Abstract
Visual inspection of R-peaks in an electrocardiogram (ECG) signal may lead to wrong diagnosis due to physiological variability and the noisy status of the QRS complexes causing its incorrect interpretation. Hence, computer-aided diagnosis (CAD) is required for better and correct diagnosis of cardiovascular diseases and interpretation of essential clinical information. Low computational cost CAD systems with good accuracy are always preferred in health informatics. Presently, performance of most of the ECG signal analysis techniques such as feature extraction, classification etc. depends heavily on the used pre-processing technique, which may consume appreciable portion of the total processing time. Therefore, it can be inferred that if pre-processing technique is skipped altogether without compromising the detection accuracy of a diagnosis, then it would save central processing unit (CPU) time reducing the overall computational time. This time saving will turn out to be very important in critical and emergency situations to save lives of several patients. Hence, the authors were motivated to propose an efficient low computational cost method based on fractional Fourier transform (FrFT) as a feature extraction technique eliminating the need of any pre-processing. In other words, the proposed technique is applied directly on the raw ECG data resulting in computational savings. Firstly, eigenvalues and eigenfunctions are proposed to be obtained using FrFT. And secondly, these are proposed to be utilized for R-peak detection using Independent Principal Component Analysis (IPCA) on the basis of kurtosis and variance of the extracted features in a noisy ECG signal with different morphologies. The proposed methodology is evaluated on the basis of various performance parameters viz. sensitivity (SE), accuracy (ACC), and positive predictive value (PPV) (of the detected ECG beats). SE of 99.97%, PPV of 99.98%, and ACC of 99.94% are obtained on MIT-BIH Arrhythmia (MIT-BIH Arr) database. All simulations are done using Intel Core i3-3240 Dual-Core Processor 3.4 GHz and 8 GB of RAM using MATLAB R2011a. The average processing time of CPU is observed to be 0.677 s with detection error rate (DER) of 0.058%. Both these values are least among other existing techniques, which establish that the proposed method incurs low computational cost. Also, consistently high values of all the performance parameters such as SE, PPV and ACC demonstrates the robustness of the proposed technique. Hence, the proposed methodology is expected to assist cardiologists for intelligent, effective, and timely diagnosis of heart rhythm irregularities.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Cardiac medicine has evocative influence on professional, technological, preventive and computational advances. Automatic detection and diagnosis are essential initiatives for identifying different pathologies from an electrocardiogram (ECG), which has been a well-established fundamental diagnostic tool used by cardiologists for the assessment of cardiac arrhythmias in clinical routine [1, 2].
An ECG signal is a combination of three waves viz. P, QRS and T waves [3, 4]. Among these waves, accurate detection of R-peak (QRS complex) is essential for successful automated diagnosis and administration of various cardiovascular diseases. Unfortunately, any acquired ECG signal is generally corrupted by various types of noise/distortion, and therefore, has distinct QRS morphologies. Timely detection of cardiac abnormalities is very important for saving the life of the subjects (patients) [5], which fundamentally depends on correct observations of R-peaks (QRS complexes) [6, 7] in terms of their specific shape and time of occurrence. The heart specialists make use of amplitude, frequency, polarity of P-QRS-T waves for detecting the underlying disease [8]. The normal heart rate lies in the range of 60–100 beats/min. Below 60 beats per minute, it is called bradycardia and above 100 beats per minute, it is known as tachycardia [9, 10].
In view of the above discussion, it can be concluded that due to involvement of various noises/distortions at the time of acquisition of ECG records, effective computational medicine is essential for detecting heart diseases by using a computer aided diagnosis (CAD) system based on an efficient feature extraction algorithm [11, 12].
In the literature, different techniques for R-peaks detection have been reported under such conditions, but it is still a challenge. Several authors have been working on pre-processing of ECG signals, but processing time, proper selection of filter parameters, overlapping of noise and artifacts with QRS complexes, selection of the frequency range of QRS complexes and temperature limits the performance of these techniques. Until now various methods such as self-convolution window (SCW) concept [13], digital differentiator (DD) [13], wavelet transform (WT) [2, 14, 15], empirical mode decomposition (EMD) [16], median filter [17], signal derivatives [18], hilbert transform [19], hidden markov model [20], phase space method [21], first derivative (FD) [22], alexander fractional differential window filter (AFDWF) [23], stockwell transform (s-transform) [24], adaptive signal processing (ASP) [25], support vector machine (SVM) [26], second order difference plot (SODP) [27] and hilbert-huang transform (HHT) [28] etc. have been attempted for pre-processing of ECG signals. In [13], a novel ECG denoising algorithm based on the concept of SCW has been proposed. It relied on Hamming window to have smaller ripples in the stop band. In [22], FD based techniques were solely adopted in a real-time scheme for large databases. These techniques did not demand training of the computational algorithms but needed corrections according to an individual object (patient). Additionally, these offered low computational cost and were faster, but resulted in more detection errors due to presence of the high-frequency noises. These detection errors are not permissible in the present scenario of sophisticated health care assembly. In [16], EMD was considered for analyzing ECG records, but there the selection of a set of intrinsic mode functions (IMFs) was essential. In [27], SODP technique was used to extract HRV features of ECG for diagnosis of different cardiac disorders, but its performance was adversely affected in the presence of noisy ECG records. The methods proposed in [19, 20] are computationally complex. In [29], an optimally designed digital differentiator (DD) has been used for precise detection of the QRS complex. In that paper, Brownian Motion Optimization (BMO) was utilized as optimization algorithm. These techniques (DD and BMO) were utilized in the pre-processing stage of the QRS detector.
Traditional ECG signal analysis techniques relied mainly on the time domain approach, but needed careful attention to analyze all its attributes. However, information about its frequency components is also required for accurate analysis. In [2], an improved QRS complex detection algorithm based on wavelet transform (WT) was presented. But this approach relies on the choice of two important parameters such as wavelet basis function and decomposition level [30,31,32,33]. These two parameters affect the performance and effectiveness of wavelet based approach. Also, WT based methods suffer due to fringing effects and phase shift problems [34].
In view of the above ECG signal analysis problems, fractional Fourier transform (FrFT) is proposed in this paper, which can provide better time–frequency localization (i.e. mixed time and frequency components) in fractional domain on the basis of correctly chosen input parameters. The novelty of this paper is to detect the R-peaks directly in raw ECG signal without using any pre-processing/filtering technique. FrFT preserves all its clinical attributes to a significant extent. Next, IPCA is used for detecting R-peak by estimating eigenvalues and eigenvectors belonging to a new orthogonal basis [35]. In IPCA, largest of the estimated variance values helped in the detection of R-peaks resulting in identification of cardiac arrhythmias. Thus a new approach by combining FrFT with IPCA is explored for detecting R-peaks by calculating sensitivity (SE), positive predictive value (PPV), accuracy (ACC), and detection error rate (DER). These parameters are then analyzed to correctly predict the health condition of the heart. The final outcomes establish the application of FrFT combined with IPCA for R-peak detection for low signal-to-noise ratio (SNR), high baseline wander (BLW) and abnormal morphologies.
The whole paper has been organized as; Sect. 2 outlines details of the considered database and methodologies under materials and methods section. Section 3 showcases and discusses some important outcomes. Finally, Sect. 4 concludes the paper.
2 Materials and Methods
Details of the considered ECG database, used feature extraction & detection techniques and performance evaluating parameters are presented in this section.
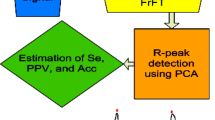
Figure 1 shows the methodology proposed in this paper.
2.1 ECG Database
Most of the researchers [2, 34, 36, 37] have used MIT-BIH Arrhythmia (MIT-BIH Arr) database for presenting their research outcomes. Therefore in this paper also, MIT-BIH Arr database is used to facilitate appropriate comparison with other existing works.
2.2 Feature Extraction Using Fractional Fourier Transform (FrFT)
Fractional Fourier transform (FrFT) has demonstrated its ability to be a splendid tool for handling the issues arising in the processing of a non-stationary signal among existing signal processing tools [38,39,40]. Mathematically, it is considered as a counter-clockwise rotation of u-axis with respect to the time axis by an angle α [40]. The value of α lies between \( 0 < \alpha < \frac{\pi }{2} \) and it depicts rotated time–frequency description of the signal as shown in Fig. 2. FrFT offers other advantages such as faster computations and better flexibility that are much needed in the domain of biomedical signal processing (BSP) [41].
Here, eigenvalues and eigenfunctions are calculated using the basic mathematical expressions of well-known Fourier transform as
where y(t) is the time domain ECG signal and Y(f) is its Fourier transform.
Hermite-Gauss functions (HGFs) are used for mathematical expression of eigenfunctions in FrFT [41], expressed as an orthonormal set by
where \( \emptyset_{{\rm n}} \left( {{\rm t}} \right) \), \( {{\rm H}}_{{\rm n}} \left( . \right) \) denotes orthonormal set and Hermite-Gauss function (HGF) for generalized sequence n, respectively.
It satisfies the standard eigenvalue equation as
where F denotes conventional Fourier transform and \( {\uplambda }_{{\rm n}} = {{\rm e}}^{{ - {{\rm jn}}\uppi/2}} \) denotes its eigenvalues, which are further modified for FrFT where all eigenvalues are expressed as \( {{\rm p}}{{\rm th}} \) power of the eigenvalues obtained from the conventional Fourier transform as
where eigenfunction \( {{\rm X}}_{{\rm n}} \) is related to time domain ECG signal, expressed as
Therefore, Eq. (4) is modified for FrFT as
The pth (p = 2α/π) order FrFT of an ECG signal y (t)∈L2 (R) is defined as [42]
where
where \( {{\rm K}}_{{\rm p}} \left( {{{\rm t}},{{\rm u}}} \right) \) denotes kernel function associated with FrFT of an ECG signal and \( {{\updelta }} \) denotes delta (impulse) function. The obtained eigenvalues from FrFT are further used in IPCA as discussed in the next subsections. IPCA visualizes the data effectively using less number of components as compared to those considered in ICA and PCA [43]. The crucial step of R-peak detection is completed in four steps as discussed in next subsection.
2.3 Independent Principal Component Analysis (IPCA)
Independent principal component analysis (IPCA) is used for R-peak detection since it possesses good characteristics of both principal component analysis (PCA) and independent component analysis (ICA) [44]. IPCA technique constitutes by applying firstly PCA using singular value decomposition (SVD) followed by ICA using joint approximate diagonalization of eigenmatrices (JADE) on the features extracted using FrFT in Sect. 2.2. IPCA is applied in the steps shown in Fig. 3 [45] and described as;
Step 1: PCA using Singular value Decomposition (SVD)
PCA is applied on extracted features obtained from FrFT i.e. \( {{\rm Y}}\left[ {{\rm i x j}} \right] \) to extract the loading vectors [46, 47]. Mathematically, it can be represented as
where \( {{\rm L}}\left( {{\rm l}} \right) \), D, \( {{\rm O}} \) denotes centered data matrix, diagonal matrix & orthogonal matrix, respectively and l denotes
R-peak locations.
Step 2: ICA using JADE algorithm
This algorithm is used on the basis of fourth order statistics to automatically filter out the background Gaussian noises. An original source signal is estimated using JADE algorithm. Additionally, it is more efficient as compared to Fast ICA [48]. The JADE algorithm is applied in the following steps;
-
(i)
Pre-whitening process
In this process source signal is assumed to have zero mean and unit variance. Mathematically, it can be expressed as
where Q denotes mixing matrix and x(t) denotes independent components.
-
(ii)
Fourth order cumulant process
The fourth order cumulant is expressed as
In (15), considered four signals are \( {{\rm y}}_{{\rm i}} ,{{\rm y}}_{{\rm j}} ,{{\rm y}}_{{\rm k}} ,{{\rm y}}_{{\rm n}} \) where \( 1 \le {{\rm i}},{{\rm j}},{{\rm k}},{{\rm n}} \le {{\rm N}} \)
where M and \( {{\rm m}}_{{\rm nk}} \) denotes arbitrary matrix and (n,k)th element, respectively, in fourth order cumulant process. The cumulant is expressed in terms of eigenvalues (\( {{\uplambda }}_{{\rm i}} ) \) as
-
(iii)
Diagonalization process
The diagonalization process is expressed as
where U is unitary matrix and Diag denotes approximate diagonalization of fourth order cumulant process.
-
(iv)
Calculation of Weight matrix (W)
The weight matrix is computed using
where U is unitary matrix and Q is mixing matrix.
Step 3: Projection of centered data matrix
In this step, mean of all independent components are calculated and subtracted from the respective components.
Here, four signals viz. \( ({{\rm y}}_{{\rm i}} - \overline{{{{\rm y}}_{{\rm i}} }} ), \) \( ({{\rm y}}_{{\rm j}} - \overline{{{{\rm y}}_{{\rm j}} }} ), \) \( ({{\rm y}}_{{\rm k}} - \overline{{{{\rm y}}_{{\rm k}} }} ), \) and \( ({{\rm y}}_{{\rm n}} - \overline{{{{\rm y}}_{{\rm n}} }} ), \) are projected.
Step 4: Estimation of kurtosis and variance values
In IPCA, kurtosis and variance values are used to choose number of independent components (ICs) and principal components (PCs), respectively [49]. The kurtosis (Ku) and variance (Va) for IPCA are given by-
where \( {{\rm y}}_{{\rm m}} \) denotes mean value of data, D denotes data size, and \( \sigma \) denotes standard deviation (calculated at N data points).
The proposed methodology has been evaluated on the basis of various parameters such as sensitivity, positive predictive value, accuracy, and detection error rate. These are defined in next subsection.
2.4 Performance Evaluating Parameters
The performance of R-peak detection in an ECG dataset has been evaluated on the basis of different parameters [26, 36, 50,51,52] defined as
where TP (True Positive) denotes correctly detected R-peaks, FN (False Negative) denotes number of R-peaks that were not detected by the proposed method, and FP (False Positive) denotes falsely detected R-peaks.
3 Results and discussion
R-peaks are detected using IPCA by estimating the eigenvalues and eigenvectors matrices alongwith variances. The average PC1 eigenvalue variance was 97.33%, which had a kurtosis value lying between 11.256 to 36.298 for the MIT-BIH Arr database. Analysis of an ECG signal heavily relies on variety of noises and artifacts that creeps in during its acquisition. Therefore, an efficient pre-processing stage is required under such circumstances which in-turn increases the computational cost of the automated ECG signal analysis further. In this paper, the proposed methodology skips the pre-processing stage and the whole operation has been performed on features extracted using FrFT from raw ECG signal followed by detection using IPCA. The raw ECG signal is shown in Fig. 4 and real & imaginary parts of FrFT are shown in Fig. 5 for 233 m and 215 m records. These datasets are selected to make suitable comparisons as most of the researchers have reported their results by considering them [3, 4, 26, 29, 46]. The peaks of the real & imaginary parts of FrFT lie at the centre (t = 4000 s.) of the duration of the recorded signal (t = 8000 s). This is similar to the characteristics of a sinc function for a rectangular input [39, 53]. Due to the noise offsets, average peak amplitudes of 87.77 mV and 329.4 mV are observed for real and imaginary FrFT parts, respectively for entire MIT-BIH Arr database. Figure 6 shows inverse FrFT where average peak amplitude of 347.77 mVolt is obtained for entire MIT-BIH Arr database. Here, it is observed that BLW noise/distortion still exists due to which even manual interpretation of R-peaks may result in wrong analysis. Therefore, IPCA is proposed to be used for R-peak detection in this paper. Finally, Fig. 7 shows detected R-peaks using IPCA for MIT-BIH Arr database (233 m and 215 m data sets) represented by light green plus sign. Using the proposed algorithm, TP of 3077 & 3376, FN of 1 &1, and FP of 0 &1 for 233 m and 215 m data sets, respectively are obtained.
Tables 1 and 2 summarize the findings and comparison with other state-of-the-art methods, respectively. For instance, obtained detection parameters for 105 m are TP = 2587, FN = 1 & FP = 1, and for 201 m are TP = 1971, FN = 0 & FP = 1. In [13, 54, 55], FN/FP values for 105 m data set were 5/4, 12/33, 0/1, respectively, and for 201 m data set, FN/FP values were 12/9, 5/0, 25/2 in that order. Table 2 shows comparison of the evaluation parameters viz. SE, PPV and ACC obtained by the proposed methodology with other existing techniques. It is observed that detection results in terms of these parameters are approximately 100%. This clearly indicates the effectiveness of the proposed methodology.
Table 3 and Table 4 present the comparison between proposed and existing methods on the basis of FN + FP for whole dataset and total beats, TP, (FN + FP), SE, PPV and ACC, respectively.
It has been observed that the proposed method performs better than existing methods and yields more accurate and fast results. This makes the proposed method a good candidate to be used as an effective tool for automatic ECG signal interpretation.
The existing techniques have SE of 99.29%, 99.90%, 99.39%, 99.80%, 99.93%, 99.90%, PPV of 99.89%, 99.10%, 99.49%, 99.90%, 99.95%, 99.88%, & ACC of 99.63%, 98.11%, 98.89%, 99.60%, 99.87%, 99.77% in Kaya and Pehlivan [56], Martis et al. [8], Qin et al. [57], Rai et al. [58], Gupta and Mittal [59] and Thakor and Zhu [51], respectively. The proposed technique has SE of 99.97%, PPV of 99.98%, & ACC of 99.94%. It is clearly observed that the proposed low computational cost CAD system results (due to eliminating the need of pre-processing altogether) are consistently higher as compared to those obtained with the existing methodologies, all of which depend upon pre-processing to produce the reported detection results. The main advantage of the proposed technique is its less detection error (FN + FP) as shown in Fig. 8. In the existing techniques, (FN + FP) of 500, 2100, 1229, 1774, 138, 249 in Kaya and Pehlivan [56], Martis et al. [8], Qin et al. [57], Rai et al. [58], Gupta and Mittal [59] and Thakor and Zhu [51], respecitlvey have been reported. In [8, 56], ACC is given but number of (FN + FP) is not mentioned, which is obtained by assuming TP of 1,10,000 using (24) for performing comparison in this paper. Rai et al. [58] have not mentioned total number of (FN + FP) in the whole database considered in their study, hence, it is scaled here for the purpose of comparison.
The average CPU processing time of 1.2 s, 0.872 s, 0.833 s, 13.7 s, 0.737 s, 0.677 s and DER of 0.44%, 1.11%, 0.49%, 0.163%, 0.125%, 0.058% have been reportedin Kaya and Pehlivan [56], Qin et al. [57], Nguyen et al. [60], Park et al. [61], Gupta and Mittal [59], and in the proposed technique, respectively. It can be observed from Fig. 9 that the proposed technique has least detection error rate and computational cost among all the techniques considered for comparison establishing its prowess.
In this paper, 17 real-time recordings are also examined using the proposed technique for validating this research work. These recordings are obtained at a sampling rate of 360 Hz using BIOPAC@MP36 equipment (@BIOPAC Systems, Inc.) [62] at NIT, Jalandhar, India. Different age-groups subjects volunteered and these records were then analyzed using BIOPAC Acqknowledge 4.0 software (@BIOPAC Systems, Inc.) [62]. Total 18,677 beats were captured in all 17 real time recordings. The results are verified by using the proposed technique. R-peaks are detected in terms of various detection parameters viz. TP of 18,669, FN of 5, FP of 4 yielding SE of 99.97%, PPV of 99.98% and ACC of 99.95%.
4 Conclusion
The proposed methodology has been implemented successfully on mostly used (in the literature) datasets of MIT-BIH Arr database to obtain SE of 99.97%, PPV of 99.98%, and ACC of 99.94%. The average processing time of CPU and detection error rate (DER) are 0.677 s and 0.058%, respectively for the proposed method. These results signify the effectiveness of the proposed method in analyzing the time-varying signals alongwith its low computational cost without compromising its accuracy. Hence, this method can be used for human stress related arrhythmia detection, in practical arrhythmia monitoring systems, in electronic cardiac pacemakers, for treatment of patients in intensive care units (ICU), to distinguish between similar cardiac arrhythmias even when the ECG signal is corrupted with noise and baseline wandering. This algorithm may also be utilized in intelligent remote health caring systems, telemedicine applications, cardiac arrhythmia measurement in varying R–R intervals/QRS complex (wide or narrow, large or small amplitudes, negative R peaks and highly noisy ECG signals) and mass screening for cardiac health. Due to low computational cost, the proposed technique (FrFT + IPCA) is likely to enhance the strength of real-time arrhythmia analysis system. It can speed up the diagnosis process without compromising with the accuracy. Finally, it may be concluded that the use of computerized analysis proposed in this paper can reduce workload of the cardiologists significantly.
As FrFT provides an alternative representation of an ECG signal in time fractional Fourier domain at selected rotation angles, it can find major applications in the domain of biomedical signal and image processing. In future, this work can be extended to use Fractional Wavelet Transform (FrWT) making best use of inherit multiresolution property of the wavelets along with its capability of representing the signal in the fractional domain similar to FrFT.
References
Marsanova, L., Ronzhina, M., Smíšek, R., Vítek, M., Němcová, A., Smital, L., et al. (2017). ECG features and methods for automatic classification of ventricular premature and ischemic heartbeats: A comprehensive experimental study. Scientific Reports, 7(1), 1–11. https://doi.org/10.1038/s41598-017-10942-6.
Bouaziz, F., Boutana, D., & Benidir, M. (2014). Multiresolution wavelet-based QRS complex detection algorithm suited to several abnormal morphologies. IET Signal Procesiing, 8(7), 774–782. https://doi.org/10.1049/iet-spr.2013.0391.
Zidelmal, Z., Amiroua, A., Ould-Abdeslamb, D., Moukademb, A., & Dieterlen, A. (2014). QRS detection using S-Transform and Shannon energy. Computers Methods and Programs in Biomedicine, 116(1), 1–9. https://doi.org/10.1016/j.cmpb.2014.04.008.
Sahoo, S., Biswal, P., Das, T., & Sabut, S. (2016). De-noising of ECG Signal and QRS Detection using Hilbert Transform and Adaptive Thresholding. Procedia Technology, 25, 68–75. https://doi.org/10.1016/j.protcy.2016.08.082.
Vimala, K. (2014). Stress causing Arrhythmia Detection from ECG Signal using HMM. International Journal of Innovative Research in Computer and Communication Engineering, 2(10), 6079–6085.
Chan, H. L., Chou, W. S., Chen, S., Fang, S. C., Liou, C. S., & Hwang, Y. S. (2005). Continuous and online analysis of heart rate variability. Journal of Medical Engineering & Technology, 29(5), 227–234. https://doi.org/10.1080/03091900512331332587.
Rabbani, H., Mahjoob, M. P., Farahabadi, E., & Farahabadi, A. (2011). R Peak detection in electrocardiogram signal based on an optimal combination of wavelet transform, hilbert transform, and adaptive thresholding. Journal of Medical Signals and Sensors, 1(2), 91–98.
Martis, R. J., Acharya, U. R., Mandana, K. M., Ray, A. K., & Chakraborty, C. (2012). Application of principal component analysis to ECG signals for automated diagnosis of cardiac health. Expert Systems with Applications, 39(14), 11792–11800.
Chandra, S., Sharma, A., & Singh, G. K. (2018). Feature extraction of ECG signal. Journal of Medical Engineering & Technology, 42(4), 306–316. https://doi.org/10.1080/03091902.2018.1492039.
Cromwell, L., Weibell, F. J., & Pfeiffer, E. A. (1980). Biomedical instrumentation and measurements (2nd ed.). Englewood Cliffs (NJ): Prentice Hall.
Sharma, A., Patidar, S., Upadhyaya, A., & Acharya, U. R. (2019). Accurate tunable-Q wavelet transform based method for QRS complex detection. Computer & Electrical Engineering, 75, 101–111. https://doi.org/10.1016/j.compeleceng.2019.01.025.
Phukpattaranont, P. (2015). QRS detection algorithm based on the quadratic filter. Expert Systems with Applications, 42(11), 4867–4877. https://doi.org/10.1016/j.eswa.2015.02.012.
Kaur, A., Agarwal, A., Agarwal, R., & Kumar, S. (2018). A Novel Approach to ECG R-Peak Detection. Arabian Journal for Science and Engineering. https://doi.org/10.1007/s13369-018-3557-8.
Leong, C. I., Mak, P. I., Lam, C. P., Dong, C., Vai, M. I., Mak, P. U., et al. (2012). A 0.83 μW QRS detection processor using quadratic spline wavelet transform for wireless ECG acquisition in 0.35 m CMOS. IEEE Transactions on Biomedical Circuits and Systems, 6(6), 586–595. https://doi.org/10.1109/TBCAS.2012.2188798.
Yochum, M., Renaud, C., & Jacquir, S. (2016). Automatic detection of P, QRS and T patterns in 12 leads ECG signal based on CWT. Biomedical Signal Processing and Control, 25, 46–52. https://doi.org/10.1016/j.bspc.2015.10.011.
Li, H., Wang, X., Chen, L., & Li, E. (2014). Denoising and R-Peak Detection of Electrocardiogram Signal Based on EMD and Improved Approximate Envelope. Circuits, Systems, and Signal Processing, 33(4), 1261–1276. https://doi.org/10.1007/s00034-013-9691-3.
Acharya, U. R., Sankaranarayanan, M., Nayak, J., Xiang, C., & Tamura, T. (2008). Automatic identification of cardiac health using modeling techniques: A comparative study. Information Sciences, 178(23), 4571–4582. https://doi.org/10.1016/j.ins.2008.08.006.
Bahoura, M., Hassani, M., & Hubin, M. (1997). DSP implementation of wavelet transform for real time ECG waveforms detection and heart rate analysis. Computers Methods and Programs in Biomedicine, 52(1), 35–44. https://doi.org/10.1016/s0169-2607(97)01780-x.
Nygåards, M. E., & Sörnmo, L. (1983). Delineation of the QRS complex using the envelope of the ECG. Medical & Biological Engineering & Computing, 21(5), 538–547. https://doi.org/10.1007/BF02442378.
Trahanias, P. E. (1993). An approach to QRS complex detection using mathematical morphology. IEEE Transactions on Biomedical Engineering, 40(2), 201–205. https://doi.org/10.1109/10.212060.
Plesnik, E., Malgina, O., Tasic, J. F., & Zajc, M. (2012). Detection of the electrocardiogram fiducial points in the phase space using the Euclidian distance measure. Medical Engineering & Physics, 34(4), 524–529. https://doi.org/10.1016/j.medengphy.2012.01.005.
Ning, X., & Selesnick, I. W. (2013). ECG enhancement and QRS detection based on sparse derivatives. Biomedical Signal Processing and Control, 8(6), 713–723. https://doi.org/10.1016/j.bspc.2013.06.005.
Verma, A. K., Saini, I., & Saini, B. S. (2018). Alexander fractional differential window filter for ECG denoising. Australasian Physical and Engineering Sciences in Medicine, 41(2), 519–539. https://doi.org/10.1007/s13246-018-0642-y.
Das, M., & Ari, S. (2013). Analysis of ECG signal denoising method based on S-transform. Innovation and Research in Biomedical Engineering (IRBM), 34(6), 362–370. https://doi.org/10.1016/j.irbm.2013.07.012.
Meireles, A.J.M.D. (2011). ECG Denoising based on Adaptive Signal Processing Technique. M.Tech dissertation, Porto, Portugal.
Mehta, S. S., & Lingayat, N. S. (2008). SVM-based algorithm for recognition of QRS complexes in electrocardiogram. Innovation and Research in Biomedical Engineering (IRBM), 29(5), 310–317. https://doi.org/10.1016/j.rbmret.2008.03.006.
Altan, G., Kutlu, Y., & Yeniad, M. (2019). ECG Based Human Identification Using Second Order Difference Plots. Computers Methods and Programs in Biomedicine, 170, 81–93. https://doi.org/10.1016/j.cmpb.2019.01.010.
Zhang, C., Li, X., & Zhang, M. (2010). A novel ECG signal denoising method based on Hilbert-Huang Transform. In IEEE conference on Computer and Communication Technologies in Agriculture Engineering (CCTAE), China (pp. 284-287). https://doi.org/10.1109/CCTAE.2010.5544365.
Nayak, C., Saha, S. K., Kar, R., & Mandal, D. (2018). An efficient QRS complex detection using optimally designed digital differentiator. Circuits, Systems, and Signal Processing, 38(5), 716–749. https://doi.org/10.1007/s00034-018-0880-y.
Dinh, A. N., Kumar, D. K., Pah, N. D., & Burton, P. (2002). Wavelet for QRS detection. Proceedings of the IEEE, 2, 1883–1887. https://doi.org/10.1109/IEMBS.2001.1020593.
Chen, S. W., Chen, C. H., & Chan, H. L. (2006). A real-time QRS method based on moving-averaging incorporating with wavelet denoising. Computers Methods and Programs in Biomedicine, 82(3), 187–195. https://doi.org/10.1016/j.cmpb.2005.11.012.
Dokur, Z., & Ölmez, T. (2001). ECG beat classification by a novel hybrid neural network. Computers Methods and Programs in Biomedicine, 66(3), 167–181. https://doi.org/10.1016/S0169-2607(00)00133-4.
Alyasseri, Z. A. A., Khader, A. T., Betar, M. A. A., & Awadallah, M. A. (2018). Hybridizing β-hill climbing with wavelet transform for denoising ECG signals. Information Sciences, 429, 229–246. https://doi.org/10.1016/J.INS.2017.11.026.
Sunkaria, R. K., Saxena, S. C., Kumar, V., & Singhal, A. M. (2010). Wavelet based R-peak detection for heart rate variability studies. Medical Engineering & Technology, 34(2), 108–115. https://doi.org/10.3109/03091900903281215.
Chawla, M. P. S. (2008). Segment Classification of ECG data and Construction of Scatter Plots Using Principal Component Analysis. Journal of Mechanics in Medicine and Biology, 8(3), 421–458. https://doi.org/10.1142/S0219519408002681.
Rodrígueza, R., Mexicanob, A., Bilac, J., Cervantesd, S., & Ponce, R. (2015). Feature extraction of electrocardiogram signals by applying adaptive threshold and principal component analysis. Journal of Applied Research and Technology, 13(2), 261–269. https://doi.org/10.1016/j.jart.2015.06.008.
Zidelmal, Z., Amirou, A., Adnane, M., & Belouchrani, A. (2012). QRS detection based on wavelet coefficients. Computers Methods and Programs in Biomedicine, 107(3), 490–496. https://doi.org/10.1016/j.cmpb.2011.12.004.
Lin, Q., Ran, T., Siyong, Z., & Yue, W. (2004). Detection and parameter estimation of multicomponent LFM signal based on the fractional Fourier transform. Science in China Series F: Information Sciences, 47(2), 184–198. https://doi.org/10.1360/02yf0456.
Ozaktas, H. M., Zalevsky, Z., & Kutay, M. A. (2001). The fractional fourier transform with applications in optics and signal processing (1st ed.). New York: Wiley.
Daamouche, A., Hamami, L., Alajlan, N., & Melgani, F. (2012). A wavelet optimization approach for ECG signal classification. Biomedical Signal Processingand Control, 7(4), 342–349. https://doi.org/10.1016/j.bspc.2011.07.001.
Lin, P. Y. (1999). The fractional fourier transform and its applications. Taipei: National Taiwan University.
Zayed, A. I. (1996). On the relationship between the Fourier and Fractional Fourier Transforms. IEEE Signal Processing Letters, 3(12), 310–311. https://doi.org/10.1109/97.544785.
Mix-Omics (2020). Omics Data Integration Project-Independent Principal Component Analysis. Resource Document. http://mixomics.org/methods/ipca/.
Yao, F., Coquery, J., & Cao, K. A. L. (2012). Independent Principal Component Analysis for biologically meaningful dimension reduction of large biological data sets. BMC Bioinformatics, 13(24), 1–15. https://doi.org/10.1186/1471-2105-13-24.
Boluwade, A., & Madramootoo, C. A. (2016). Independent principal component analysis for simulation of soil water content and bulk density in a Canadian Watershed. International Soil and Water Conservation Research, 4(3), 151–158. https://doi.org/10.1016/j.iswcr.2016.09.001.
Kaur, H., & Rajni, R. (2017). On the detection of Cardiac Arrhythmia with Principal Component Analysis. Wireless Personal Communications, 97, 5495–5509. https://doi.org/10.1007/s11277-017-4791-1.
Chawla, M. P. S., Verma, H. K., & Kumar, V. (2008). A new statistical PCA–ICA algorithm for location of R-peaks in ECG. International Journal of Cardiology, 129(1), 146–148.
Zhang, K., Tian, G., &Tian, L. (2015). Blind source separation based on JADE algorithm and application. In Proceedings of 3rd International Conference on Mechatronics, Robotics and Automation, Atlantis Press, (Vol. 15, pp. 252-255). https://doi.org/10.2991/icmra-15.2015.50.
Gupta, V., & Mittal, M. (2019). A Comparison of ECG signal pre-processing using FrFT, FrWT and IPCA for improved analysis. Innovation and Research in Biomedical Engineering (IRBM), 40(3), 145–156. https://doi.org/10.1016/j.irbm.2019.04.003.
Shia, H., Wanga, H., Zhanga, F., Huanga, Y., Zhaob, L., & Liu, C. (2019). Inter-patient heartbeat classification based on region feature extraction and ensemble classifier. Biomedical Signal Processing and Control, 51, 97–105. https://doi.org/10.1016/j.bspc.2019.02.012.
Thakor, N. V., & Zhu, Y. S. (1991). Applications of Adaptive Filtering to ECG Analysis: Noise Cancellation and Arrhythmia Detection. IEEE TransactionsonBiomedical Engineering, 38(8), 785–794. https://doi.org/10.1109/10.83591.
Gu, X., Hu, J., Zhang, L., Ding, J., & Yan, F. (2020). An improved method with high anti-interference ability for R peak detection in wearable devices. Innovation and Research in Biomedical Engineering (IRBM), 41(3), 172–183. https://doi.org/10.1016/j.irbm.2020.01.002.
Mendlovic, D., & Ozaktas, H. M. (1993). Fractional Fourier transforms and their optical implementation. Journal of the Optical Society of America A, 10(9), 1875–1881. https://doi.org/10.1364/JOSAA.10.001875.
Barhatte, A., Dale, M., & Ghongade, R. (2019). Cardiac events detection using curvelet transform. Sadhana., 44(47), 1–10. https://doi.org/10.1007/s12046-018-1046-0.
Hussein, E. A. R., Hassooni, A. S., & Al-Libawy, H. (2019). Detection of electrocardiogram QRS complex based on modified adaptive threshold. International Journal of Electrical and Computer Engineering, 9(5), 3512–3521. https://doi.org/10.11591/ijece.v9i5.pp3512-3521.
Kaya, Y., & Pehlivan, H. (2015). Classification of premature ventricular contraction in ECG. International Journal of Advanced Computer Science and Applications (IJACSA), 6(7), 34–40. https://doi.org/10.14569/ijacsa.2015.060706.
Qin, Q., Li, J., Yue, Y., & Liu, C. (2017). An adaptive and time- efficient ECG R-Peak detection algorithm. Journal of Healthcare Engineering, 17, 1–14. https://doi.org/10.1155/2017/5980541.
Rai, H.M., Trivedi, A., Chatterjee, K., & Shukla, S. (2014). R-Peak Detection using Daubechies Wavelet and ECG Signal Classification using Radial Basis Function Neural Network. Journal of The Institution of Engineers (India): Series B, 95: 63–71. https://doi.org/10.1007/s40031-014-0073-4.
Gupta, V., Mittal, M., & Mittal, V. (2020). Detection of R-peaks using Fractional Fourier Transform and Principal Component Analysis. Innovation and Research in Biomedical Engineering (IRBM), in press. https://www.sciencedirect.com/journal/irbm.
Nguyen, T., Qin, X., Dinh, A., & Bui, F. (2019). Low resource complexity R-peak detection based on triangle template matching and moving average filter. Sensors, 19(18), 1–17. https://doi.org/10.3390/s19183997.
Park, J. S., Lee, S. W., & Park, U. (2017). R peak detection method using wavelet transform and modified shannon energy envelope. Journal of Healthcare Engineering, 17, 1–14. https://doi.org/10.1155/2017/4901017.
Biopac Systems. (2020). The Premier Data Acquisition & Analysis Program. Resource Document. https://www.biopac.com/wp-content/uploads/AcqKnowledge-Products.pdf.
Afonso, V. X., Tompkins, W. J., Nguyen, T. Q., & Luo, S. (1999). ECG beat detection using filter banks. IEEE Transactions on Biomedical Engineering, 46(2), 192–202. https://doi.org/10.1109/10.740882.
Hamilton, P. (2002). Open source ECG analysis. Computers in Cardiology, 29, 101–104.
Kaya, Y., & Pehlivan, H. (2015). Feature selection using genetic algorithms for premature ventricular contraction classification. In IEEE conference on Electrical and Electronics Engineering, Turkey (pp. 1229-1232). https://doi.org/10.1109/ELECO.2015.7394628.
Kaya, Y., Pehlivan, H., & Tenekeci, M. E. (2017). Effective ECG beat classification using higher order statistic features and genetic feature selection. Biomedical Research, 28(17), 7594–7603.
Pan, J., & Tompkins, W. J. (1985). A Real-Time QRS Detection Algorithm. IEEE Transactions on Biomedical Engineering, 32(3), 230–236. https://doi.org/10.1109/tbme.1985.325532.
Liu, X., Yang, J., Zhu, X., Zhou, S., Wang, H., & Zhang, H. (2014). A novel R-Peak Detection Method Combining Energy and Wavelet Transform in Electrocardiogram Signal. Biomedical Engineering: Applications, Basis and Communications, 26(1), 1–9. https://doi.org/10.4015/S1016237214500070.
Kim, J., Shin, H. S., Shin, K., & Lee, M. (2009). Robust algorithm for arrhythmia classification in ECG using extreme learning machine. BioMedical Engineering OnLine, 8(31), 1–12. https://doi.org/10.1186/1475-925X-8-31.
Aqil, M., Jbari, A., & Bourouhou, A. (2016). Adaptive ECG Wavelet analysis for R-peaks Detection In IEEE conference on Electrical and Information Technologies, Tangiers, Morocco (pp. 1–4). https://doi.org/10.1109/EITech.2016.7519582.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gupta, V., Mittal, M. & Mittal, V. An Efficient Low Computational Cost Method of R-Peak Detection. Wireless Pers Commun 118, 359–381 (2021). https://doi.org/10.1007/s11277-020-08017-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11277-020-08017-3