Abstract
Aquaporins belong to the highly conserved major intrinsic protein family and are involved in the transcellular membrane transport of water and other small solutes. However, there has been little work on cloning aquaporin (AQP) family genes and characterizing their functions in plants under various environmental stimuli. In this study, a total of 18 full-length AQP genes were identified in Tamarix hispida, a woody halophyte. Sequence analysis showed that most of these AQP proteins have six transmembrane domains connected by five loops. Phylogenetic analysis revealed that the members of the AQP family can be divided into four groups based on their structural characteristics, including plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), NOD26-like intrinsic proteins (NIPs), and small basic intrinsic proteins (SIPs). Furthermore, the expression profiles of AQP genes were analyzed in the roots, stems, and leaves under salinity, drought, heavy metal, and abscisic acid (ABA) treatments using real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) assays. The results demonstrated that the AQP were involved in abiotic stress responses, indicating that they play important roles in response to abiotic stress and are involved in ABA-dependent stress-signaling pathways. These data will be useful to elucidate the complexity of the AQP gene family and the molecular mechanisms of abiotic stress tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants constantly experience complex and variable stresses during their growth and development, including salinity, drought, heavy metals, and extreme temperatures. Therefore, plants have evolved a series of protective mechanisms for adequate growth and reproduction. The effectiveness of water transporters, such as the aquaporins (AQPs), is an important component of the plant response to stress (Kaldenhoff and Fischer 2006). The AQPs belong to the highly conserved major intrinsic protein family, also called water channels, and interact with the cell membrane system to facilitate the transport of water and a variety of low-molecular weight solutes, such as glycerol and volatile substances (Maurel et al. 2008; Mitani-Ueno et al. 2011). Currently, large numbers of AQP genes have been identified in different plant species, including 35 homologs in Arabidopsis thaliana (Johanson et al. 2001), 31 in Zea mays (Chaumont et al. 2001), 33 in Oryza sativa (Sakurai et al. 2005), and 71 in Gossypium hirsutum (Park et al. 2010). On the basis of subcellular localization and sequence homology, plant AQPs can be largely divided into at least four different subfamilies, including plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), nodulin-26 intrinsic proteins (NIPs), and small basic intrinsic proteins (SIPs) (Gupta and Sankararamakrishnan 2009).
All members of the AQP family are conserved in plants and characterized by six transmembrane domains consisting of α-helices (TM1–TM6) that are connected by five interhelical loops (A–E). The cytosolic loop (loop B) between the second and third transmembrane domain and the extra-cytosolic loop (loop E) between the fifth and sixth transmembrane domain form the aqueous pore. Loops B and E also contain the highly conserved NPA (Asn-Pro-Ala) motifs, which overlap the middle of the lipid bilayer of the membrane forming a 3-D “hourglass” structure where the water flows through (Tajkhorshid et al. 2002). The second constriction is known as the aromatic/arginine (ar/R) selectivity filter and is formed on the extracellular side of the pore by four residues from transmembrane helices (H2 and H5) and loop E (LE1 and LE2). The ar/R filter is able to selectively bind water molecules and prevent other molecules from entering the aqueous pore. Based on the specific structural features, plant AQPs may confer a broad spectrum of selectivity for water and/or solutes (Törnroth-Horsefield et al. 2006).
Although the PIP, TIP, NIP, and SIP subfamilies are conserved in plants, plant AQPs have divergent functions. For example, it is generally known that AQPs play a major role in maintaining water balance by regulating the water transport system in the root to protect against a variety of environmental stimuli and facilitate water transport through inner leaf tissues during transpiration (Maurel et al. 2008; Nguyen et al. 2013). Recently, increasing evidence has demonstrated that plant AQPs play several important roles in numerous physiological processes, including seed germination (Liu et al. 2013), plant reproduction (Bots et al. 2005; Kaldenhoff and Fischer 2006), cell division and elongation (Okubo-Kurihara et al. 2009), fruit ripening (Mut et al. 2008), and leaf movements (Uehlein and Kaldenhoff 2008; Heinen et al. 2009). Furthermore, AQPs promote carbon and nitrogen fixation through their involvement in the passage of many substrates, such as H2O2, urea, silicic acid, methylated arsenic species, and ammonia (Bienert et al. 2006; Maurel et al. 2008; Li et al. 2009). However, there have been a limited number of studies on the expression of AQPs during the growth and development of native plants or in plant organs over their lifespan, especially in woody halophyte plant species.
Tamarix (Tamaricaceae) is a family of woody halophyte species, which is widely distributed in the saline soils of drought-stricken areas of central Asia and China. Tamarix hispida, a species of the genus Tamarix, is highly tolerant to salinity, drought, and extreme temperatures. These characteristics make the species suitable for investigation of stress tolerance mechanisms. In this study, we screened the functional annotation of non-redundant unigenes in seven transcriptomes and identified 18 AQP genes representing four subfamilies. Then, a comprehensive analysis of their structure and phylogeny was performed. The expression of 18 AQPs in the roots, stems, and leaves of T. hispida under abiotic stresses (salt, drought, and heavy metal) and with exogenous application of ABA was investigated using real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR). These systemic overviews are the first comprehensive study of the AQP gene family in T. hispida and may provide valuable information for further exploring the functions of genes mediated by the water stress-responsive AQP genes.
Materials and methods
Identification of AQP family genes from transcriptomes of T. hispida
In a previous study, seven transcriptomes were built from T. hispida roots treated with NaHCO3 for 0, 12, 24, and 48 h and leaves treated with NaHCO3 for 0, 12, and 24 h by RNA-seq using high-throughput Solexa sequencing technology. The raw reads from each library were respectively assembled into contigs using the SOAPdenovo software; the contigs were further assembled into scaffolds using paired-end joining and gap filling, and tentative unigenes (TUGs) were generated by assembling these scaffolds from each library. Sequence overlaps in the sorted results files are filtered using user-defined criteria: the minimum length of the overlap (default 40 bp), the minimum percent of repeat stringency (default 95%), and the maximum unmatched overhangs (the default starts at 30 nucleotides). The general characteristics of the seven transcriptomes and the length distributions of the unigenes are described in Wang et al. (2014a). A total of 94,359 non-redundant unigenes were generated using the TGI clustering tools (Pertea et al. 2003), and these unigenes were subjected to BLASTX (http://blast.ncbi.nlm.nih.gov/) analysis against the NR and Swiss-Prot protein databases to search for functional annotation. The unigenes with a threshold E value of more than 1 × 10−5 were discarded. The key word “aquaporin” was used as a query to search against the functional annotation of non-redundant unigenes for identifying the AQP genes. The open reading frame (ORF) of each AQP gene was resolved using ORF Finder from NCBI (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Then, the primers for amplifying the ORF of AQP genes were designed according to these sequences. The PCR products were ligated into the pMD18-T Easy vector (TaKaRa) and subsequently sequencing were used to confirm the AQP sequences. Primer sequences are shown in Table S1.
Bioinformatics analysis of the AQP family genes
The isoelectronic point (pI) and theoretical molecular weight (M W) predictions of deduced AQPs were calculated with the ExPASy compute pI/M W tool (http://www.expasy.org/tools/protparam.html). Sequence identity among the AQPs was calculated using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/). The transmembrane helical domain was predicted using the TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) (Krogh et al. 2001). Prediction of the subcellular localization of putative AQPs was performed using the WoLF PSORT algorithm (http://www.genscript.com/wolf-psort.html) (Horton et al. 2007). Sequence alignments of related proteins were performed with ClustalW using BioEdit software and adjusted manually. The unrooted phylogenetic tree was constructed with MEGA 5.05 using the neighbor-joining (NJ) method, and the internal branch support was estimated with 1000 bootstrap replicates (Tamura et al. 2011). Next, the conserved motifs in the full-length protein sequences were identified using the MEME online program (http://meme-suite.org) with the following parameters: number of repetitions = any, maximum number of motifs = 20, and optimum motif width = 30 to 70 residues (Bailey et al. 2009).
Plant growth and stress treatments
Seeds of T. hispida were grown in pots containing a mixture of turf peat and sand (2:1 v/v) under controlled greenhouse conditions of 70–75% relative humidity, 14 h light, 10 h dark, and an average temperature of 24 °C. Two-month-old seedlings were exposed to 300 mM NaHCO3 for 12, 24, and 48 h. Following the treatment, roots, and leaves from at least 24 seedlings of each group were harvested and stored at −80 °C prior to RNA extraction for building transcriptomes. In addition, the seedlings were exposed to the following solutions: 400 mM NaCl (salt stress), 25% (w/v) PEG6000 (PEG-simulated drought stress), 150 μM CdCl2 (heavy metal stress), or 150 μM ABA for 0, 3, 6, 9, 12, or 24 h. A fresh water-only control was conducted in parallel. Following these treatments, the leaves, stems, and roots of seedlings from each sample (at least 10 seedlings) were harvested at the indicated times and stored at −80 °C until required.
RNA isolation and real-time quantitative reverse transcription polymerase chain reaction
Total RNA was extracted from each sample by the CTAB method with minor modifications and treated with RQ1 RNase-free DNase (Promega, Madison, WI, USA) to remove genomic DNA contamination. The first-strand cDNA was synthesized with approximately 0.5 μg of purified total RNA using the PrimeScript™ RT reagent kit (TaKaRa, Dalian, China) according to the manufacturer’s protocol. The synthesized cDNAs were diluted to 100 μL with sterile water and were used as the template for real-time qRT-PCR. qRT-PCR was performed on the Opticon 2 System (Bio-Rad, Hercules, CA) using SYBR Green Real-time PCR Master Mix (Toyobo) and specific primers (Table S1). The reaction mixture (20 μL) contained 10 μL 2 × SYBR Green Real-time PCR Master Mix, 0.5 μM of each of the forward and reverse primers, and 2 μL of cDNA template. The amplification was completed with the following cycling parameters: 94 °C for 30 s; followed by 45 cycles at 94 °C for 12 s, 58 °C for 30 s, 72 °C for 40 s, and 1 s at 79 °C for plate reading. A melting curve analysis of the product was performed immediately after the final PCR cycle by increasing the temperature from 55 to 90 °C at 0.5 °C s−1. To ensure the reproducibility of the results, at least three biological replicates, each with three technical replicates, were performed for each sample. The relative expression ratios were calculated from the threshold cycle according to the 2-ΔΔCt method (Livak and Schmittgen 2001). The relative expression (ΔΔCT) of each gene was normalized to the geometric mean of α-tubulin, β-tubulin, and β-actin as internal control genes (Vandesompele et al. 2002). We used 2-ΔΔCt > 2 and p-values <0.05 as a threshold to significance of gene expression differences. All of the relative expression levels were log2 transformed.
Results and discussion
Identification and classification of the AQP genes in T. hispida
Previously, we generated seven transcriptomes from the roots and leaves of T. hispida treated with NaHCO3 at various time intervals. The general characteristics of the seven transcriptomes and the length distributions of the unigenes are described in Wang et al. (2014a). After functional annotation, a total of 34 non-redundant unigenes were identified as AQP family genes. However, because genome sequencing of T. hispida is incomplete, we cannot determine the exact number of AQP genes in T. hispida. To further confirm the reliability of the identified genes, a BLASTP and BLASTX program search of the NR protein and Swiss-Prot databases and an online domain batch search with an E-value of more than 1 × 10−5 were performed. Among them, 18 non-redundant unigenes with full open reading frames (ORFs) were identified in the NR protein database. The genes with 6 homologs classified as plasma membrane intrinsic proteins (PIPs), 5 homologs classified as tonoplast intrinsic proteins (TIPs), 5 homologs classified as NOD26-like intrinsic proteins (NIPs) and 2 homologs classified as small basic intrinsic proteins (SIPs), respectively. The GenBank accession numbers of ThAQPs are shown in Table S1. A comparison of 18 deduced AQP protein sequences and those of other plants revealed a high sequence identity at the protein level (67–91%) and at the nucleotide level (55–82%). The amino acid number varied from a minimum of 240 to a maximum of 307, with predicted sizes ranging from a minimum of 25.47 to 31.59 kDa and pI values ranging from 4.8 to 9.76 (Table 1).
Structure and phylogenetic analysis of the AQP proteins in T. hispida
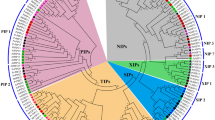
Sequence homology analysis of the predicted peptide sequences of these AQPs showed that high sequence diversity exists among the four subfamilies of AQPs in T. hispida. The highest sequence identity of 27.46–36.73% was found between PIPs and TIPs, and the lowest sequence identity of 16.36–21.76% was found between NIPs and SIPs (Table S2). To examine the phylogenetic relationships between the AQP proteins in T. hispida and Populus trichocarpa, the AQP protein sequences from the T. hispida transcriptomes were subjected to BLAST against the Populus trichocarpa proteome (v3.0), which was downloaded from the MIPModDB database (http://bioinfo.iitk.ac.in/MIPModDB/) (Gupta and Sankararamakrishnan 2009; Gupta et al. 2012). Then, the 53 putative Populus AQP protein sequences were collected for phylogenetic analysis (Fig. 1). Based on the predicted amino acid sequences, the ThAQPs could be classified into four groups of PIPs, TIPs, NIPs, and SIPs, with different cellular localizations and functions (Zardoya 2005), and designated as ThPIP1;1, ThPIP2;1 to ThPIP2;5, ThTIP1;1 to ThTIP1;3, ThTIP2;1, ThTIP4;1, ThNIP1;1 to ThNIP1;2, ThNIP3;1 to ThNIP3;3, ThSIP1; and 1 to ThSIP1;2. This observation is consistent with previous phylogenetic analyses of plant AQPs (Gupta and Sankararamakrishnan 2009; Rasheed-Depardieu et al. 2012). In addition, we further analyzed the motif distributions among these 18 ThAQP proteins using the MEME program, and a total of 14 conserved motifs were identified and named 1–14 (Fig. 2a). The consensus sequences of these motifs are given in Fig. 2b. Of these, motif 4 was found to be in all the ThAQPs. With the exception of the ThSIPs, the remaining ThAQPs possessed motifs 1, 2, and 3. The four motifs correspond to the main transmembrane domain regions. Motif 5 was mainly found in the ThPIPs and ThTIPs, and motif 6 was only present in the ThPIPs and ThNIPs. The remaining 8 motifs were specifically found in the ThNIPs and ThSIPs. Prediction of TMDs (transmembrane domains) by TMHMM Server v. 2.0 showed that most identified putative AQPs contained a typical topology of six TMDs with the exception of ThTIP1;1, ThTIP1;2, ThTIP2;1, ThNIP3;1, ThNIP3;3, and ThSIP1;2. Of these, ThTIP1;1, ThTIP1;2, and ThTIP2;1 had seven TMDs, whereas ThNIP3;1 and ThNIP3;3 had five TMDs, and ThSIP1;2 had four TMDs (Fig. 3, Table 2). The cellular localization prediction analysis showed that the six PIP subfamily members were localized to plasma membranes, which is in agreement with the current literature (Table 2). The TIP members were diverse and predicted localizations were in the cytosol and vacuolar membrane. NIP members were generally predicted to be localized to the plasma membrane. The two members of the SIP subfamily were predicted to be localized to the cytosol and vacuolar membrane.
Phylogenetic analysis of aquaporin proteins from T. hispida and Populus. Predicted amino acid sequences were aligned using ClustalW2, and the unrooted neighbor-joining tree was constructed using MEGA 5.05 with 1000 bootstrap replicates. Bootstrap support values are indicated on each node. The sequences of the poplar aquaporin proteins were downloaded from the MIPModDB database (http://bioinfo.iitk.ac.in/MIPModDB/) (Gupta and Sankararamakrishnan 2009; Gupta et al. 2012)
Multiple sequence alignment of T. hispida aquaporins. Predicted amino acid sequences were aligned using ClustalW with BioEdit software. The two conserved NPA motifs (NPAI and NPAII), ar/R selectivity filters (H2, H5, LE1, and LE2) and Froger’s positions (P1–P5) are boxed and marked in bold below the alignment, and the transmembrane domains (TM1–TM6) are underlined
We further analyzed several features, including the highly conserved dual NPA motifs (NPAI and NPAII), the nature of the ar/R selective filter (H2, H5, LE1, and LE2), and 5 Froger’s positions (P1–P5). Multiple sequence alignments of the predicted amino acid sequences of the 18 Tamarix AQPs and their dual NPA motifs, ar/R selective filter and Froger’s positions are illustrated in Fig. 3, Table 2. In PIPs, the dual NPA motifs and ar/R selective filter (F-H-T-R) are highly conserved across all plant species, the Froger’s positions P2 to P5 are strictly conserved and consist of S-A-F-W, and only the P1 position is more variable and can be Met (M), Gln (Q), Gly (G), or Tyr (Y) (Venkatesh et al. 2013). Members of the PIP subfamily in other plant species have been shown to be positively regulated in their water transport activity via phosphorylation (Azad et al. 2008; Whiteman et al. 2008). All of the ThPIP members in T. hispida contained the highly conserved dual NPA motifs, exhibited identical amino acids in the ar/R selectivity filter (F-H-T-R) with the exception of ThPIP2;2, in which Thr (T) was replaced with Cys (C) in the LE1 position, and had either Met (M) or Gln (Q) in the P1 position. In the TIPs, dual NPA motifs and the P3, P4, and P5 positions are highly conserved, but the selectivity filter (ar/R) is highly variable across plant species (Venkatesh et al. 2013). However, among the five ThTIPs, dual NPA motifs, ar/R and Froger’s positions exhibited no variation as the expected amino acids were identified, such as His (H), Thr (T), Ser (S), and Ala (A) in the H2, P1, P2, and P3 positions, respectively, and the TIPs are characterized by Ala (A) in the LE1 and an unusual Val (V) instead of Arg (R) in the LE2. However, ThTIP2;1 deviated from this pattern and had G (LE1) and R (LE2) residues. ThNIP1;1 and ThNIP1;2 showed typical dual NPA motifs, while in ThNIP3;1, ThNIP3;2, and ThNIP3;3, Ala (A) is replaced with Ser (S) in the first NPA motif and Val (V) in second NPA motif. Similar features were observed in the Arabidopsis NIP5;1 and NIP6;1 (Miwa et al. 2010). With the exception of the LE2 position, plant NIPs are highly variable in the ar/R selectivity filter. The five ThNIPs showed the expected Trp (W)/Gly (G)/Ala (A)/Thr (T) residues in the H2 position with the exception of ThNIP3;1, which had Ser (S). The H5 position is conserved and was either Ile (I) or Val (V). ThNIP3;2 and ThNIP3;3 had Gly (G) instead of Ala (A) in LE1 position, similar to the Arabidopsis NIP7 and potato NIP7;1 (Venkatesh et al. 2013). In Froger’s positions, all the ThNIPs showed the expected amino acids. For example, Ala (A) and Tyr (Y) were in the P3 and P4 positions and Leu (L) or Ile (I) in the P5. Plant SIPs show a highly variable first NPA motif with the Ala (A) residue replaced by Leu (L), Ala (A), Thr (T), or Cys (C), but the second NPA motif is conserved. However, ThSIP1;1 showed variation in the first NPA motif, in which Ala (A) is replaced by Thr (T), and in the second NPA motif, in which Ala (A) is replaced by Thr Val (V). All ar/R and Froger’s positions (P1 and P2) are highly variable. In contrast, highly conserved Ala (A), Tyr (Y) and Trp (W) residues were observed in the P3, P4, and P5 positions.
Relative abundance of the 18 AQP genes in the roots, stems, and leaves of T. hispida
The expression profiling of AQPs in the roots, stems, and leaves of T. hispida under normal growth conditions was determined by the real-time RT-PCR analysis. The geometric transcript levels of the three internal control genes were used as a calibrator (its expression was set as 1) to normalize the expression levels of AQP genes. The results showed that the level of expression varied greatly from gene to gene in each tissue (Fig. 4). ThPIP1;1 and ThPIP2;1 were highly abundant in root, stem, and leaf tissues. ThPIP2;3, ThPIP2;4, ThTIP1;1, ThTIP1;2, ThTIP2;1, and ThTIP4;1 were relatively highly expressed in leaves compared with root and stem tissues, especially ThTIP1;1. The remaining genes were expressed at lower levels in root, stem, and leaf tissues.
Expression patterns of the AQP genes under different abiotic stresses
First, we analyzed the expression of these AQP genes in response to NaHCO3 by comparing RPKM (reads per kilobase of exon model per million mapped reads) values obtained from transcriptomes (Mortazavi et al. 2008). The results showed that most of these AQP genes were differentially regulated by NaHCO3 stress and displayed different expression patterns in response to NaHCO3 (Table S3). We further studied the expression profile of each ThAQP in the roots, stems, and leaves of T. hispida under different abiotic stresses, including high salinity, PEG-simulated drought and heavy metal stresses. Two-month-old uniform seedlings were subjected to the indicated treatments, and the phenotype was observed, as described previously (Wang et al. 2014a). The expression patterns are shown in the following figures.
Under salt stress, the expression of ThPIP2;4, ThPIP1;1, ThPIP2;2, ThPIP2;5, ThTIP1;2, ThTIP2;1, ThTIP4;1, ThNIP1;2, ThNIP3;1, ThNIP3;3, and ThSIP1;1 displayed similar expression patterns in the roots of the plants (Fig. 5a). They were all rapidly downregulated after 3 h of stress, gradually increased to their peak expression level at 9 h, and then decreased during the subsequent time points. The remaining genes were downregulated at 3 h, ThPIP2;3, ThTIP1;2, and ThTIP1;3 were upregulated during the 6 to 24 h time period, but ThTIP1;1 was downregulated, and the others did not markedly change during salt stress. In the stems and leaves, the members of the ThPIP, ThTIP, ThNIP, and ThSIP subfamilies displayed approximately similar expression patterns (Fig. 5b, c). ThPIP members were upregulated with the exception of several individual time points, especially the ThPIP2;5 gene. The expression of ThTIP1;2 was markedly upregulated, and ThTIP1;1 was noticeably downregulated during salt stress. The remaining genes did not significantly change. However, the members of the ThNIP subfamily were upregulated under salt stress. The ThSIP genes were up- or downregulated in response to salt stress but did not markedly change.
Expression profiles of the aquaporin genes in the roots, stems, and leaves of 2-month-old T. hispida seedlings subjected to salt stress (400 mM NaCl). Relative expression level = log2 (transcription level under stress treatment/transcription level under control conditions). The error bars indicate standard deviations, which were calculated from multiple replicates of real-time PCR
Under PEG-simulated drought stress conditions, all of the ThAQPs, with the exception of ThPIP2;5, ThTIP1;2 and ThTIP1;3, were rapidly downregulated in the roots of plants at 3 h, gradually increased to their peak expression levels at a later time point but were then downregulated (Fig. 6a). The expression of ThPIP2;5 was upregulated (nearly sixfold induced) after 6 and 9 h of stress, and ThTIP1;2 and ThTIP1;3 were upregulated in the roots during the 6 to 12 h time points. In the stems, most of ThAQPs were up- or downregulated but did not markedly change during the stress (Fig. 6b). However, ThPIP2;5 was rapidly upregulated and increased sixfold at 3 h and recovered its relatively low expression at subsequent time points. The expressions of ThTIP1;2 and ThTIP1;3 were markedly upregulated during the drought stress. In contrast, ThPIP2;2, ThTIP2;1, and ThSIP1;2 expressions were downregulated by approximately twofold. In the leaves, ThPIP2;2, ThPIP2;3, ThPIP2;4, ThTIP2;1, ThNIP1;1, and ThNIP1;2 displayed similar expression patterns (Fig. 6c). They were all initially downregulated after 3 h of stress, gradually decreased to the lowest expression levels at a later time point (6, 9, or 12 h), but they were all upregulated at 24 h of stress. ThPIP2;1, ThTIP1;1, ThNIP3;1, ThNIP3;3, and ThSIP1;2 were downregulated during the drought stress, while ThPIP2;5, ThTIP1;2, and ThTIP1;3 were upregulated under drought stress, and respectively reached their peak levels at 3 or 24 h (nearly fivefold induced).
Expression profiles of the aquaporin genes in the roots, stems, and leaves of 2-month-old T. hispida seedlings subjected to PEG-simulated drought stress (25% PEG6000) treatment. Relative expression level = log2 (transcription level under stress treatment/transcription level under control conditions). The error bars indicate standard deviations, which were calculated from multiple replicates of real-time PCR
Heavy metals, such as cadmium, copper, lead, chromium, and mercury are major environmental pollutants and affect various physiological and biochemical processes in plants (Nagajyoti et al. 2010; Villiers et al. 2011; Datta et al. 2011; Shen et al. 2011; Choudhary et al. 2012). In this study, the expression of ThAQPs in response to heavy metal (cadmium) stress was monitored. The results showed that these ThAQP genes can roughly be divided into two groups according to their expression patterns (Fig. 7a). The first group included the six members of ThPIP subfamily, ThTIP1;2, ThNIP1;3, and ThNIP3;3, whose expression was initially downregulated after 3 h of stress, gradually increased to their peak expression level at a later stress time point (6, 9 or 12 h), and then rapidly decreased at 24 h of stress. The second group included the ThTIP1;2, ThTIP2;1, ThTIP4;1, ThNIP3;1, ThSIP1;1, and ThSIP1;2 genes that were downregulated during the heavy metal stress. In addition, ThNIP3;2 and ThNIP1;1 expressions were initially downregulated at 3 h and upregulated at subsequent time points. In the stems, with the exception of ThPIP2;5, the members of ThPIPs did not noticeably change during the heavy metal stress. ThPIP2;5 expression rapidly increased to the peak level after 3 h of stress and recovered relatively low expression levels at subsequent time points (Fig. 7b). The ThPIPs were all downregulated during the heavy metal stress, especially ThTIP1;2. The expressions of ThNIP1;1, ThNIP1;2, and ThNIP3;2 were upregulated, but the ThSIPs were downregulated under the stress. In the leaves, with the exception of ThPIP2;1, the members of ThPIPs were upregulated during salt stress (Fig. 7c). However, ThPIP2;4 was downregulated at 24 h of stress and decreased more than twofold. ThPIP2;1 expression was downregulated after 3 h of stress and gradually increased at the subsequent time points. The expressions of ThTIP1;2, ThTIP1;3, and ThTIP2;1 were downregulated in response to the heavy metal stress; notably, ThTIP1;2 was markedly decreased fourfold to sixfold in the leaves. The ThNIPs were significantly up- or downregulated (> twofold) at individual time points and did not significantly change during the stress. ThSIP1;1 was noticeably downregulated during 3 to 24 h.
Expression profiles of the aquaporin genes in the roots, stems, and leaves of 2-month-old T. hispida seedlings subjected to heavy metal stress (150 μM CdCl2). Relative expression level = log2 (transcription level under stress treatment/transcription level under control conditions). The error bars indicate standard deviations, which were calculated from multiple replicates of real-time PCR
Plants sense various signals from the surrounding environment and adjust their water transport properties accordingly (Li et al. 2014). Recent studies have shown that a large number of AQP genes were induced by different abiotic stresses, such as salt, drought, and cold. Furthermore, overexpression of some AQP genes significantly increased the abiotic stress tolerance in transgenic plants (Matsumoto et al. 2009; Hu et al. 2012; Zhou et al. 2012; Ahamed et al. 2012; Reuscher et al. 2013; Rodrigues et al. 2013; Sreedharan et al. 2013; Wang et al. 2014b). In this study, most of the AQP genes are differentially regulated by salt, drought, or heavy metal stress, indicating that they may play important roles in response to abiotic stress. For example, the ThPIP2;2, ThPIP2;5, ThTIP1;1, ThTIP1;2, and ThTIP1;3 genes are significantly induced by NaCl and PEG stresses, indicating that they are involved in salt and drought stress tolerance. BLASTX analysis revealed that ThPIP2;2 and ThPIP2;5 had high sequence identity with TaTIP2;2 from Triticum aestivum and PIP2-5 from Prunus mum, and ThTIP1;1, ThTIP1;2, and ThTIP1;3 had high sequence identity with TIP1–1 from Nelumbo nucifera and TIP1–3 from Eucalyptus grandis. The wheat TaAQP7 was very close to the PIP2 subfamily and was induced by PEG and ABA stresses. Overexpression of TaAQP7 in tobacco increased drought tolerance by enhancing the antioxidant system, increasing the ability to retain water, and reducing ROS accumulation and membrane damage (Zhou et al. 2012). The TaTIP2;2 gene is downregulated by salinity and drought stress. Heterologous expression of the wheat AQP gene, TaTIP2;2, compromises the drought and salinity tolerance of transgenic Arabidopsis via an ABA-independent pathway (Xu et al. 2013). The Thellungiella salsuginea TsTIP1;2 gene is expressed highly in the leaves and is induced by several abiotic and hormonal stresses. Ectopic overexpression of TsTIP1;2 significantly increases tolerance to drought, salt, and oxidative stresses in Arabidopsis (Wang et al. 2014b). These results will be useful for elucidating the functions of same ThAQP genes under different environment stresses.
Effect of abscisic acid on the expression of the AQP genes in T. hispida
Abscisic acid (ABA) is an important plant hormone that has crucial roles in root and seed development, seed germination, and biotic and abiotic stress responses. ABA interacts with other phytohormones to mediate plant response to stress conditions and modulates expression of many genes at the transcriptional and post-transcriptional levels (Finkelstein et al. 2002; Cutler et al. 2010). Therefore, we measured the expression level of each ThAQP gene in T. hispida seedlings treated with ABA. In the roots, the expression patterns of ThAQPs can be roughly divided into three groups (Fig. 8a). The first group, including the ThPIP2;2, ThPIP2;3, ThTIP2;1, ThTIP4;1, ThNIP1;1, ThNIP1;2, ThNIP3;3, ThSIP1;1, and ThSIP1;2 genes, was downregulated during the ABA treatment, especially after 3 or 24 h of stress. The second group included ThPIP2;4, ThPIP2;5, ThTIP1;2, ThTIP1;3, and ThNIP3;1, whose expression was initially downregulated at 3 h and then gradually increased to their peak expression level at 6 or 9 h and finally decreased again at 24 h. The third group included the ThPIP1;1, ThPIP2;1, and ThNIP3;2 genes that were rapidly decreased after 3 h of ABA treatment and then recovered normal levels. In the stems, the ThPIPs, ThTIP2;1, ThNIP1;2, ThNIP3;1, ThNIP3;3, and ThSIP1;2 were initially downregulated at 3 h and gradually increased to their peak expression level at 9 or 12 h and then rapidly increased (> twofold) at 24 h (Fig. 8b). ThPIP2;3, ThTIP1;3, and ThSIP1;2 were initially upregulated and decreased to their lowest levels at 12 h, then rapidly increased to peak levels at 24 h. However, ThPIP2;5 and ThTIP1;2 were upregulated during the ABA treatment. In the leaves, all of the ThAQPs, with the exception of ThPIP2;5, ThTIP1;1, ThNIP3;3, and ThSIP1;2, did not significantly change during the ABA treatment (Fig. 8c). ThPIP2;5 and ThTIP1;2 were rapidly increased and had high expression levels after 3 h of treatment; ThPIP2;5 gradually decreased at subsequent time points, but ThTIP1;2 maintained high levels. In contrast, ThPIP2;5 and ThTIP1;1 showed the opposite pattern. ThNIP3;3 and ThSIP1;2 were downregulated under ABA treatment. Overall, some AQP genes, such as ThPIP2;5 and ThNIP3;3, were significantly induced and suppressed in different tissues by ABA treatment, indicating that these AQP genes participate or might play roles in the ABA-dependent pathways. Previous studies described the effects of exogenous ABA on AQP gene expression in different plant species. Following ABA application, TsTIP1;2 expression was induced, with the highest levels occurring at 1–3 h (Wang et al. 2014b). The TaNIP expression was induced by NaCl, drought, and ABA treatments, overexpression of TaNIP in transgenic Arabidopsis enhanced higher salt tolerance than wild-type (WT) plants (Gao et al. 2010). Lian et al. reported that the expression of OsPIP1;2, OsPIP2;5, and OsPIP2;6 in roots and OsPIP1;2, OsPIP2;4, and OsPIP2;6 in leaves of upland rice was enhanced under ABA treatment. Jang et al. also investigated the expressions of 13 Arabidopsis PIPs under ABA treatment. However, the responsiveness of PIP genes to ABA was different, implying that the regulation of PIP genes involves both ABA-dependent and ABA-independent signaling pathways during water deficit (Jang et al. 2004; Lian et al. 2006).
Effect of ABA on the expression of the aquaporin genes in the roots, stems, and leaves of the 2-month-old T. hispida seedlings. Relative expression level = log2 (transcription level under stress treatment/transcription level under control conditions). The error bars indicate standard deviations, which were calculated from multiple replicates of real-time PCR
Conclusion
This study presents a comprehensive analysis of AQP genes in T. hispida, including their sequence, structural properties, phylogeny, and expression patterns. Figure 9 illustrates the involvement of the ThAQP genes in different stress-related physiological processes. All the ThAQP genes can be differentially regulated by NaCl, PEG, CdCl2, or ABA stress in the leaves, stems, or roots of T. hispida, especially in the roots. Roots are the primary uptake organ for different salt ions. It is conceivable that AQPs play an important role in response to abiotic stress. ThPIP2;2, ThPIP2;5, ThTIP1;1, ThTIP1;2, ThTIP1;3, and ThSIP1;1 were highly induced in response to high salinity, drought, heavy metal stresses, and ABA treatment, indicating that they may play roles in response to abiotic stress and were closely regulated by the ABA-dependent stress-signaling pathway. These results will provide further insight into the roles of AQP genes in abiotic stress tolerance in plants.
Involvement of the aquaporin genes in T. hispida response to abiotic stresses and ABA treatment. The aquaporin genes are grouped based on their stress responses. Arrows indicate the genes that are upregulated or downregulated by at least twofold with the indicated treatments in the roots, stems, and leaves of the T. hispida plants. P ThPIP, T ThTIP, N ThNIP, S ThSIP
Abbreviations
- AQP:
-
Aquaporin
- ABA:
-
Abscisic acid
- NJ:
-
Neighbor-Joining
- ORF:
-
Open reading frame
- RPKM:
-
Reads per kilobase exon model per million mapped reads
- QRT-PCR:
-
Quantitative reverse transcription polymerase chain reaction
- PIPs:
-
Plasma membrane intrinsic proteins
- TIPs:
-
Tonoplast membrane intrinsic proteins
- NIPs:
-
NOD26-like intrinsic proteins
- SIPs:
-
Small basic intrinsic proteins
References
Ahamed A, Murai-Hatano M, Ishikawa-Sakurai J, Hayashi H, Kawamura Y, Uemura M (2012) Cold stress-induced acclimation in rice is mediated by root-specific aquaporins. Plant Cell Physiol 53(8):1445–1456
Azad AK, Katsuhara M, Sawa Y, Ishikawa T, Shibata H (2008) Characterization of four plasma membrane aquaporins in tulip petals: a putative homolog is regulated by phosphorylation. Plant Cell Physiol 49(8):1196–1208
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208
Bienert GP, Schjoerring JK, Jahn TP (2006) Membrane transport of hydrogen peroxide. BBA-Biomembranes 1758(8):994–1003
Bots M, Vergeldt F, Wolters-Arts M, Weterings K, van As H, Mariani C (2005) Aquaporins of the PIP2 class are required for efficient anther dehiscence in tobacco. Plant Physiol 137(3):1049–1056
Chaumont F, Barrieu F, Wojcik E, Maarten J, Chrispeels MJ, Jung R (2001) Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol 125(3):1206–1215
Choudhary SP, Kanwar M, Bhardwaj R, Yu JQ, Tran LSP (2012) Chromium stress mitigation by polyamine-brassinosteroid application involves phytohormonal and physiological strategies in Raphanus sativus L. PLoS One 7:e33210
Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Ann Rev Plant Biol 61:651–679
Datta JK, Bandhyopadhyay A, Banerjee A, Mondal NK (2011) Phytotoxic effect of chromium on the germination, seedling growth of some wheat (Triticum aestivum L.) cultivars under laboratory condition. J Agr Sci Tech 7(2):395–402
Finkelstein RR, Gampala SSL, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14(suppl 1):S15–S45
Gao Z, He X, Zhao B, Zhou C, Liang Y, Ge R, Shen Y, Huang Z (2010) Overexpressing a putative aquaporin gene from wheat, TaNIP, enhances salt tolerance in transgenic Arabidopsis. Plant Cell Physiol 51(5):767–775
Gupta AB, Sankararamakrishnan R (2009) Genome-wide analysis of major intrinsic proteins in the tree plant Populus trichocarpa: characterization of XIP subfamily of aquaporins from evolutionary perspective. BMC Plant Biol 9(1):134
Gupta AB, Verma RK, Agarwal V, Vajpai M, Bansal V, Sankararamakrishnan R (2012) MIPModDB: a central resource for the superfamily of major intrinsic proteins. Nucleic Acids Res 40(D1):D362–D369
Heinen RB, Ye Q, Chaumont F (2009) Role of aquaporins in leaf physiology. J Exp Bot 60(11):2971–2985
Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35(suppl 2):W585–W587
Hu W, Yuan Q, Wang Y, Cai R, Deng X, Wang J, Zhou S, Chen M, Chen L, Huang C, Ma Z, Yang G, He G (2012) Overexpression of a wheat aquaporin gene, TaAQP8, enhances salt stress tolerance in transgenic tobacco. Plant Cell Physiol 53(12):2127–2141
Jang JY, Kim DG, Kim YO, Kim JS, Kang H (2004) An expression analysis of a gene family encoding plasma membrane aquaporins in response to abiotic stresses in Arabidopsis thaliana. Plant Mol Biol 54(5):713–725
Johanson U, Karlsson M, Johansson I, Gustavsson S, Sjövall S, Fraysse L, Weig AR, Kjellbom P (2001) The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol 126(4):1358–1369
Kaldenhoff R, Fischer M (2006) Functional aquaporin diversity in plants. BBA-Biomembranes 1758(8):1134–1141
Krogh A, Larsson BÈ, Von Heijne G, Sonnhammer E (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305(3):567–580
Li G, Santoni V, Maurel C (2014) Plant aquaporins: roles in plant physiology. BBA-Gen Subjects 1840(5):1574–1582
Li RY, Ago Y, Liu WJ, Mitani N, Feldmann J, McGrath PS, Ma JF, Zhao FJ (2009) The rice aquaporin Lsi1 mediates uptake of methylated arsenic species. Plant Physiol 150(4):2071–2080
Lian HL, Yu X, Lane D, Sun WN, Tang ZC, Su WA (2006) Upland rice and lowland rice exhibited different PIP expression under water deficit and ABA treatment. Cell Res 16(7):651–660
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25(4):402–408
Liu C, Fukumoto T, Matsumoto T, Gena P, Frascaria D, Kaneko T, Katsuhara M, Zhong S, Sun X, Zhu Y, Iwasaki I, Ding X, Calamita G, Kitagawa Y (2013) Aquaporin OsPIP1; 1 promotes rice salt resistance and seed germination. Plant Physiol Bioch 63:151–158
Matsumoto T, Lian HL, Su WA, Tanaka D, Liu C, Iwasaki I, Kitagawa Y (2009) Role of the aquaporin PIP1 subfamily in the chilling tolerance of rice. Plant Cell Physiol 50(2):216–229
Maurel C, Verdoucq L, Luu DT, Santoni V (2008) Plant aquaporins: membrane channels with multiple integrated functions. Annu Rev Plant Biol 59:595–624
Mitani-Ueno N, Yamaji N, Zhao FJ, Ma JF (2011) The aromatic/arginine selectivity filter of NIP aquaporins plays a critical role in substrate selectivity for silicon, boron, and arsenic. J Exp Bot 62(12):4391–4398
Miwa K, Tanaka M, Kamiya T, Fujiwara T (2010) Molecular mechanisms of boron transport in plants: involvement of Arabidopsis NIP5; 1 and NIP6; 1[M]//Mips and their role in the exchange of metalloids. Springer New York 83–96
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNASeq. Nat Methods 5(7):621–628
Mut P, Bustamante C, Martinez G, Alleva K, Sutka M, Civello M, Amodeo G (2008) A fruit-specific plasma membrane aquaporin subtype PIP1; 1 is regulated during strawberry (Fragaria × ananassa) fruit ripening. Physiol Plantarum 132(4):538–551
Nagajyoti PC, Lee KD, Sreekanth TVM (2010) Heavy metals, occurrence and toxicity for plants: a review. Environ Chem Lett 8(3):199–216
Nguyen MX, Moon S, Jung KH (2013) Genome-wide expression analysis of rice aquaporin genes and development of a functional gene network mediated by aquaporin expression in roots. Planta 238(4):669–681
Okubo-Kurihara E, Sano T, Higaki T, Kutsuna N, Hasezawa S (2009) Acceleration of vacuolar regeneration and cell growth by overexpression of an aquaporin NtTIP1; 1 in tobacco BY-2 cells. Plant Cell Physiol 50(1):151–160
Park W, Scheffler BE, Bauer PJ, Campbell BT (2010) Identification of the family of aquaporin genes and their expression in upland cotton (Gossypium hirsutum L.) BMC Plant Biol 10(1):142
Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, Lee Y, White J, Cheung F, Parvizi B (2003) TIGR Gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19(5):651–652
Rasheed-Depardieu C, Parent C, Crèvecoeur M, Parelle J, Tatin-Froux F, Provost GL, Capelli N (2012) Identification and expression of nine oak aquaporin genes in the primary root axis of two oak species, Quercus petraea and Quercus robur. PLoS One 7(12):e51838
Reuscher S, Akiyama M, Mori C, Aoki K, Shibata D, Shiratake K (2013) Genome-wide identification and expression analysis of aquaporins in tomato. PLoS One 8(11):e79052
Rodrigues MI, Bravo JP, Sassaki FT, Severino FE, Maia IG (2013) The tonoplast intrinsic aquaporin (TIP) subfamily of Eucalyptus grandis: characterization of EgTIP2, a root-specific and osmotic stress-responsive gene. Plant Sci 213:106–113
Sakurai J, Ishikawa F, Yamaguchi T, Uemura M, Maeshima M (2005) Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol 46(9):1568–1577
Shen QI, Jiang M, Li HUA, Che LL, Yang ZM (2011) Expression of a Brassica napus heme oxygenase confers plant tolerance to mercury toxicity. Plant Cell Environ 34(5):752–763
Sreedharan S, Shekhawat UKS, Ganapathi TR (2013) Transgenic banana plants overexpressing a native plasma membrane aquaporin MusaPIP1; 2 display high tolerance levels to different abiotic stresses. Plant Biotechnol J 11(8):942–952
Tajkhorshid E, Nollert P, Jensen MO, Miercke LJ, O’Connell J, Stroud RM, Schulten K (2002) Control of the selectivity of the aquaporin water channel family by global orientational tuning. Science 296(5567):525–530
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Törnroth-Horsefield S, Wang Y, Hedfalk K, Johanson U, Karlsson M, Tajkhorshid E, Neutze R, Kjellbom P (2006) Structural mechanism of plant aquaporin gating. Nature 439(7077):688–694
Uehlein N, Kaldenhoff R (2008) Aquaporins and plant leaf movements. Ann Bot-London 101(1):1–4
Vandesompele J, De Preter K, Pattyn F, Poppe B, Roy NV, Paepe AD, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7):research0034
Venkatesh J, Yu JW, Park SW (2013) Genome-wide analysis and expression profiling of the Solanum tuberosum aquaporins. Plant Physiol Bioch 73:392–404
Villiers F, Ducruix C, Hugouvieux V, Jarno N, Ezan E, Garin J (2011) Investigating the plant response to cadmium exposure by proteomic and metabolomic approaches. Proteomics 11(9):1650–1663
Wang L, Wang C, Wang D, Wang Y (2014a) Molecular characterization and transcript profiling of NAC genes in response to abiotic stress in Tamarix hispida. Tree Genet Genomes 10(1):157–171
Wang LL, Chen AP, Zhong NQ, Liu N, Wu XM, Wang F, Yang CL, Romero MF, Xia GX (2014b) The Thellungiella salsuginea tonoplast aquaporin TsTIP1; 2 functions in protection against multiple abiotic stresses. Plant Cell Physiol 55(1):148–161
Whiteman SA, Nühse TS, Ashford DA, Sanders D, Maathuis FJM (2008) A proteomic and phosphoproteomic analysis of Oryza sativa plasma membrane and vacuolar membrane. Plant J 56(1):146–156
Xu C, Wang M, Zhou L, Qua T, Xia G (2013) Heterologous expression of the wheat aquaporin gene TaTIP2;2 compromises the abiotic stress tolerance of Arabidopsis thaliana. PLoS One 8(11):e79618
Zardoya R (2005) Phylogeny and evolution of the major intrinsic protein family. Biol Cell 97(6):397–414
Zhou S, Hu W, Deng X, Ma Z, Chen L, Huang C, Wang C, Wang J, He Y, Yang G, He G (2012) Overexpression of the wheat aquaporin gene, TaAQP7, enhances drought tolerance in transgenic tobacco. PLoS One 7(12):e52439
Acknowledgments
The work was financially supported by National Natural Science Foundation of China (No. 31300571) and the Fundamental Research Funds for the Central Universities (2572014CA12). We also thank the editor and reviewers for their critical comments and thoughtful suggestions.
Data archiving statement
All the identified AQP genes were deposited with GenBank with the GenBank accession numbers: ThPIP1;1 (KT951251); ThPIP2;1 (ACB71281); ThPIP2;2 (KT951253); ThPIP2;3 (KT951254); ThPIP2;4 (KT951250); ThTIP1;1 (KT951258); ThTIP1;2 (ACB71282); ThTIP1;3 (KT951255); ThTIP2;1 (KT951256); ThTIP4;1 (KT951257); ThNIP3;1 (KT951260); ThNIP3;2 (KT951262); ThNIP3;3 (KT951261); ThPIP2;5 (KT951252); ThNIP1;1 (KT951263); ThNIP1;2 (KT951259); ThSIP1;1 (KT951264); and ThSIP2;2 (KT951265).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by C. Dardick
Rights and permissions
About this article
Cite this article
Wang, C., Wang, L., Yang, C. et al. Identification, phylogeny, and transcript profiling of aquaporin genes in response to abiotic stress in Tamarix hispida . Tree Genetics & Genomes 13, 81 (2017). https://doi.org/10.1007/s11295-017-1163-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-017-1163-7