi:10.1093/bioinformatics/btm404
Lee SC, Luan S (2012) ABA signal transduction at the cross road of biotic and abiotic stress responses. Plant Cell Environ 35:53–60. doi:10.1111/j.1365-3040.2011.02426.x
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi:10.1006/meth.2001.1262
Lopez-Molina L, Mongrand S, Chua NH (2001) A post germination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci U S A 98:4782–4787. doi:10.1073/pnas.081594298
Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324:1064–1068. doi:10.1126/science.1172408
Miyakawa T, Fujita Y, Yamaguchi-Shinozaki K, Tanokura M (2013) Structure and function of abscisic acid receptors. Trends Plant Sci 18:259–266. doi:10.1016/j.tplants.2012.11.002
Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, Shinozaki K, Yamaguchi-Shinozaki K (2009) Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 50:1345–1363. doi:10.1093/pcp/pcp083
Oh SK, Kang H, Shin DH, Yang J, Chow KS, Yeang HY, Wagner B, Breiteneder H, Han KH (1999) Isolation, characterization, and functional analysis of a novel cDNA clone encoding a small rubber particle protein from Hevea brasiliensis. J Biol Chem 274:17132–17138. doi:10.1074/jbc.274.24.17132
Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu JK, Schroeder JI, Volkman BF, Cutler SR (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324:1068–1071. doi:10.1126/science.1173041
Peng SQ, Xu J, Li HL, Tian WM (2009) Cloning and molecular characterization of HbCOI1 from Hevea brasiliensis. Biosci Biotechnol Biochem 73:665–670. doi:10.1271/bbb.80721
Pirrello J, Leclercq J, Dessailly F, Rio M, Piyatrakul P, Kuswanhadi K, Tang C, Montoro P (2014) Transcriptional and post-transcriptional regulation of the jasmonate signaling pathway in response to abiotic and harvesting stress in Hevea brasiliensis. BMC Plant Biol 14:341. doi:10.1186/s12870-014-0341-0
Raghavendra AS, Gonugunta VK, Christmann A, Grill E (2010) ABA perception and signalling. Trends Plant Sci 15:395–401. doi:10.1016/j.tplants.2010.04.006
Rahman AYA, Usharraj AO, Misra BB, Thottathil GP, Jayasekaran K, Feng Y et al (2013) Draft genome sequence of the rubber tree (Hevea brasiliensis). BMC Genomics 14:75. doi:10.1186/1471-2164-14-75
Robert X, Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi:10.1093/nar/gku316
Sah SK, Reddy KR, Li J (2016) Abscisic acid and abiotic stress tolerance in crop plants. Front Plant Sci 7:571. doi:10.3389/fpls.2016.00571
Saha J, Chatterjee C, Sengupta A, Gupta K, Gupta B (2014) Genome-wide analysis and evolutionary study of sucrose non-fermenting 1-related protein kinase 2 (SnRK2) gene family members in Arabidopsis and Oryza. Comput Biol Chem 49:59–70. doi:10.1016/j.compbiolchem. 2013.09.005
Shao Y, Qin Y, Zou Y, Ma F (2014) Genome-wide identification and expression profiling of the SnRK2 gene family in Malus prunifolia. Gene 552:87–97. doi:10.1016/j.gene.2014.09.017
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi:10.1093/molbev/mst197
Tang C, Qi J, Li H, Zhang C, Wang Y (2007) A convenient and efficient protocol for isolating high quality RNA from latex of Hevea brasiliensis (para rubber tree). J Biochem Biophys Methods 70:749–754. doi:10.1016/j.jbbm.2007.04.002
Tang C, Yang M, Fang Y, Luo Y, Gao S, Xiao X et al (2016) The rubber tree genome reveals new insights into rubber production and species adaptation. Nature Plants 16073. doi:10.1038/NPLANTS.2016.73
Tian WW, Huang WF, Zhao Y (2010) Cloning and characterization of HbJAZ1 from the laticifer cells in rubber tree (Hevea brasiliensis Muell. Arg.) Trees 24:771–779. doi:10.1016/j.plaphy.2015.10.023
Tungngoen K, Kongsawadworakul P, Viboonjun U, Katsuhara M, Brunel N, Sakr S, Narangajavana J, Chrestin H (2009) Involvement of HbPIP2;1 and HbTIP1;1 aquaporins in ethylene stimulation of latex yield through regulation of water exchanges between inner liber and latex cells in Hevea brasiliensis. Plant Physiol 151:843–856. doi:10.1104/pp.109.140228
Tungngoen K, Viboonjun U, Kongsawadworakul P, Katsuhara M, Julien JL, Sakr S, Chrestin H, Narangajavana J (2011) Hormonal treatment of the bark of rubber trees (Hevea brasiliensis) increases latex yield through latex dilution in relation with the differential expression of two aquaporin genes. J Plant Physiol 168:253–262
Vilela B, Pagès M, Riera M (2015) Emerging roles of protein kinase CK2 in abscisic acid signaling. Front Plant Sci 6:966. doi:10.3389/fpls.2015.00966
Wang L, Hu W, Sun J, Liang X, Yang X, Wei S, Wang X, Zhou Y, Xiao Q, Yang G, He G (2015) Genome-wide analysis of SnRK gene family in Brachypodium distachyon and functional characterization of BdSnRK2.9. Plant Sci 237:33–45. doi:10.1016/j.plantsci.2015.05.008
Wasilewska A, Vlad F, Sirichandra C, Redko Y, Jammes F, Valon C, Frei dit Frey N, Leung J (2008) An update on abscisic acid signaling in plants and more. Mol Plant 2:198–217. doi:10.1093/mp/ssm022
Yoo MJ, Ma T, Zhu N, Liu L, Harmon AC, Wang Q, Chen S (2016) Genome-wide identification and homeolog-specific expression analysis of the SnRK2 genes in Brassica napus guard cells. Plant Mol Biol 91:211–227. doi:10.1007/s11103-016-0456-9
Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K (2006) The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem 281:5310–5318. doi:10.1074/jbc.M509820200
Yoshida T, Mogami J, Yamaguchi-Shinozaki K (2014) ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr Opin Plant Biol 21:133–139. doi:10.1016/j.pbi.2014.07.009
Zhang XL, Jiang L, Xin Q, Liu Y, Tan JX, Chen ZZ (2015) Structural basis and functions of abscisic acid receptors PYLs. Front Plant Sci 6:88. doi:10.3389/fpls.2015.00088
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273. doi:10.1146/annurev.arplant.53.091401.143329
Zhu J, Zhang Z (2009) Ethylene stimulation of latex production in Hevea brasiliensis. Plant Signal Behav 4:1072–1074
Acknowledgements
This study was supported by National Natural Science Foundation of China (No. 31471169) and Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (No. 1630052016003) and the earmarked fund for China Agriculture Research System (CARS-34-ZP1).
Data archiving statement
Nucleotide sequences were deposited with the GenBank HbSnRK2.1 (GenBank accession no. KY211982), HbSnRK2.2 (GenBank accession no. KY211983), HbSnRK2.3 (GenBank accession no. KY211984), HbSnRK2.4 (GenBank accession no. KY211985), HbSnRK2.5 (GenBank accession no. KY211986), HbSnRK2.6 (GenBank accession no. KY211987), HbSnRK2.7 (GenBank accession no. KY211988), HbSnRK2.8 (GenBank accession no. KY211989), HbSnRK2.9 (GenBank accession no. KY2119990), and HbSnRK2.10 (GenBank accession no. KY211991).
Author information
Authors and Affiliations
Contributions
Shi-Qing Peng and Dong Guo conceived and designed the experiments and drafted the manuscript; Dong Guo, Hui-Liang Li, Jia-Hong, Zhu Ying Wang, An Feng, and Gui-Shui Xie carried out the gene isolation, sequence analysis, and gene expression analysis. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by W. Ratnam
Electronic supplementary material
Table S1
(DOCX 14 kb).
Rights and permissions
About this article
Cite this article
Guo, D., Li, HL., Zhu, JH. et al. Genome-wide identification, characterization, and expression analysis of SnRK2 family in Hevea brasiliensis . Tree Genetics & Genomes 13, 86 (2017). https://doi.org/10.1007/s11295-017-1168-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-017-1168-2