Abstract
The transition to flowering is a major developmental switch in flowering plants. The nuclear RNA-binding protein FCA responds to seasonal signals and abscisic acid (ABA), which can control the flowering time via ambient temperature and autonomous pathways. Citrus FCA ortholog (PtFCA) has been isolated and characterized from precocious trifoliate orange (Poncirus trifoliata L. Raf). Three alternatively spliced transcripts of PtFCA (PtFCA1, PtFCA2, and PtFCA3) were isolated. The expression pattern of PtFCA indicated that it may be involved in phase transition in precocious trifoliate orange. A functional complementation experiment of PtFCA indicated that PtFCA1 partially rescued the late-flowering phenotype of the fca-1 mutant. There was no influence on flowering time of transgenic Arabidopsis by PtFCA3 as compared with PtFCA2, which exhibits delayed flowering time in a fca-1 background. Meanwhile, these three transcripts also showed different abilities to regulate root development in the fca-1 background. The study of protein–protein interactions suggested that PtFCA may form higher order complexes with PtFY and PtNF-YA7 to regulate timing of the transition from the vegetative to reproductive phase in precocious trifoliate orange. ABA and ambient temperature treatments changed the expression of PtFCA and interaction protein. These findings reveal that PtFCA may play important roles in flowering time and root development of precocious trifoliate orange through the formation of multiple protein complexes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The major developmental transition in flowering plants is the switch from vegetative to reproductive development (Srikanth and Schmid 2011). Previous studies reported that flowering time is regulated principally by internal genetic factors and environmental signals, such as temperature, light intensity, day length, gibberellins, aging by sequentially operating miRNAs, and body size (Amasino and Michaels 2010; Jaeger et al. 2006). So far, the autonomous, gibberellin, photoperiod, vernalization, and aging pathways have been shown to regulate flowering in Arabidopsis, and several genes related to the five response networks have been identified by molecular analysis of Arabidopsis mutants (Amasino and Michaels 2010; Jaeger et al. 2006; Khan et al. 2014; Srikanth and Schmid 2011). The autonomous pathway refers to endogenous regulators that are independent of the photoperiod and gibberellin pathways (Srikanth and Schmid 2011). However, the genes of the autonomous pathway are involved in mediating the effects of the ambient temperature and circadian clock pathways (Abou-Elwafa et al. 2011; Blazquez et al. 2003). The autonomous pathway includes FCA, FY, LUMINIDEPENDENS (LD), FPA, FLOWERING LOCUS D (FLD), FLOWERING LOCUS VE (FVE), and RELATIVE OF EARLY FLOWERING 6 (Khan et al. 2014; Liu et al. 2007). Decades of studying Arabidopsis have revealed that FCA and FVE are involved in the perception of ambient temperatures. In contrast to Arabidopsis, many perennial plants with different growth forms show a diversity in flowering responses that are not easily explained based solely on studies of flowering in Arabidopsis, thus illustrating the value of model tree systems (Mouradov et al. 2002; Amasino and Michaels 2010; Khan et al. 2014; Lee et al. 2005).
FCA protein consists of two RNA recognition motifs (RRMs) and a protein interactor WW domain (Macknight 1997). In Arabidopsis, FCA is involved in controlling flowering time and plays more general roles in RNA-mediated chromatin silencing (Jang et al. 2009b). This process is conserved and diverse in Arabidopsis (dicot) and rice (monocot), and it occurs through alternative splicing and alternative polyadenylation resulting in different transcripts. Only one transcript forms a functional nuclear protein that can promote flowering in Arabidopsis, whereas alternative processing of OsFCA transcript is more complex than its Arabidopsis counterpart (Jang et al. 2009a; Kumar et al. 2011; Lee et al. 2005; Macknight 2002). FCA protein was found to interact with various proteins having different functional domains, although several of the interactors contain XPXPP motif. For example, FCA protein interacts physically and genetically with the RNA 3′-end processing factor FY containing two PPLP motifs via its WW domain (Simpson et al. 2003). This interaction is required both for correct processing of transcripts derived from FCA itself and for the down-regulation of transcript (directly or indirectly) at the expense of full-length FCA mRNA, thus limiting the expression of active FCA in Arabidopsis and rice (Jang et al. 2009b; Sarnowski et al. 2002; Simpson et al. 2003). FCA-RRM domains are known to mediate RNA-binding functions in numerous proteins. Many of the reports on FCA-RRMs were related to their roles in floral development (Liu and Cai 2013; Quesada 2003; Sun et al. 2011). Indeed, FCA research has provided hints about additional role(s) in the induction of dormancy and the germination process and its effects on abscisic acid (ABA)-responsive promoters of Em and VP1 (Kumar et al. 2011; Ruttink et al. 2007). In addition, a recent study indicated that FCA also binds to pri-miR172 transcripts and promotes their processing in response to ambient temperature changes (Jung et al. 2012). However, most information about FCA functions and expression regulation has come from studies of model annual plants (typically Arabidopsis), with relatively few reports regarding woody plants.
Citrus is one of the most economically important evergreen fruit crops for the production of fresh fruit and juice (Tan and Swain 2007). However, the phase transition in most citrus plants requires 4–8 years and is much longer for some species, such as seedling oranges and grapefruit, which require 8–10 years to flowering (Davenport 1990). Precocious trifoliate orange was derived from trifoliate orange (Poncirus trifoliata L. Raf.); about 20 % of the seedlings from precocious trifoliate orange seeds first flowered in the next year after the spring planting; the juvenile period of precocious trifoliate orange has been greatly reduced to 1–2 years as compared with other citrus plants which has a juvenile period of 6 to 8 years. Therefore, precocious trifoliate orange provides good material for studying the molecular mechanism of the seasonal response and flowering in citrus plants (Zhang et al. 2011a). For most citrus plants, there are three important types of shoots produced during the growing season: spring shoots, summer shoots, and autumn shoots, with spring shoots being the most important for flower formation and flowering (Liang et al. 1999; Zhang et al. 2014). All three types of citrus shoots typically cease growth temporarily by abortion of the shoot tips. This physiological phenomenon is called “self-pruning.” Previous cytological studies revealed that the floral buds of spring shoots in trifoliate orange initiated differentiation immediately after self-pruning (Zhang et al. 2014). Therefore, self-pruning appears to be a demarcation point for the meristem to initiate leaf bud or floral bud development (Li et al. 2010; Zhang et al. 2014). In addition, flower induction and flowering of citrus plants are also regulated by temperature in subtropical regions. Previous studies reported that citrus trees continue vegetative growth without producing flower buds at temperatures above 25 °C, whereas the trees produce flower buds at temperatures below 25 °C (Nishikawa et al. 2007). This phenomenon indicated that cool ambient temperatures may play an important role in floral induction of citrus plants.
Based on these observations, it appears that the genes of the autonomous and thermosensory flowering pathway may be involved in the unique flowering behaviors of citrus plants. Therefore, we have isolated and characterized a homolog to the Arabidopsis FCA gene from precocious trifoliate orange to study the molecular mechanism of the seasonal signal response and flower formation in perennial woody plants.
Materials and methods
Plant material and growth conditions
The samples of precocious trifoliate orange were collected from the experimental fields of the National Citrus Breeding Center at Huazhong Agricultural University (30° 35′ N, 114° 17′ E). Seeds were planted in 20-cm pots containing commercial medium (PeiLei, China) for 2 months in a greenhouse maintained at 25 °C with 16-h light/8-h dark photoperiod. Seedlings were then transplanted and grown under natural environmental conditions.
To investigate the expression of PtFCA in spring shoots, the terminal bud and the following five buds (the major node positions for flower formation) from the spring shoots of adult trees (7–8 years old and had flowered several times) were collected at three distinct phases (15 days before self-pruning, beginning of self-pruning, and 20 days after self-pruning). To examine different developmental stages, juvenile samples from shoot apical meristems of 6-month-old trees, phase transition samples from 12-month-old trees, and adult samples from the terminal bud and the following five buds from 7-year-old trees after self-pruning of spring shoots were collected. To analyze the spatial expression, roots, shoots, leaves, flowers at full bloom, and the whole fruits at 1 month after flowering were collected.
To examine the effects of ambient temperatures on gene expression, 3-month-old seedlings were grown at either 23 or 27 °C with a photoperiod of 16-h light/8-h dark for 2 weeks. Leaves were collected for further gene expression analysis. To examine the effects of ABA on gene expression, 15 μmol/L ABA was sprayed on the spring shoots of adult trees from shoot sprouting to flowering at 5 p.m. every day in the field. Shoots were collected according to the developmental stages. At the sprouting stage, the entire spring shoots were collected when they had reached a length of 2–3 cm. For the phase transition stage, the terminal bud and the following five buds of spring shoots were collected during self-pruning. In this study, all plant tissues were sampled according to the requirement of each experiment from the three groups of trees (with each group containing three trees) used as biological repeats, immediately frozen in liquid nitrogen, and stored at −80 °C until further analysis.
Gene isolation and sequence analysis
The primers were designed for reverse transcription (RT)-PCR amplification (Table S1). PCR amplification was conducted in 25 μL reactions containing 50 ng of cDNA template, 2.5 μL 10 × PCR buffer, 0.5 mM each primer, 0.5 U LA Taq DNA polymerase (TaKaRa, Japan), and 2.5 mM dNTPs. RT-PCR was performed with the following protocol: denaturing at 95 °C for 5 min; 36 cycles of annealing at 95 °C for 35 s, 52 °C for 35 s, and 72 °C for 2 min; and extension at 72 °C for 10 min. The PCR product was cloned into pMD18-T vector (TaKaRa, Japan) and sequenced (BGI Tech, China). Sequence analysis was performed by the National Center for Biotechnology Information (NCBI) and GeneScan (http://genescan.info/). The sequence alignment was carried out using ClustalX (ver. 2.1), and the phylogenetic tree was constructed by the neighbor-joining method using MEGA 4.0 (Tamura et al. 2007). Bootstrap values were derived from 1000 replicate runs. The amino acid sequence of PtAGL24 was aligned with homologous protein sequences from various plants through BLASTN. All the sequences were downloaded from the NCBI database.
Gene expression analysis by real-time PCR
Real-time PCR was applied to evaluate transcription levels of PtFCA by using the SYBR Green PCR master mix (Roche Applied Science, Germany). Total RNA was extracted by using Trizol reagent (TaKaRa) according to the manufacturer’s instructions. About 1 μg total RNA after treatment with RNase-free DNase I was synthesized into cDNA with the Reverse First Strand cDNA Synthesis Kit (Invitrogen, USA). Real-time PCR was conducted on the LightCyclerW480 Detection System (Roche Applied Science) as described previously (Zhang et al. 2009). Real-time PCR was normalized with the results of β-actin.
Three biological replicates and four technical replicates were assayed in this study, and all biological replicates showed similar trends. Data from one biological replicate are presented. The data were processed using one-way analysis of variance (ANOVA), and statistical differences were compared based on Student’s t test, taking P < 0.01 as significant.
Subcellular localization analysis
For localization studies in onion epidermal cells, the open reading frame (ORF) of PtFCA without the terminator codon of different transcripts was constructed to the pCAMBIA1302 vector fused to green fluorescent protein (GFP) by using restriction enzymes. The PtFCA-GFP expression vector was precipitated on gold particles and bombarded onto the onion epidermal layer tissue (von Arnim 2007). The transformed onion cells for GFP (excitation 488 nm) were observed with a universal fluorescence microscope (90i, Nikon).
Arabidopsis transformation and phenotype analysis
PtFCA transcripts containing ORF were ligated into the binary plant transformation vector pCAMBIA1301 driven by CaMV35S and then transferred into Agrobacterium strain EH105. Arabidopsis (fca-1) plants were transformed by the floral dip method (Clough and Bent 1998). Positive lines were selected as described by Zhang’s method (Zhang et al. 2011a). Transgenic plants (T1 and T2) were confirmed by PCR amplification. To investigate flowering time, the days to flowering and the number of rosette leaves were counted when plants had 1-cm-long inflorescence. To investigate root development, the length of transgenic plant roots was measured at 15 days of age. The root tip phenotype of Arabidopsis 15-day-old plants analyzed in this study was observed on a Nikon 90i stereomicroscope. Statistical analysis was performed by using SPSS, taking P < 0.01 as significant.
Yeast two-hybrid analysis
For yeast two-hybrid screening, a cDNA library was constructed from precocious trifoliate orange. The conserved domain of PtFCA was cloned into pGBKT7. A yeast two-hybrid experiment was performed by the Matchmaker Yeast Two-Hybrid System (Clontech). Yeast cells were transformed by the LiAc/DNA/PEG method according to the Yeast Protocols Handbook from Clontech (http://www.clontech.com). The transformed cells growing on SD/Leu-Trp-His-Ade/X-α-Gal media were transferred to selective medium containing X-α-Gal (20 mg/ml), and the blue positive colonies were characterized. The screening of interaction clones was carried out according to the manufacturer’s instructions. Plasmids were isolated from colonies showing a positive (blue) reaction and introduced into AH109 for the further confirmation of protein interaction. The one-to-one hybrid experiment was performed on selective SD/-Trp/-Leu/-His/-Ade/X-α-gal (40 μg/ml) media supplemented with 5 mM 3-amino-triazole (3-AT). Inserts of the plasmids were also sequenced by the Beijing Genomics Institute (Wuhan, China).
Bimolecular fluorescence complementation analysis
The full-length ORF of PtFCA and PtFY without a stop codon were cloned into the BiFC vectors (Walter et al. 2004) pUC-SPYNE and pUC-SPYCE by using BamHI and KpnI, respectively. Each fusion construct was verified by sequencing. The BY-2 cell lines (Nicotiana tabacum L. cv. Bright Yellow 2) were transformed with each of the above constructs by particle bombardment (von Arnim 2007). The yellow fluorescent protein (YFP, excitation 514 nm) signal was observed under a confocal laser scanning microscope (LSM510 Meta, Zeiss, Germany) and recorded by LSM Image Examiner software (Zeiss).
Results
Isolation and sequence analysis of PtFCA
To understand the molecular mechanism of flowering in citrus plants, a previous transcriptional study was executed at the phase transition of precocious trifoliate orange by transcriptome sequencing (Zhang et al. 2011b). Five Arabidopsis FCA homologous unigenes were found by de novo analysis, but only three transcripts (PtFCA1, PtFCA2, and PtFCA3) were left after excluding the redundant sequences by PCR analysis (Fig. 1a and S1B). These transcripts could be localized into the same genome DNA sequence, suggesting that they arise via an alternative splicing process. Structure analysis revealed that PtFCA1 contained 19 exons and 18 introns (Fig. 1b). The relative sizes of exons and introns were also generally well conserved as compared with Arabidopsis (Macknight 1997). The third intron (3741 bp) was determined to be the largest PtFCA1 intron, in accord with Arabidopsis, Brassica napus, Oryza sativa, and pea (Macknight 2002). The ORF of PtFCA1 was 2016 bp, containing 700 amino acids with two RRMs and a WW protein interaction domain. The PtFCA2 was formed from cleavage and polyadenylation within intron 3 and contained intron 1, 2, and 309 bp of intron 3, while 60 bp of the 5′ region in the retained portion of intron 3 was spliced (Fig. 1b). In contrast, PtFCA3 contained 369 bp of intron 3 without a splice occurring in the 5′ region (Fig. S1A). In addition, four kinds of transcripts (α, β, γ, and δ) could be generated from FCA in Arabidopsis (Macknight 2002). Therefore, to find other alternative splicing of PtFCA, a 3′-RACE experiment was preformed based on 3′ conserved sequences of PtFCA, but no new alternative splicing was found.
a The amino acid sequences of PtFCA proteins. The different conserved domains of FCA protein are represented by shaded regions with their names above every domain. PtFCA1, PtFCA2, PtFCA3 from Poncirus trifoliata; HvFCA from Hordeum vulgare; PsFCA from Pisum sativum; AtFCA from Arabidopsis; OsFCA from Oryza sativa. b Structures of the genomic sequence of PtFCA. The black boxes represent exons, and the gray boxes represent introns
The deduced PtFCA1 protein showed only 53 and 43 % similarity to Arabidopsis and rice, respectively. However, the two RRMs and the WW domain exhibited high similarity to Arabidopsis (83 and 79 %, respectively) and to rice (83 and 72 %, respectively). Previous studies reported that all the monocot FCAs contained glycine-rich regions at the protein N-terminus corresponding to a GC-rich region at the 5′ end of the nucleic acid region (Kumar et al. 2011). However, this region was lacking in Arabidopsis as well as precocious trifoliate orange (Fig. 1). PtFCA1 has a proline-rich region behind the RRMs domain, which was absent from all the other FCA proteins. There was a longer PolyQ region in the N-terminus than that in Arabidopsis. PtFCA2 and PtFCA3 were truncated and only comprised a partial RRM1 domain without RRM2 and the WW domain. PtFCA3 consisted of 101 amino acid residues, whereas PtFCA2 with only one amino acid. Their C-terminus was slightly divergent by several amino acids (Fig. 1). Experiments indicated that the alternative processing of PtFCA is conserved between citrus and Arabidopsis. A phylogenetic tree based on a sample of known FCA protein sequences from monocots and dicots was generated (Fig. S1C). Protein-based phylogenetic comparison exhibited clear divergence between monocot and dicot FCA. Observations revealed that PtFCA is most closely related to Arabidopsis FCA in dicots.
Localization of PtFCA proteins
Sequence analysis revealed that PtFCA1 possesses a nuclear localization signal “KRPRP” at the 293rd amino acid position. But the nuclear localization signal (NLS) was absent in PtFCA2 and PtFCA3. The PtFCA-GFP fusion protein was constructed to examine whether PtFCA transcripts were localized in the nucleus. PtFCA1, PtFCA2, and PtFCA3 ORFs (without the stop codon) were fused to N-terminus of the GFP reporter gene under the control of CaMV35S. The fusion plasmids and the control (GFP) were transiently expressed in onion epidermal cells and monitored for green fluorescence with a microscope. Transient expression assays indicated that GFP, PtFCA2-GFP, and PtFCA3-GFP were detected in the cytoplasm and nucleus, as seen in the control, whereas the PtFCA1-GFP fusion protein was exclusively restricted in the nucleus (Fig. 2). The nuclear localization of PtFCA1-GFP also suggests that PtFCA1 is a nuclear protein.
Localization of PtFCA-GFP in onion epidermal cells. A: Fluorescent image of 35S::GFP for positive control. B: Fluorescent image of 35S::PtFCA1:GFP. C: Fluorescent image of 35S::PtFCA2:GFP. D: Fluorescent image of 35S::PtFCA3:GFP. a, b, c, and d indicated bright-field image of onion epidermal cells corresponding to A, B, C, and D, respectively. Bar = 50 μm
Expression pattern of PtFCA in precocious trifoliate orange
To understand the role of PtFCA in the seasonal periodicity of flowering in precocious trifoliate orange, the dynamic expression of three PtFCA transcripts and total PtFCA (tPtFCA: including PtFCA1, PtFCA2, and PtFCA3) were investigated during different developmental stages of spring shoots of precocious trifoliate orange by real-time PCR. tPtFCA and PtFCA1 had the same expression pattern during the flower development process of spring shoots. A high transcript level of PtFCA1 was seen before self-pruning, decreased as the spring shoots began to self-pruning, followed by maintenance of a high level of expression after self-pruning. PtFCA2 was present at higher levels during the self-pruning process. Interestingly, the high transcript level of PtFCA3 was also seen before self-pruning of spring shoots and the amount decreased at the beginning of self-pruning.
To understand the relationship between phase transition and the expression of PtFCA, we investigated the expression level of PtFCA at three key developmental stages (juvenile, phase transition, and adult) of precocious trifoliate orange. The expression of tPtFCA1 showed higher levels at the phase transition stage as compared with the juvenile stage; PtFCA1 showed higher levels at the adult stage as compared with the juvenile and phase transition stages; PtFCA2 increased at the phase transition stage and remained at a high level of expression at the adult stage. However, PtFCA3 showed the highest transcript level at the phase transition stage compared with the levels in other two stages (Fig. 3b). In addition, the expression of PtFCA was also investigated in different tissues including leaves, roots, spring shoots, flowers, and fruits of adult trees by real-time PCR. PtFCA exhibited broad expression patterns, with transcripts detected in all the plant organs. The expression levels of tPtFCA and PtFCA3 were particularly higher in roots as compared with other tissues (Fig. 3c). The expression of tPtFCA and PtFCA2 was strongly detected in fruits; the PtFCA1 and PtFCA3 were detected strongly in the flowers and leaves, slightly in shoots, and scarcely in fruits.
The temporal and spatial expression pattern of PtFCA (tPtFCA, PtFCA1, PtFCA2, and PtFCA3). a The relative expression of PtFCA during reproductive transition process of spring shoot. Stage 1: 15 days before self-pruning; stage 2: beginning of self-pruning; stage 3: 20 days after self-pruning. b The relative quantities of PtFCA genes during different developmental stages (asterisks indicate significant difference from stage 1.*P < 0.05, **P < 0.01). Juvenile: 6-month-old seedling; transition: 12-month-old precocious trifoliate orange; adult: 7-year-old precocious trifoliate orange. c The relative expression of PtFCA in different tissues including roots, shoots, leaves, flowers at anthesis, and whole fruits at 1 month after flowering (asterisks indicate significant difference from root.*P < 0.05, **P < 0.01). β-actin was used as a control. Real-time PCR experiments were conducted using the primers displayed in Supplementary Table S1
Functional analysis of PtFCA in transgenic Arabidopsis
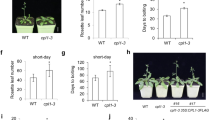
To evaluate the function of PtFCA in flowering regulation of precocious trifoliate orange, PtFCA1, PtFCA2, and PtFCA3 were ectopically expressed in Arabidopsis fca-1 mutant driven by the 35S promoter, 15, 18, and 20 independent kanamycin-resistant plants were obtained in the T1 generation, respectively. In order to further analyze PtFCA function, three independent transgenic lines were randomly selected for phenotypic observation by PCR detection using DNA templates. We selected 15 T3 transgenic plants for each transgenic line. The results showed that overexpression of PtFCA1 can partially rescue the late-flowering and short-root phenotype in fca-1 mutants (Fig. 4a and b). Three 35S::PtFCA1 transgenic lines flowered significantly earlier than the fca-1 mutants (Student’s t test, P < 0.05) in terms of both days to flowering and number of leaves (Table 1). The average time to flowering of three transgenic lines ranged from 18.1 to 24.1 days, whereas that of the fca-1 and wild-type plants was 42.8 and 19.8 days (Table 1). The average number of rosette leaves at flowering ranged from 11 to 14 in three transgenic lines and was 21 and 9.5 in the fca-1 and wild-type plants (Table 1). No difference in the appearance of flowers and inflorescences was observed among 35S::PtFCA1 and wild-type and fca-1 plants. In addition, the meristem and elongation zone of transgenic Arabidopsis primary roots were also different (Fig. 4c). These results indicated that PtFCA1 might promote flowering and root development.
Interestingly, PtFCA2 was capable of conferring a late-flowering phenotype and rescue short-root phenotype in the fca-1 background (Fig. 4a, b, Table 1). Three 35S::PtFCA2 transgenic lines flowered significantly later than all other transgenic plants (Table 1). The average time to flowering ranged from 47 to 60 days, and the average number of leaves at flowering ranged from 20.3 to 30.7 in three transgenic lines (Table 1). The average length of roots from 17.8 to 21.0 mm in three transgenic lines. However, it is worth noting that the average length of roots was 13.1 and 17.2 mm in the fca-1 and wild-type plants, respectively (Table 1). However, overexpression of PtFCA3 did not exhibit any effect on flowering time and root development (Fig. 4). Moreover, these results also reveal that PtFCA transcripts has functional similarity and difference with Arabidopsis FCA with regard to flowering time regulation and root development.
Isolation of PtFCA interaction protein by yeast two-hybrid system
To better understand the role of PtFCA during the development of precocious trifoliate orange, we attempted to identify proteins that interact with PtFCA. A yeast two-hybrid system was used to screen a cDNA expression library of precocious trifoliate orange. The coding sequence of WW and RRM domains was cloned into pGBKT7 as baits, respectively. There was no positive clone obtained by using the RRM domain when screening the library. Various kinds of proteins were found on the bases of screens conducted with WW domain baits (Table 2). Approximately 1 × 106 yeast colonies were screened, and 131 positive clones were identified from the cDNA library. DNA sequencing showed that the 131 clones represented 11 proteins. Further analysis was also performed for interaction between the WW domain bait and the 11 proteins by one-to-one hybrid, but APX2 does not interact with FCAWW. One possible explanation is that the selection pressure was increased during interaction verification process by add 3-AT in media. These interacting proteins were found to be involved in translation, metabolism, and cell division processes (Fig. 5a). Among these interaction proteins, one transcript factor, named PtNF-YA7, was a homolog of NUCLEAR FACTOR-Y SUBUNIT A 7 (NF-YA7), which plays an important role in the integration of vernalization and photoperiod seasonal signals along with regulation of flowering initiation in temperate cereals (Siefers et al. 2009; Zhang et al. 2011a). In addition, it is also able to regulate ABA signaling components during germination and other ABA-mediated developmental responses in Arabidopsis (Siriwardana et al. 2014). Another interesting interaction protein was PtELIP2, a homolog of early light inducible proteins (ELIP2), which was transiently induced by light, ABA, and other environmental stresses, such as heat, cold, salinity, and desiccation.
The isolated interaction protein with PtFCA protein. a Yeast two-hybrid analysis. b PtFCA/PtFY interactions examined by one-to-one hybrid. c PtFCA/PtFY interaction in tobacco BY-2 cells by bimolecular fluorescence complementation. BY-2 cells were transformed by particle bombardment with a set of constructs for PtFCA-YFPN and PtFY-YFPC. Bar = 10 μm
On the other hand, the FY/FCA interaction is clearly needed in Arabidopsis for FCA autoregulation (Simpson et al. 2003), and this has also been demonstrated in rice (Jang et al. 2009b). However, we did not obtain the homolog of FY by screening a yeast two-hybrid library. To further investigate the PtFY/FCA interaction, a homolog of FY′ from precocious trifoliate orange was isolated according to the Citrus genome database (http://citrus.hzau.edu.cn/orange/). The FY WD domain and PPLPP motifs were cloned into pGADT7, respectively. The result revealed that PtFCA interacts with PtFY via the PtFCA-WW domain and PtFY-PPLPP motifs (Fig. 5b). To verify the PtFY/FCA interaction in plants, a bimolecular fluorescence complementation (BiFC) experiment was carried out in tobacco Bright Yellow 2 (BY-2) cell lines by particle bombardment. After the fusion of PtFCA and PtFY to the N- or C-terminal YFP fragment and co-expression in tobacco cells, strong BiFC signals were observed in the transformed cells (Fig. 5c).
Expression profiles of PtFCA in response to ABA and ambient temperature
PtFCA has diverse domains and interacts with several ABA and ambient temperature-related proteins. Thus, the expression pattern of PtFCA was detected in precocious trifoliate orange under ABA and ambient temperature treatments. The effect of ABA was investigated during the flowering developmental process of spring shoots, which can flower after sprouting without dormancy. Length, number of nodes per shoot, and the flower number to shoot number ratio were reduced (Fig. 6a). A similar expression pattern of tPtFCA and PtFLC was detected under ABA treatment: it increased at phase transition and remained at a high level of expression at the flowering stage. The same expression pattern of PtFCA3 and PtNFYA7 was also observed at these development stages either with or without ABA treatments (Fig. 6a), and the two genes tended to decrease from the sprouting stage to the flowering stage. A significant difference in the transcript level was seen between the treatment and control in PtFCA2: it showed the highest transcript level at flowering stage compared with the levels in the control. At the flowering stage, PtFCA1 showed the opposite trend to PtFCA3 with ABA treatment. However, compared with the levels in treated tissue, the two genes were detected and present at high levels in the control tissue.
Effect of ABA and ambient temperature on precocious trifoliate orange seedlings. a The length, node number per shoot, and ratio of flowering number to shoot number of spring shoots with H2O or 15 μm/ml ABA treatment, respectively. Asterisks indicate significant differences between H2O and 15um/ml ABA treatment (*P < 0.05, **P < 0.01). b The expression pattern of PtFCA, PtFY, PtNF-YA7, and PtFLC during the development stages of spring shoots with ABA treatment. Stage 1: sprouting stage; stage 2: transition stage; stage 3: flowering stage. b Expression patterns of PtFCA, PtFY, PtNF-YA7, and PtFLC under ambient temperature treatment (*P < 0.05, **P < 0.01). The 3-month-old seedlings were grown under a photoperiod of 16-h light/8-h dark at 23 or 27 °C. Total RNA was extracted after 2 weeks of treatment
To investigate the effect of low temperature, 3-month-old seedlings were treated at 27 or 23 °C for 2 weeks. The results indicated that PtFCA1 and PtFY in seedlings kept at higher ambient temperature were reduced greatly after 2 weeks, contrary to PtFCA2 and PtFCA3 (Fig. 6b). PtNF-YA7 exhibited higher expression at 27 °C, whereas PtFY showed higher expression at 23 °C.
Discussion
The molecular mechanism for flowering has been studied in citrus plants, and several genes involved in flowering time regulation have been identified and characterized (Nishikawa et al. 2007; Tan and Swain 2007; Zhang et al. 2011b; Zhang et al. 2009). According to previous reports regarding flowering pathway in Arabidopsis, these genes appear to be involved in the photoperiod, vernalization, and gibberellic pathways (Zhang et al. 2011b). However, findings about other flowering pathways (autonomous and thermosensory) have not yet been reported (Khan et al. 2014). In the current study, we reported on the molecular identification and characterization of a citrus FCA homolog (PtFCA) from precocious trifoliate orange. PtFCA exhibited high homology with Arabidopsis, especially in the RRM and WW domains, and contains an additional feature of proline-rich and a longer PolyQ region (Fig. 1). The proline-rich domain is known to display competitive interaction with the WW domain. Their diverse interactions are critically important in the transcription and signal transduction pathways (Gao et al. 2014). Longer PolyQ expansions can retard FCA homolog autoregulation and affect FCA–FY interaction (Lindqvist et al. 2007). It seems that the FCA has sequence conservation between trifoliate orange and Arabidopsis, which may be related to their conserved functions in different species.
In Arabidopsis, FCA contains two RNA recognition motifs and a WW domain, suggesting a role in posttranscriptional RNA modifications. On the other hand, FCA mRNA was also shown to be subject to alternative splicing, and different transcripts were expressed at different levels in various tissues (Jang et al. 2009a; Simpson et al. 2003). In Arabidopsis, four kinds of transcripts can be generated from FCA: α, β, γ, and δ (Macknight 2002). In this study, we demonstrated that the alternative splicing of the PtFCA was also evolutionarily conserved; transcripts that were equivalent to FCA-β and FCA-γ could be detected in citrus, whereas those equivalent to FCA-α and FCA-δ could not be detected. These results indicated that the alternative processing of PtFCA has diverged between citrus and Arabidopsis. Meanwhile, our findings suggest also that PtFCA might be a candidate FCA gene in precocious trifoliate orange. In addition, we identified two alternative splicing transcripts that lacked the nuclear localization signal, and the subcellular localization analysis dovetailed nicely with bioinformatics prediction (Fig. 4). In Arabidopsis, there are several examples where protein isoforms from alternatively spliced pre-mRNAs are targeted to different cellular locations and affected functions (Kriechbaumer et al. 2012; Papadopoulos et al. 2011; Remy et al. 2013). Our observations seem to suggest that PtFCA may display different functions in citrus plants by altering the subcellular localization of proteins. In this study, PtFCA2 and PtFCA3 were not contained RRM2, NLS and WW domains compared with PtFCA1. The WW domain of FCA protein is critical for the interaction between FCA and FY to occur in Arabidopsis and rice (Jang et al. 2009b; Macknight 1997). In addition, the WW domain could also interact with components of the multiprotein complexes involved in transcription, RNA processing, chromatin remodeling, and actin polymerization by location in the nucleus (Ingham et al. 2005). These results further indicated that PtFCA1 may play more important and extensive role compared with PtFCA2 and PtFCA3 during the development process of precocious trifoliate orange.
FCA is a transcription factor and regulates transition to flowering by interacting with FY in Arabidopsis and rice (Jang et al. 2009b; Simpson et al. 2003). Arabidopsis fca-1 mutant exhibited a strong late-flowering and significantly short-root phenotypes (Macknight 2002). The transcript γ can complement the fca-1 late-flowering phenotype, but transcripts FCA-β and FCA-δ do not (Macknight 1997; Macknight 2002; Quesada 2003). FCA was expressed during all development stages and in all tissues in Arabidopsis and rice, although it was expressed at lower levels in root than in other tissues (Lee et al. 2005; Macknight 1997). Likewise, PtFCA transcripts accumulated in all tissues examined in these experiments. In contrast, PtFCA were observed at higher levels in root than other tissues (Fig. 3c). These results suggested that PtFCA transcripts may be associated with a function in root development. Therefore, whether PtFCA regulates root development or not need to be further investigated in precocious trifoliate orange. For alternatively spliced transcripts of PtFCA, the expression of PtFCA1 that was equivalent to functional FCA-γ transcript presents at higher levels in the adult compared with the juvenile stage (Fig. 3b). It also exhibited higher levels in the stages of summer shoots as compared with spring shoots (Fig. 3a). Based on the expression pattern of PtFCA1, we propose that PtFCA1 may involve in root development and reproductive growth. On the other hand, FCA appears to be a component of a posttranscriptional cascade involved in the control of flowering time (Macknight 1997). In this study, ectopic expression of different PtFCA transcripts affected the flowering time and root development in transgenic Arabidopsis. PtFCA1 partially complemented the fca-1 late-flowering and short-root phenotype. This result coincides with the reports in transgenic OsFCA as compared to Arabidopsis (Lee et al. 2005). These results indicated that PtFCA1 performed the same functions with Arabidopsis FCA. Thus, we conclude that PtFCA1 regulates flowering time, which is functionally conserved to FCA in Arabidopsis. However, ectopic overexpression of PtFCA2 in fca-1 Arabidopsis delayed the flowering time and promoted root development (Fig. 4). The 35S::PtFCA3 transgenic Arabidopsis plants did not show any aberrant phenotype (Fig. 4). This finding suggested that expressions of PtFAC2 may control flowering time and root development. Meanwhile, PtFCA2 is a new transcript with several amino acids different from PtFCA3, but the function is different. One possible explanation for this observation is that these different amino acids may lead to differences in the crystal structure of PtFCA2 and PtFCA3. Further studies were required by solving high-resolution crystal structures of the two proteins because this was a preliminary, inconclusive deduction on our part. However, the Arabidopsis FCA-α, FCA-β, and FCA-δ transcripts being nonfunctional did not complement the fca-1 phenotype (Macknight 2002). This evidence further supports the hypothesis that citrus plants’ FCA may have divergent functions as compared with herbaceous FCA.
Interestingly, FCA is autoregulated by its own transcript levels through its interaction with FY (Quesada 2003; Simpson et al. 2003). PtFCA has conserved and divergent functions of herbaceous FCA and contains diverse domains, so the molecular mechanism of PtFCA to regulate trifoliate orange development may be variable. The Arabidopsis FCA protein interacts with FY and AtSWI3B (Sarnowski et al. 2002; Simpson et al. 2003). The regulation of FCA transcript levels by interaction with FY is an important determinant in regulating FLC and controlling the floral transition (Jang et al. 2009b; Sarnowski et al. 2002; Simpson et al. 2003). In this study, the WW domain of PtFCA could interact with the components of multiprotein complexes involved in multiple biological processes (Fig. 5), consistent with the results of FCA in rice (Ingham et al. 2005). This may explain why PtFCA could interact through its WW domain with various proteins that had different functions. We have verified that the PtFY interaction with PtFCA was conserved in citrus. This result suggested that PtFCA was regulating the flowering time in a FY-dependent manner in citrus, which may share the same mechanism with herbaceous plants. Through interaction with FY, FCA affects alternative polyadenylation of many transcripts including flowering repressor FLC (Simpson et al. 2010). Our lab reported that the 5′-untranslated region of the PtFLC gene has alternate splicing (Zhang et al. 2009). These reports indicated that the PtFCA/PtFY complex may regulate the alternate splicing of PtFLC in trifoliate orange. In addition, PtFCA also interacted with PtNF-YA7, a homolog of NF-YA7, which is another interaction protein in the NF-Y family (Siriwardana et al. 2014; Sorin et al. 2014). The competition of NF-YA7 with VERNALIZATION2 (VRN2) and CONSTANS2 (CO2) proteins for interaction plays an important role in the integration of vernalization and photoperiod seasonal signals by regulating flowering initiation in temperate cereals (Zhang et al. 2011a). Thus, PtFCA may regulate flowering time by forming higher order complexes to integrate various flowering signaling pathways in citrus. Both the special proline-rich domain and the interactor of PtFCA are involved in the perception of ABA-mediated developmental responses and ambient temperature (Table 2) (Blazquez et al. 2003; Jung et al. 2012; Siriwardana et al. 2014).
In Arabidopsis, Razem et al. reported that FCA protein is a receptor for ABA (Razem et al. 2006). However, Joanna et al. find that ABA affects FCA autoregulation but does not bind FCA protein (Risk et al. 2008). Our expression analysis showed that PtFCA expression was more abundant in root and was regulated by exogenous ABA treatment (Figs. 3c and 6b), the FY interaction with FCA was also conserved in trifoliate orange (Fig. 5). Therefore, ABA may indirectly regulate PtFCA expression via FCA–FY interaction complex. PtFCA3 lacking the WW domain responded to 27 °C ambient temperature more strongly than to 23 °C. However, PtFCA1 responding to higher ambient temperature was contrary to the situation in PtFCA2 and PtFCA3 (Fig. 6b). Furthermore, the PtFCA2 and PtFCA3 did not contain RRM2 and WW domains compared with PtFCA1. These results suggested that the RRM2 and WW domains may be critical for the response to higher ambient temperature in trifoliate orange. Different experiments have been conducted to study the effect of FCA responses to ABA and ambient temperature in Arabidopsis (Kumar et al. 2011; Lee et al. 2014). FCA seems to control flowering time in response to temperature changes mostly through an FLC-independent pathway (Blazquez et al. 2003). Different expression patterns of ABA treatment and the ambient temperature response indicate that PtFCA may respond to seasonal environmental inputs during plant developmental stages. However, more analysis is needed to better define the PtFCA response to seasonal environmental inputs through its domain or protein interactions.
References
Abou-Elwafa SF, Buttner B, Chia T, Schulze-Buxloh G, Hohmann U, Mutasa-Gottgens E, Jung C, Muller AE (2011) Conservation and divergence of autonomous pathway genes in the flowering regulatory network of Beta vulgaris. J Exp Bot 62:3359–3374
Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14:S111–S130
Amasino RM, Michaels SD (2010) The timing of flowering. Plant Physiol 154:516–520
Blazquez MA, Ahn JH, Weigel D (2003) A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet 33:168–171
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Davenport T (1990) Citrus flowering. Hortic Rev 12:349–408
Gao YG, Yang H, Zhao J, Jiang YJ, Hu HY (2014) Autoinhibitory structure of the WW domain of HYPB/SETD2 regulates its interaction with the proline-rich region of huntingtin. Structure 22:378–386
Ingham RJ, Colwill K, Howard C, Dettwiler S, Lim CS, Yu J, Hersi K, Raaijmakers J, Gish G, Mbamalu G, Taylor L, Yeung B, Vassilovski G, Amin M, Chen F, Matskova L, Winberg G, Ernberg I, Linding R, O’Donnell P, Starostine A, Keller W, Metalnikov P, Stark C, Pawson T (2005) WW domains provide a platform for the assembly of multiprotein networks. Mol Cell Biol 25:7092–7106
Jaeger KE, Graf A, Wigge PA (2006) The control of flowering in time and space. J Exp Bot 57:3415–3418
Jang YH, Lee JH, Park H-Y, Kim S-K, Lee B-Y, Suh MC, Kim J-K (2009a) OsFCA transcripts show more complex alternative processing patterns than its Arabidopsis counterparts. J Plant Biol 52:161–166
Jang YH, Park HY, Kim SK, Lee JH, Suh MC, Chung YS, Paek KH, Kim JK (2009b) Survey of rice proteins interacting with OsFCA and OsFY proteins which are homologous to the Arabidopsis flowering time proteins, FCA and FY. Plant Cell Physiol 50:1479–1492
Risk JM, Macknight RC, Da CL (2008) FCA does not bind abscisic acid. Nature 456:E5–E6
Jung JH, Seo PJ, Ahn JH, Park CM (2012) Arabidopsis RNA-binding protein FCA regulates microRNA172 processing in thermosensory flowering. J Biol Chem 287:16007–16016
Khan MR, Ai XY, Zhang JZ (2014) Genetic regulation of flowering time in annual and perennial plants. Wiley Interdiscip Rev RNA 5:347–359
Kriechbaumer V, Wang P, Hawes C, Abell BM (2012) Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation. Plant J 70:292–302
Kumar S, Jiang S, Jami SK, Hill RD (2011) Cloning and characterization of barley caryopsis FCA. Physiol Plant 143:93–106
Lee JH, Cho YS, Yoon HS, Suh MC, Moon J, Lee I, Weigel D, Yun CH, Kim JK (2005) Conservation and divergence of FCA function between Arabidopsis and rice. Plant Mol Biol 58:823–838
Lee HJ, Jung JH, Llorca LC, Kim SG, Lee S, Baldwin IT, Park CM (2014) FCA mediates thermal adaptation of stem growth by attenuating auxin action in Arabidopsis. Nat Commun 5:5473–5473
Li ZM, Zhang JZ, Mei L, Deng XX, Hu CG, Yao JL (2010) PtSVP, an SVP homolog from trifoliate orange (Poncirus trifoliata L. Raf.), shows seasonal periodicity of meristem determination and affects flower development in transgenic Arabidopsis and tobacco plants. Plant Mol Biol 74:129–142
Liang S, Zhu W, Xiang W (1999) Precocious trifoliate orange (Poncirus trifoliata (L.) Raf.) biology characteristic and its stock experiment. ZheJiang Citrus 16:2–4
Lindqvist C, Laakkonen L, Albert VA (2007) Polyglutamine variation in a flowering time protein correlates with island age in a Hawaiian plant radiation. BMC Evol Biol 7:105
Liu D, Cai X (2013) OsRRMh, a spen-like gene, plays an important role during the vegetative to reproductive transition in rice. J Integr Plant Biol 55:876–887
Liu F, Quesada V, Crevillén P, Bäurle I, Swiezewski S, Dean C (2007) The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol Cell 28:398–407
Macknight R (1997) FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89:737–745
Macknight R (2002) Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell 14:877–888
Nishikawa F, Endo T, Shimada T, Fujii H, Shimizu T, Omura M, Ikoma Y (2007) Increased CiFT abundance in the stem correlates with floral induction by low temperature in Satsuma mandarin (Citrus unshiu Marc.). J Exp Bot 58:3915–3927
Papadopoulos C, Arato K, Lilienthal E, Zerweck J, Schutkowski M, Chatain N, Muller-Newen G, Becker W, de la Luna S (2011) Splice variants of the dual specificity tyrosine phosphorylation-regulated kinase 4 (DYRK4) differ in their subcellular localization and catalytic activity. J Biol Chem 286:5494–5505
Quesada V (2003) Autoregulation of FCA pre-mRNA processing controls Arabidopsis flowering time.pdf. EMBO J 22:3142–3152
Razem FA, El-Kereamy A, Abrams SR, Hill RD (2006) The RNA-binding protein FCA is an abscisic acid receptor. Nature 439:290–294
Remy E, Cabrito TR, Baster P, Batista RA, Teixeira MC, Friml J, Sa-Correia I, Duque P (2013) A major facilitator superfamily transporter plays a dual role in polar auxin transport and drought stress tolerance in Arabidopsis. Plant Cell 25:901–926
Ruttink T, Arend M, Morreel K, Storme V, Rombauts S, Fromm J, Bhalerao RP, Boerjan W, Rohde A (2007) A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell 19:2370–2390
Sarnowski TJ, Swiezewski S, Pawlikowska K, Kaczanowski S, Jerzmanowski A (2002) AtSWI3B, an Arabidopsis homolog of SWI3, a core subunit of yeast Swi/Snf chromatin remodeling complex, interacts with FCA, a regulator of flowering time. Nucleic Acids Res 30:3412–3421
Siefers N, Dang KK, Kumimoto RW, Bynum WE, Tayrose G, Holt BF 3rd (2009) Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol 149:625–641
Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C (2003) FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113:777–787
Simpson GG, Laurie RE, Dijkwel PP, Quesada V, Stockwell PA, Dean C, Macknight RC (2010) Noncanonical translation initiation of the Arabidopsis flowering time and alternative polyadenylation regulator FCA. Plant Cell 22:3764–3777
Siriwardana CL, Kumimoto RW, Jones DS, Holt BF (2014) Gene family analysis of the Arabidopsis NF-YA transcription factors reveals opposing abscisic acid responses during seed germination. Plant Mol Biol Report 32:971–986
Sorin C, Declerck M, Christ A, Blein T, Ma L, Lelandais-Briere C, Fransiska Njo M, Beeckman T, Crespi M, Hartmann C (2014) A miR169 isoform regulates specific NF-YA targets and root architecture in Arabidopsis. New Phytol 202:1197–1211
Srikanth A, Schmid M (2011) Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci 68:2013–2037
Sun F, Liu C, Zhang C, Qi W, Zhang X, Wu Z, Kong D, Wang Q, Shang H, Qian X, Li F, Yang J (2011) A conserved RNA recognition motif (RRM) domain of Brassica napus FCA improves cotton fiber quality and yield by regulating cell size. Mol Breed 30:93–101
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tan FC, Swain SM (2007) Functional characterization of AP3, SOC1 and WUS homologues from citrus (Citrus sinensis). Physiol Plant 131:481–495
von Arnim A (2007) Subcellular localization of GUS- and GFP-tagged proteins in onion epidermal cells. Cold Spring Harb Protoc 2007: pdb. prot4689
Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40:428–438
Zhang J-Z, Ai X-Y, Sun L-M, Zhang D-L, Guo W-W, Deng X-X, Hu C-G (2011a) Molecular cloning and functional characterization of genes associated with flowering in citrus using an early-flowering trifoliate orange (Poncirus trifoliata (L.) Raf.) mutant. Plant Mol Biol 76:187–204
Zhang J-Z, Ai X-Y, Sun L-M, Zhang D-L, Guo W-W, Deng X-X, Hu C-G (2011b) Transcriptome profile analysis of flowering molecular processes of early flowering trifoliate orange mutant and the wild-type [Poncirus trifoliata (L.) Raf.] by massively parallel signature sequencing. BMC Genomics 12:63
Zhang J-Z, Li Z-M, Mei L, Yao J-L, Hu C-G (2009) PtFLC homolog from trifoliate orange (Poncirus trifoliata) is regulated by alternative splicing and experiences seasonal fluctuation in expression level. Planta 229:847–859
Zhang JZ, Zhao K, Ai XY, Hu CG (2014) Involvements of PCD and changes in gene expression profile during self-pruning of spring shoots in sweet orange (Citrus sinensis). BMC Genomics 15:892
Acknowledgments
We are grateful to Prof. Jia-Ling Yao for her helpful discussion in this manuscript. This research was supported financially by the National Natural Science Foundation of China (grant nos. 31130046, 31471863, 31360469, 31372046, and 31521092) and Fundamental Research Funds for the Central Universities (Program No. 2662016PY037).
Author contributions
X-YA carried out the yeast two-hybrid, bimolecular fluorescence complementation, and PCR experiments; X-YA and T-JL prepared the plant material and obtained the transgenic plants; J-ZZ and C-GH designed the experiments and the study; X-YA and J-ZZ wrote the paper. All authors discussed the data obtained. All authors reviewed and provided comments upon preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Data archiving statement
The PtFCA1, PtFCA2, and PtFCA3 have been deposited in GenBank under accession nos. KX440390, KX440391, and KX440392, respectively.
Additional information
Communicated by W.-W. Guo
Xiao-Yan Ai and Jin-Zhi Zhang contributed equally to this work.
Electronic supplementary material
Figure S1
Gene sequences, structures, and phylogenetic relationship of PtFCA. A: Sequence alignment between PtFCA2 and PtFCA3. The gray box represents the sequence differences between PtFCA2 and PtFCA3. B: Alternatively spliced transcripts were validated in precocious trifoliate orange by RT-PCR, M: Marker (DL2000). Lanes 1–4: PtFCA1 in trifoliate orange, 5–7: PtFCA2/3 in precocious trifoliate orange. C: Phylogenic tree of FCA showing evolutionary relationships. The sequences with the highest percent sequence similarity are grouped together. The accession numbers of FCA proteins used in this analysis are HvFCA (FJ188402.2), VvFCA (GU300763.1), RcFCA (XM_002519206.1), GmFCA-like (XM_003529468.1), PsFCAgamma (AY805329.1), MtFCA (XM_003609755.1), FCAalpha (Z82993.1), FCAbeta (Z82991.1), FCAdelta (Z82990.1), FCAgamma (Z82989.1), OsFCA-1 (AY274928.1), OsFCA-2 (AY311344.1), OsFCA-3 (AY311343.1), OsFCA-4 (AY331574.1). (JPEG 4617 kb)
Table S1
List of primer sequences used in this study. (XLS 25 kb)
Rights and permissions
About this article
Cite this article
Ai, XY., Zhang, JZ., Liu, TJ. et al. PtFCA from precocious trifoliate orange is regulated by alternative splicing and affects flowering time and root development in transgenic Arabidopsis . Tree Genetics & Genomes 12, 85 (2016). https://doi.org/10.1007/s11295-016-1035-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-016-1035-6