Abstract
Since chloroplasts are maternally inherited and have unique features in evolution, their genome sequences have been broadly used in phylogenetic studies of plants. Here, we assembled the chloroplast genome sequence of cultivated pineapple (Ananas comosus (L.) Merr.) that is the most economically significant plant in the Bromeliaceae using next-generation sequencers. The genome length was 159,636 bp and included a pair of inverted repeats of 26,774 bp separated by a small single-copy region of 18,622 bp and a large single-copy region of 87,466 bp. The genome contained 113 unique genes (79 protein-coding, 4 rRNA, and 30 tRNA genes), 19 of which were duplicated in the inverted repeats, giving a total of 132 genes. We identified a total of 65 simple sequence repeats of >10 bp in length. Phylogenetic tree identified Ananas as a basal member of the Poales, closer to Musa (Musaceae, Zingiberales) than to species of the Poaceae. The genes, indels, and simple sequence repeats identified in this study will provide tools for use in evolutionary studies at both intra- and interspecific levels.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The monocot order Poales comprises 16 families and approximately 18,000 species. Relationships among families are generally well resolved and supported (Guisinger et al. 2010). The largest family within the Poales, the Poaceae, has provided the basis for many studies, including complete chloroplast genome sequencing, owing to its ecological, economic, and evolutionary importance (Guisinger et al. 2010).
Cultivated pineapple (Ananas comosus (L.) Merr.) belongs to the family Bromeliaceae in the Poales. Pineapple is the third most important tropical fruit in world production after banana and citrus (Rohrbach et al. 2002). It has been cultivated for more than 500 years in the Americas. Domesticated pineapple was already widely distributed in the Americas and the Caribbean prior to the arrival of Columbus on 1493 (Rohrbach et al. 2002). Pineapple has the crassulacean acid metabolism (CAM) photosynthetic pathway (Malézieux et al. 2002). CAM plants conserve water by conducting most of their gas exchange in the relatively cool atmosphere at night, and CAM plants can grow in strongly water-limited semidesert habitats (West-Eberhard et al. 2011). Such strong drought tolerance also enables months-long storage of vegetative propagules of pineapple (Hepton 2002). Strong drought tolerance itself and ease of transport due to long-life vegetative propagules facilitated its wide diffusion throughout the tropics. Ananas includes two major species, A. macrodontes and A. comosus. The latter has five botanical varieties: A. comosus var. bracteatus, var. parguazensis, var. comosus, var. ananassoides, and var. erectifolius (Coppens d Eeckenbrugge et al. 2002). Only var. comosus has edible cultivars. To clarify the phylogeny of Ananas, analyses of morphological characters and DNA markers have been performed (Coppens d’Eeckenbrugge et al. 1997; Duval et al. 2001, 2003; Paz et al. 2005, 2012; Hamdan et al. 2013). Mexican and Cuban pineapples were characterized by using an amplified-fragment-length polymorphism method (Paz et al. 2005, 2012). Phylogenetic analysis of Malaysian cultivars was performed using the chloroplast-encoded rbcL sequence (Hamdan et al. 2013). Both restriction-fragment-length polymorphism markers and chloroplast genotypes were used to study genetic diversity in Ananas (Duval et al. 2001, 2003). So far, however, few chloroplast-derived markers have been used to study evolutionary relationships of Ananas (Hamdan et al. 2013).
In this study, we determined the complete nucleotide sequence of the chloroplast genome of A. comosus var. comosus, using next-generation sequencers. We compared it with other sequenced chloroplast genomes and discuss structural differences in the form of indels and microsatellites.
Materials and methods
Plant materials and DNA extraction
The pineapple cultivar ‘N67-10’ grown at Nago Branch of Okinawa Prefectural Agricultural Research Center (Nago, Okinawa, Japan) was used. Unexpanded young leaves in the crown were collected and applied for DNA extraction. Total DNA was extracted with a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
DNA sequencing
For the 454 GS FLX+ genome sequencer (Roche Diagnostics, Basel, Switzerland), the total genomic DNA of ‘N67-10’ was sheared by nebulization (600–900 bp in length). A rapid library was prepared with a GS FLX Titanium Rapid Library Preparation Kit (Roche) using the sheared DNA fragments. Then, a library was clonally amplified with emulsion PCR with GS FLX Titanium LV emPCR kit (Roche). Purified beads with an amplified library were applied to DNA sequencing by the 454 GS FLX+ genome sequencer. Two runs of single-read pyrosequencing were performed. For the HiSeq 2500 sequencer (Illumina, San Diego, CA, USA), the total genomic DNA of ‘N67-10’ was fragmented to 350 bp using a Covaris M220 (Covaris, Woburn, MA) and a paired-end library was prepared with a TruSeq DNA LT Sample Prep Kit (Illumina). A library was sequenced by the HiSeq2500. The paired-end read length was 100 bp. All experiments were performed according to the manufacturer’s instructions.
De novo assembly of 454 GS FLX+ data
The 454 GS FLX+ sequence reads were assembled in the CLC Genomics Workbench 7.0 de novo assembly program (Qiagen). We sorted the assembled contiguous sequences (contigs) by depth of coverage to distinguish the chloroplast genome (>50×) from the mitochondrial and nuclear genomes (<50×). We confirmed the high-coverage (>50×) contigs as chloroplast genome by BLAST search against the nucleotide collection database (nr/nt) of the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/). In addition, we confirmed that the Typha latifolia chloroplast genome (GI: 289065068) showed the highest similarity to that of pineapple among the registered chloroplast genomes. We mapped the >50× contig sequences against the T. latifolia sequence by BLASTN. Fourteen contigs were mapped on the T. latifolia chloroplast genome, and gaps were filled in with sequence reads with at least 50 bp of continuous perfect match from both ends.
Read-mapping and correction of draft genome sequences
After the circular draft genome was assembled, we mapped the Illumina reads to it in CLC Genomic Workbench software to find and correct ambiguous nucleotides. The HiSeq 2500 reads used the mapping parameters length fraction = 1.00 and similarity fraction = 1.00. Low-coverage sites were assumed to be errors, and these misassembled sites were manually corrected against HiSeq 2500 sequence reads. After all of the ambiguous nucleotides were corrected, encoded genes were annotated by using DOGMA (Dual Organellar GenoMe Annotator, http://dogma.ccbb.utexas.edu; Wyman et al. 2004). The circular map of the chloroplast genome was drawn by the GenomeVx program (Conant and Wolfe 2008).
Comparative analysis of organelle genomes
The chloroplast genome sequences of T. latifolia and Musa acuminata (GI: 525312436) were compared with that of A. comosus. Dot-plot analysis in PipMaker software (Schwartz et al. 2000) used default settings. The lengths of indels were assessed by creating alignments of complete chloroplast genome sequences in CLC Genomics Workbench.
Phylogenetic analyses
Phylogenetic analyses were performed on an aligned data matrix of 62 angiosperm taxa and 76 protein-coding genes (atpA, atpB, atpE, atpF, atpH, atpI, ccsA, cemA, clpP, infA, matK, ndhA, ndhB, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK, petA, petB, petD, petG, petL, petN, psaA, psaB, psaC, psaI, psaJ, psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ, rbcL, rpl14, rpl16, rpl2, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36, rpoA, rpoB, rpoC1, rpoC2, rps11, rps12, rps14, rps15, rps16, rps18, rps19, rps2, rps3, rps4, rps7, rps8, ycf3, ycf4). Amino acid sequences were aligned by using the Multiple Sequence Web Viewer and Alignment Tool (http://mswat.ccbb.utexas.edu), manually adjusted, and then manually concatenated. The best-scoring maximum likelihood tree was constructed from the sequences in RAxML ver. 8.0.19 software with the PROTCATWAG model (Stamatakis 2014). The likelihood bootstrap probability of each branch was calculated in the “rapid bootstrap” algorithm of RAxML using 1000 replicates.
Simple sequence repeats
Simple sequence repeats (SSRs) were searched for by using the microsatellite search tool MISA (http://pgrc.ipk-gatersleben.de/misa/misa.html).
Results
Genome assembly and validation
Sequencing on the 454 GS FLX+ system generated a total of 1,340,605 reads with an average length of 568 bases that covered 761 Mb. After cleaning and trimming, the remaining reads (1,163,292 reads with an average length of 466 bases) were assembled. Fourteen generated contigs were mapped on the Typha chloroplast genome, and gaps were filled with sequence reads. Mapping the HiSeq 2500 reads onto the resultant supercontig and treating low-coverage sites as sequence errors detected 85 errors, which were corrected.
Size and gene content of the Ananas chloroplast genome
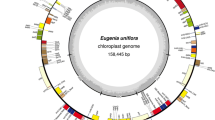
The total length of the constructed Ananas chloroplast genome was 159,636 bp and included a large single-copy (LSC) region of 87,466 bp, a small single-copy (SSC) region of 18,622 bp, and a pair of inverted repeats (IRa and IRb) of 26,774 bp each (Fig. 1). The genome contained 113 unique genes, 19 of which were duplicated in the IRs, giving a total of 132 genes (Table 1). Among 4 rRNA and 30 tRNA genes identified, all 4 rRNA and 8 tRNA genes were duplicated in the IR. The tRNA genes were identical to those of well characterized vascular plants. The genome consisted of 59.80 % coding regions and 40.20 % noncoding regions, including both intergenic spacers and introns. It had a GC content of 37.37 % and an AT content of 62.63 %.
Gene map of the Ananas comosus chloroplast genome. The thick line indicates the inverted repeats (IRa and IRb), which separate the genome into small single-copy (SSC) and large single-copy (LSC) regions. Genes on the outside of the map are transcribed in the clockwise direction and genes on the inside in the counterclockwise direction. Genes containing introns are marked with an asterisk
Simple sequence repeats
We identified 65 SSR regions with ≥10 repeated nucleotides (Table 2): 9 with dinucleotide repeat motifs of AT or TA, 23 A stretches (10–17 bases), and 33 T stretches (10–16 bases), but no C or G stretches. Of the 65 SSR regions, 46 were in intergenic spacers, 12 within introns, and 7 in gene-coding regions.
Phylogenetic analysis
Phylogenetic analyses were performed on an aligned data matrix of 62 angiosperm taxa and 76 protein-coding genes with a total length of 20,038 amino acids aligned. Bootstrap analysis indicated that 41 out of the 57 nodes were significantly supported (≥95 %; Fig. 2). The results were in good accordance with previously reported relationships among the major groups of vascular plants (Jansen et al. 2007). The analysis suggests that Ananas (Poales) shows a close phylogenetic relationship to Typha (Poales) and Musa (Zingiberales).
Comparison of the Ananas chloroplast genome with those of Typha and Musa
Dot-plot analysis showed similar gene order and organization in Ananas and Typha (Fig. 3a). It revealed three insertions and four deletions of >200 bp in Ananas (shown as breakpoints in Fig. 3a), all in intergenic spacer regions. Coding regions had only two deletions of >10 bp: 15 bp in accD and 24 bp in ycf2 (Table 3). No missense indels between Typha and Ananas were found.
Compared with Musa, however, there were four insertions and six deletions of >200 bp in Ananas (Fig. 3b). Three common indels in Ananas differed from those in Typha and Musa: insertions in rpoB–trnC_GCA and ndhF–rpl32 and a deletion in trnE_UUC–trnT_GGU. A large deletion of 7807 bp in the Ananas–Musa dot-plot shows as a large disconnection (Fig. 3b). Coding regions had 30 indels of >10 bp (Table 3). Two 5-bp missense indels were found in rpl16 and rps19 (Table 3). The 5-bp insertion in rpl16 at positions 6–10 from the 3′ end of the coding sequence in Ananas created a termination signal (TAG) 8 bp earlier than in Musa (Fig. 4). The 5-bp deletion in rps19 of Ananas corresponding to positions 2–6 in Musa changed the initiation codon (ATG) to GTG in Ananas (Fig. 5).
Figure 6 shows details of IR–SC border positions with respect to adjacent genes in Ananas, Typha, and Musa. Lengths of LSC, IR, and SSC were similar in Ananas and Typha. On the other hand, whereas lengths of LSC were similar in Ananas and Musa, IR of Ananas was 8659 bp shorter and SSC of Ananas was 7854 bp longer than those of Musa (Fig. 6). In Ananas, the IRa/SSC border occurred in the 3′ region of ycf1 and created a ycf1 pseudogene of 1089 bp. Typha showed a similar structure. In Musa, the IRa/SSC border occurred in the 3′ region of ndhA and created an ndhA pseudogene. In Ananas, Typha, and Musa, the IRa/LSC border occurred downstream of the noncoding region of psbA, and the IRb/LSC border occurred upstream of the noncoding region of rpl22.
Discussion
We determined the complete nucleotide sequence of the Ananas chloroplast genome using only next-generation sequencers instead of Sanger sequencing method. Since no perfect assembler program has been created so far, de novo assembly always generates misassembled contigs, and thus assembled contigs must be checked by read-mapping and be scanned for any gaps of lower coverage (Naito et al. 2013). Most of the errors corrected by the HiSeq 2500 sequencing were homopolymer stretches (data not shown), which are likely when the 454 GS FLX system is used (Gilles et al. 2011).
We identified 65 SSRs in the Ananas chloroplast genome. To date, chloroplast SSRs have been detected in Pinus radiata (Cato and Richardson 1996; Powell et al. 1995), Oryza sativa (Ishii et al. 2001), Panax ginseng (Kim and Lee 2004), Cucumis sativus (Kim et al. 2006), Vigna radiata (Tangphatsornruang et al. 2010), and Pyrus pyrifolia (Terakami et al. 2012). These SSRs can be useful in evolutionary studies because of their variability at the inter- and intrapopulation levels. We could not indicate phylogenetic data of Ananas for validation of SSRs here. Future research will need to focus on the validity of SSRs to phylogenetic and ecological studies of Ananas.
There has been a rapid increase in the number of studies using DNA sequences from completely sequenced chloroplast genomes for estimating phylogenetic relationships among angiosperms (Goremykin et al. 2005; Leebens-Mack et al. 2005; Bausher et al. 2006; Jansen et al. 2006, 2007; Ravi et al. 2006; Ruhlman et al. 2006). Our phylogenetic tree indicates a close relationship between Ananas and Typha with high bootstrap support (100 %). The phylogenetic tree identified Ananas as a basal member of the Poales, closer to Musa than to species of the Poaceae. These results are in good accordance with data revealed by phylogenetic methods based on the rbcL sequence (Bremer 2000).
The Ananas chloroplast genome structure is similar to that of Typha. Within the Poales, members of the Poaceae have a smaller chloroplast genome size, with several alterations such as large inversions in the LSC and indels, than that of Typha (Katayama and Ogihara 1996; Guisinger et al. 2010). The similar LSC, SSC, and IR sizes of Ananas to those of Typha and the absence of an inversion in the LSC of Ananas strongly indicate that Ananas and Typha are closely related among the Poales and are phylogenetically far from the Poaceae. On the other hand, the chloroplast genomes of Ananas and Musa show many structural differences. That of Ananas has an 8659-bp shorter IR and a 7854-bp longer SSC than that of Musa and is 10 kb smaller overall than that of Musa. Martin et al. (2013) suggested that the expansion of IRa to the SSC junction resulted in the incorporation of ycf1, rps15, ndhH, and ndhA in IRa of Musa. An idea that occurrence of deletion of IRb and the change of IRa to SSC in Ananas and Typha was not supported because such extreme IR expansion was not observed in other species and might have occurred independently only in the Musaceae (Martin et al. 2013).
Most indels of >200 bp between Ananas and Typha or Musa were located in intergenic spacer regions. Insertions in rpoB–trnC_GCA and ndhF–rpl32 and a deletion in trnE_UUC–trnT_GGU in Ananas appeared in both comparisons. Therefore, these large indels seem to be specific to Ananas. A large (291-bp) deletion from Musa occurred in the coding region of accD. The accD protein in most monocots is around 500 amino acids, for example, 491 in Phoenix (GenBank ID: ADF28155.1) and Cocos (AGS43475.1), 482 in Oncidium (ACT83118.1), and 489 in Lilium (AGQ55767.1). The Ananas accD comprised 488 amino acids, whereas the Musa accD comprised 599 (CCW72384.1). Thus, the length of the Ananas accD is consistent with that in other monocots, but the Musa accD is much longer, suggesting a Musa-specific DNA insertion. Indels in coding regions between Ananas and Musa occurred especially frequently in ycf1 and ycf2 (Table 3). ycf1 and ycf2 show a wide range of length variation among species and are absent in the Poaceae (Asano et al. 2004; Chang et al. 2006; Hiratsuka et al. 1989; Leebens-Mack et al. 2005; Maier et al. 1995; Ogihara et al. 2000). These results suggest that alterations to ycf1 and ycf2 are nonfatal, and that indels occur comparatively easily in ycf1 and ycf2. Missense indels in rpl16 and rps19 were found in Ananas. The insertion in the 3′ end of the rpl16 coding sequence seemed not to influence the protein function because the region is not conserved region among species. The deletion in rps19 changed the initiation codon, ATG, to GTG in Ananas. GTG occurs in rps19 in various seed plant species (Raubeson et al. 2007), and among the monocot species used in the phylogenic tree, only Musa has an ATG in rps19 (Asano et al. 2004; Chang et al. 2006; Hiratsuka et al. 1989; Leebens-Mack et al. 2005; Maier et al. 1995; Martin et al. 2013; Ogihara et al. 2000). Therefore, this alteration was not critical to protein function either.
The complete chloroplast nucleotide sequence of Ananas and the structural and sequence differences between Ananas and other species that we present here will contribute to ecological and evolutional studies.
References
Asano T, Tsudzuki T, Takahashi S, Shimada H, Kadowaki K (2004) Complete nucleotide sequence of the sugarcane (Saccharum officinarum) chloroplast genome: a comparative analysis of four monocot chloroplast genomes. DNA Res 11:93–99
Bausher MG, Singh ND, Mozoru J, Lee SB, Jansen RK, Daniell H (2006) The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var. ‘Ridge Pineapple’: organization and phylogenetic relationships to other angiosperms. BMC Plant Biol 6:21
Bremer K (2000) Early cretaceous lineages of monocot flowering plants. Proc Natl Acad Sci U S A 97:4707–4711
Cato SA, Richardson TE (1996) Inter- and intraspecific polymorphism at chloroplast SSR loci and the inheritance of plastids in Pinus radiata D. Don. Theor Appl Genet 93:587–592
Chang CC, Lin HC, Lin IP, Chow TY, Chen HH, Chen WH, Cheng CH, Lin CY, Liu SM, Chaw SM (2006) The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol Biol Evol 23:279–291
Conant GC, Wolfe KH (2008) GenomeVx: simple web-based creation of editable circular chromosome maps. Bioinformatics 24:861–862
Coppens d’Eeckenbrugge G, Leal F (2002) Morphology, anatomy and taxonomy. In: Bartholomew DP, Paull RE, Rohrbach KG (eds) The pineapple: botany, production and uses. CAB International, Wallingford, pp 13–32
Coppens d’Eeckenbrugge G, Leal F, Duval MF (1997) Germplasm resources of pineapple. In: Janick J (ed) Horticultural reviews, vol 21. John Wiley and Sons, Hoboken, pp 133–175
Duval MF, Noyer JL, Perrier X, Coppens d’Eeckenbrugge G, Hamon P (2001) Molecular diversity in pineapple assessed by RFLP markers. Theor Appl Genet 102:83–90
Duval MF, Buso GS, Ferreira FR, Noyer JL, Coppens d’Eeckenbrugge G, Hamon P, Ferreira ME (2003) Relationships in Ananas and other related genera using chloroplast DNA restriction site variation. Genome 46:990–1004
Gilles A, Meglécz E, Pech N, Ferreira S, Malausa T, Martin JF (2011) Accuracy and quality assessment of 454 GS-FLX Titanium pyrosequencing. BMC Genomics 12:245
Goremykin VV, Holland B, Hirsch-Ernst KI, Hellwig FH (2005) Analysis of Acorus calamus chloroplast genome and its phylogenetic implications. Mol Biol Evol 22:1813–1822
Guisinger MM, Chumley TW, Kuehl JV, Boore JL, Jansen RK (2010) Implications of the plastid genome sequence of Typha (Typhaceae, Poales) for understanding genome evolution in Poaceae. J Mol Evol 70:149–166
Hamdan N, Samad AA, Hidyat T, Salleh FM (2013) Phylogenetic analysis of eight Malaysian pineapple cultivars using a chloroplast marker (rcbL gene). J Teknol 64:29–33
Hepton A (2002) Culture system. In: Bartholomew DP, Paull RE, Rohrbach KG (eds) The pineapple: botany, production and uses. CAB International, Wallingford, pp 109–142
Hiratsuka J, Shimada H, Whittier R, Ishibashi T, Sakamoto M, Mori M, Kondo C, Honji Y, Sun CR, Meng BY et al (1989) The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet 217:185–194
Ishii T, Xu Y, McCouch SR (2001) Nuclear and chloroplast microsatellite variation in a genome species of rice. Genome 44:658–666
Jansen RK, Kaittanis C, Saski C, Lee SB, Tomkins J, Alverson AJ, Daniell H (2006) Phylogenetic analyses of Vitis (Vitaceae) based on complete chloroplast genome sequences: effects of taxon sampling and phylogenetic methods on resolving relationships among rosids. BMC Evol Biol 6:32
Jansen RK, Cai Z, Raubeson LA, Daniell H, dePamphilis CW, Leebens-Mack J, Muller KF, Guisinger-Bellian M, Haberle RC, Hansen AK et al (2007) Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A 104:19369–19374
Katayama H, Ogihara Y (1996) Phylogenetic affinities of the grasses to other monocots as revealed by molecular analysis of chloroplast DNA. Curr Genet 29:572–581
Kim KJ, Lee HL (2004) Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res 11:247–261
Kim JS, Jung JD, Lee JA, Park HW, Oh KH, Jeong WJ, Choi DW, Liu JR, Cho KY (2006) Complete sequence and organization of the cucumber (Cucumis sativus L. cv. Baekmibaekdadagi) chloroplast genome. Plant Cell Rep 25:334–340
Leebens-Mack J, Raubeson LA, Cui L, Kuehl J, Fourcade M, Chumley T, Boore JL, Jansen RK, dePamphilis CW (2005) Identifying the basal angiosperms in chloroplast genome phylogenies: sampling one’s way out of the Felsenstein zone. Mol Biol Evol 22:1948–1963
Maier RM, Neckermann K, Igloi GL, Kossel H (1995) Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J Mol Biol 251:614–628
Malézieux E, Côte F, Bartholomew DP (2002) Crop environment, plant growth and physiology. In: Bartholomew DP, Paull RE, Rohrbach KG (eds) The pineapple: botany, production and uses. CAB International, Wallingford, pp 69–108
Martin G, Baurens FC, Cardi C, Aury JM, D’Hont A (2013) The complete chloroplast genome of banana (Musa acuminata, Zingiberales): insight into plastid monocotyledon evolution. PLoS One 8:e67350
Naito K, Kaga A, Tomooka N, Kawase M (2013) De novo assembly of the complete organelle genome sequences of azuki bean (Vigna angularis) using next-generation sequencers. Breed Sci 63:176–182
Ogihara Y, Isono K, Kojima T, Endo A, Hanaoka M, Shiina T, Terachi T, Utsugi S, Murata M, Mori N et al (2000) Chinese spring wheat (Triticum aestivum L.) chloroplast genome: complete sequence and contig clones. Plant Mol Biol Rep 18:243–253
Paz EY, Gil K, Rebolledo L, Rebolledo A, Uriza D, Martinez O, Isidron M, Simpson J (2005) AFLP characterization of the Mexican pineapple germplasm collection. J Am Soc Hort Sci 130:575–579
Paz EY, Gil K, Rebolledo L, Rebolledo A, Uriza D, Martinez O, Isidron M, Diaz L, Lorenzo JC, Simpson J (2012) Genetic diversity of Cuban pineapple germplasm assessed by AFLP markers. Crop Breed Appl Biotech 12:104–110
Powell W, Morgante M, McDevitt R, Vendramin GG, Rafalski JA (1995) Polymorphic simple sequence repeat regions in chloroplast genomes: applications to the population genetics of pines. Proc Natl Acad Sci U S A 92:7759–7763
Raubeson LA, Peery R, Chumley T, Dziubek C, Fourcade HM, Boore JL, Jansen RK (2007) Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics 8:174
Ravi V, Khurana JP, Tyagi AK, Khurana P (2006) The chloroplast genome of mulberry: complete nucleotide sequence, gene organization and comparative analysis. Tree Genet Genomes 3:49–59
Rohrbach KG, Leal F, Coppens d’Eeckenbrugge G (2002) History, distribution and world production. In: Bartholomew DP, Paull RE, Rohrbach KG (eds) The pineapple: botany, production and uses. CAB International, Wallingford, pp 1–12
Ruhlman T, Lee SB, Jansen RK, Hostetler JB, Tallon LJ, Town CD, Daniell D (2006) Complete plastid genome sequence of Daucus carota: implications for biotechnology and phylogeny of angiosperms. BMC Genomics 7:224
Schwartz S, Zhang Z, Frazer K, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W (2000) PipMaker: a web server for aligning two genomic DNA sequences. Genome Res 10:577–586
Stamatakis A (2014) RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Tangphatsornruang S, Sangsrakru D, Chanprasert J, Uthaipaisanwong P, Yoocha T, Jomchai N, Tragoonrung S (2010) The chloroplast genome sequence of mungbean (Vigna radiata) determined by high-throughput pyrosequencing: structural organization and phylogenetic relationships. DNA Res 17:11–22
Terakami S, Matsumura Y, Kurita K, Kanamori H, Katayose Y, Yamamoto T, Katayama H (2012) Complete sequence of the chloroplast genome from pear (Pyrus pyrifolia): genome structure and comparative analysis. Tree Genet Genomes 8:841–854
West-Eberhard MJ, Smith JA, Winter K (2011) Photosynthesis, reorganized. Science 332:311–312
Wyman SK, Boore JL, Jansen RK (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255
Acknowledgments
This study was partially supported by an Okinawa special promotion grant.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. L. Wegrzyn
Data Archiving Statement
Ananas comosus chloroplast DNA complete sequence is submitted to DDBJ; the accession number of the sequence is AP014632. All the sequence data (.fastq files) were deposited in the DDBJ Sequence Read Archive (accession: DRA002476).
This article is part of the Topical Collection on Genome Biology
Rights and permissions
About this article
Cite this article
Nashima, K., Terakami, S., Nishitani, C. et al. Complete chloroplast genome sequence of pineapple (Ananas comosus). Tree Genetics & Genomes 11, 60 (2015). https://doi.org/10.1007/s11295-015-0892-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-015-0892-8