Abstract
The process of biofilm formation is intricate and multifaceted, requiring the individual cells to secrete extracellular polymeric substances (EPS) that subsequently aggregate and adhere to various surfaces. The issue of biofilms is a significant concern for public health due to the increased resistance of microorganisms associated with biofilms to antimicrobial agents. The current study describes the whole genome and corresponding functions of a biofilm inhibiting and eradicating actinobacteria isolate identified as Nocardiopsis lucentensis EMB25. The N. lucentensis EMB25 has 6.5 Mbp genome with 71.62% GC content. The genome analysis by BLAST Ring Image Generator (BRIG) revealed it to be closely related to Nocardiopsis dassonvillei NOCA502F. Interestingly, based on orthologous functional groups reflected by average nucleotide identity (ANI) analysis, it was 81.48% similar to N. arvandica DSM4527. Also, it produces lanthipeptides and linear azole(in)e-containing peptides (LAPs) akin to N. arvandica. The secondary metabolite search revealed the presence of major gene clusters involved in terpene, ectoine, siderophores, Lanthipeptides, RiPP-like, and T1PKS biosynthesis. After 24 h of treatment, the cell-free extract effectively eradicates the pre-existing biofilm of P. aeruginosa PseA. Also, the isolated bacteria exhibited antibacterial activity against MRSA, Staphylococcus aureus and Bacillus subtilis bacteria. Overall, this finding offers valuable insights into the identification of BGCs, which contain enzymes that play a role in the biosynthesis of natural products. Specifically, it sheds light on the functional aspects of these BGCs in relation to N. lucentensis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The global spread of antimicrobial resistance (AMR) is widely recognized as a significant challenge in healthcare and disease management. The World Health Organization has warned that the rise of multidrug-resistant bacteria is putting the effectiveness of antibiotics in jeopardy. Antibiotics have played a crucial role in advancing medical science, but their usefulness is now being threatened. The involvement of biofilms in antimicrobial resistance (AMR) is a crucial factor that substantially contributes to the development of resistance (Costerton et al. 2003). The thick layer of the extracellular polymeric substance (EPS) could potentially hinder the effectiveness of antibiotics that try to penetrate biofilms through diffusion-reaction inhibition. This could occur through the antibiotics binding with EPS to form complexes, or through enzymatic degradation (Billings et al. 2015; Lahiri et al. 2019; Sharma et al. 2019). Therefore, finding biofilm inhibitors that can inhibit the bacterial biofilm mode of AMR could be an important scientific venture. Although chemical synthesis has significantly contributed to antimicrobial compounds, nature remains the richest and primary gateway for discovering new antimicrobial compounds (Samrot et al. 2021).
Actinobacteria are a valuable source of bioactive compounds with antibiotic properties that are highly significant in the pharmaceutical industry (De Simeis & Serra. 2021). Actinobacteria of terrestrial origin have been essential in the quest for novel antibiotics for more than half a centuary. However, the possibility of re-discovering the same compounds presents a significant limitation (Chen et al. 2021). Lately, there has been a lot of interest in screening marine actinobacteria due to their abundance and their potential for producing unique bioactive compounds with diverse structures and properties (Eliwa et al. 2017).
Nocardiopsis is a genus that holds significant importance in natural product research within the pharmaceutical and biotechnological industries (Hamed et al. 2018). It is an aerobic, Gram-positive, and halo-tolerant actinobacteria. These organisms can be discovered in a variety of habitats such as terrestrial areas as epiphytes or endophytes, desert soils, and even marine environments. They are typically found in environments that range from moderate to hypersaline (Bennur et al. 2016). In order to survive in harsh environments, they have the ability to produce various extremozymes, surfactants, compatible solutes, and bioactive compounds (Chen et al. 2021).
In recent years, it has become clear that the genus Nocardiopsis has a lot of potential for producing therapeutic leads. This is due to their many different chemotypes (Hadj Rabia-Boukhalfa et al. 2017). There are currently almost 22 sequenced genomes from the Nocardiopsis genus that have been submitted to NCBI from different marine and terrestrial environments (Eliwa et al. 2017). To predict and discover new secondary metabolites with different frameworks efficiently, it’s best to use genomic screening and chemical investigation through chromatographic methods on microbial strains like Nocardiopsis (Ibrahim et al. 2018).
Nocardiopsis is responsible for producing around 3% of marine actinomycetes compounds, making it the third largest contributor among actinomycetes. Studies on this genus have found a variety of secondary metabolites with potential bioactivity. These include dopsisamine (Takahashi et al. 1986), lucentamycins A–D (Cho et al. 2007), griseusin D (Li et al. 2007), nocapyrones A–D (Schneemann et al. 2010), nocazoline A (Fu et al. 2011), nocapyrones H–J (Kim et al. 2013), and nocardiopsins A and B (Wu et al. 2013). Recent studies have also discovered other compounds that may have bioactive properties (Shi et al. 2022).
Our previous study concentrated on N. lucentensis EMB25’s ability to prevent biofilm formation in P. aeruginosa strains by analyzing the organic extract and docking identified compounds with RhlR and LasR (Goel et al. 2023). In this study, we have performed the genomic investigation of the N. lucentensis EMB25 strain, the first biofilm-inhibiting and eradicating strain. This data has the potential to generate new knowledge in various areas related to medicinal chemistry. These areas may include, the identification and validation of new drug targets in human pathogens, and the discovery of new chemical entities.
Materials and methods
The strain
The bacteria was isolated from salt pan, Pen, Maharashtra, India (18.7358°N, 73.0947° E) and identified as N. lucentensis EMB25 by 16S rRNA sequencing. The gene sequence was submitted to NCBI GenBank with the accession number MW582546.1. Currently, our laboratory maintains the strain as EMB25 for continued research and analysis.
The other test pathogenic strains used in the study are following: Gram-positive (Staphylococcus aureus ATCC 23235, MRSA ATCC 43300, Bacillus subtilis, a lab isolate) and Gram-negative bacteria (P. aeruginosa PseA MTCC 10634).
Extraction of genomic DNA of N. lucentensis EMB25
A single colony of N. lucentensis EMB25 was inoculated in International Streptomyces Project-2 Medium (ISP-2) broth and incubated at 30 °C. The cells were harvested after 7 days of incubation by centrifuging at 8000 x g for 10 min. The pellets were suspended, and genomic DNA was isolated using the Quick-DNA Fungal/Bacterial Miniprep Kit (Zymo Research, USA). The extraction process was carried out in accordance with the manufacturer’s instructions. The quality and quantity were accessed using agar gel electrophoresis and Nanodrop 2000 (Thermo Scientific, USA).
Carbon source utilization by N. lucentensis EMB25
Carbon source utilization was investigated using KB009 TM HiCarbo Kit (Himedia, India). Inoculation and interpretation of the results were made as per the guidelines of the kit.
Scanning electron microscopy
The morphological characteristics of N. lucentensis EMB25 were recorded through scanning electron microscopy (SEM) following the protocol described by Ali et al. (2021). Images were acquired and processed using FESEM (FEI Company, Netherland, Model: FEI Quanta 200 F SEM).
Whole genome sequencing
The whole genome sequencing was performed using Illumina sequencing and the GridION X5, the long-read platforms at Genotypic Technology, Bangalore, India.
The Illumina library preparation was carried out using the QIASeq FX DNA kit as per the instructions mentioned by the manufacturer. Briefly, enzymatically fragmentation of genomic DNA, end-repairment and a-tailing, all three reactions were simultaneously carried out utilizing the FX Enzyme Mix followed by adapter ligation to generate sequencing libraries. The obtained libraries were then subjected to Indexing-PCR and final extension to enrich the adapter-tagged fragments. After the purification of generated libraries, sequencing was carried out on the Illumina HiSeq X Ten sequencer for 150 cycles.
The genomic DNA was purified, end-paired, and cleaned up, and native barcode ligation was done using NEB blunt/ TA ligase to prepare the library for nanopore sequencing. The DNA was quantified, and adaptors were ligated for 15 min using NEBNext Quick Ligation Module and cleaned up using 0.6X AmPure beads. Finally, the library prepared to be sequenced was eluted in 15 μL of elution buffer. Using a SpotON flow cell R9.4 (FLO-MIN106), a 48-hour sequencing technique was used on the GridION X5. Guppy v2.3.4 was used to base-call and de-multiplex nanopore raw reads (‘fast5’ format).
Data analysis and genome comparison
The generated raw reads were assembled using a unicycler hybrid assembler generated using SPAdes 3.6.2 (Wick et al. 2017) from both Illumina and nanopore platforms. The assembled genome sequence was annotated using the NCBI Prokaryotic Genomes Annotation Pipeline (PGAP) v5.2 (Tatusova et al. 2016). The predicted genes were subjected to pathway analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa 2000). The genomic comparison was carried out using BLAST Ring Image Generator (BRIG) v0.95, which uses CGview to generate the circular image and BLAST for genome similarity comparison (Alikhan et al. 2011) and Orthologous Average Nucleotide Identity Tool (OAT) v0.93.1 analysis tool, which uses only orthologous fragments to calculate ANI values to generate heatmap (Lee et al. 2016).
RAST annotation and secondary metabolites prediction
RAST (Rapid Annotation using Subsystem Technology), which was released in 2007, is a fully annotated service for full or almost complete bacterial and archaeal genomes. The genome sequence is submitted either in FASTA or Genbank format to the RAST server. The server takes around 12–24 h to annotate the genome. In the current study, version 2.0 was used to annotate the genome, and for further comparison with other genomes, the SEED platform was used, also to know the subsystem category distribution (Overbeek et al. 2014). Furthermore, anti-SMASH (version 7.0), an online database, was used to predict the secondary metabolites in N. lucentensis EMB25. The genome was run to the web based online tool to identify BGCs encoding secondary metabolites (Blin et al. 2023). Based on the findings, inferences were drawn regarding the gene sequence and percent identity. In addition, BActeriocin GEnome mining tooL (BAGEL4), a web service, was used to identify Ribosomally synthesised and Post Translationally Modified Peptides (RiPPs) and antimicrobial peptides (bacteriocins) (van Heel et al. 2018).
Antibacterial activity
The antibacterial activity was performed against Gram-positive (Staphylococcus aureus, MRSA, Bacillus subtilis) and Gram-negative bacteria (Pseudomonas aeruginosa PseA) using agar-plug assay (Ortlieb et al. 2021). The actinobacteria EMB25 was streaked tightly on ISP2 agar media and incubated at 30 °C for seven days. Afterwards, test microorganisms were spread onto nutrient agar media, and a well is then cut to add EMB25 bacteria after sporulation. The plates were incubated at 30 °C for 24 h, and the zone of inhibition was measured. In addition, disc diffusion assay was performed against S. aureus and MRSA with the organic extract prepared as mentioned by Goel et al. (2023).
Biofilm eradication activity of N. lucentensis EMB25 cell-free extract and confocal microscopy
The biofilm-forming bacteria, P. aeruginosa PseA was used to explore the biofilm eradication potential of N. lucentensis EMB25. After 24 h of biofilm formation, cell-free extract of N. lucentensis EMB25 was added in various volumes (50, 100, 150 and 200 μL) and incubated at 30 °C for 24 h. Thereafter, biofilm quantification was done using crystal violet (CV) assay as described by Balasubramanian et al. (2017), and cell counts were performed to check cell viability. Along with the CV assay, confocal microscopy was performed, as mentioned by Goel et al. (2023). The adherent cells control and test (150 μL) were stained with the Filmtracer™ LIVE/DEAD™ Biofilm Viability Kit (Invitrogen, ThermoFisher Scientific, USA) and visualized under the confocal microscope (Laser Scanning Confocal Microscope: Leica TCS SP8, Germany).
Results
Biochemical characterization and morphology study of N. lucentensis EMB25
The N. lucentensis EMB25 exhibited a wide range of qualitative carbon source utilization. Of 35 biochemical tests, 32 expressed positive, and 3 expressed negative results. It grew optimally on lactose, xylose, fructose, dextrose, sucrose, and inositol. It was cable of utilizing some of the other carbon sources viz. maltose, galactose, trehalose, melibiose, mannose, adonitol, arabitol, erythritol, alpha-Methyl-D-glucoside, but with relatively less growth. However, it failed to grow on raffinose, al-arabinose, and malonate (Supplementary Table S1).
The SEM images of N. lucentensis EMB25 grown on ISP2 agar clearly show the typical actinobacteria-like hyphae network (Fig. 1).
Sequencing, assembly, and genomic comparisons
N. lucentensis EMB25 genome is of linear size 6.5 Mbp with 71.6% GC content. Out of 5775 protein-coding sequences, 5393 (93.38%) proteins were annotated using the PROKKA tool (Seemann 2014). The assembly statistics of the obtained sequences are summarized in Table 1. Genomic comparison analysis revealed that N. lucentensis EMB25 isolate is closely related to the Nocardiopsis dassonvillei NOCA502F (NZ_CP017965.1) (Supplementary Figure S1), which is known to be active against Gram-positive bacteria Bacillus subtilis, yeast Candida albicans and produces enzymes protease, β-galactosidase, and glucosidase (Yu et al. 2017).
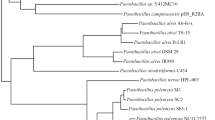
The orthoANI values for each genome comparison with the N. lucentensis EMB25 are mentioned in Supplementary Table S2. The maximum value is 81.48 inferring the highest similarity with the Nocardiopsis arvandica DSM45278 based on orthologous groups match (Fig. 2).
Genomic prediction and genome annotation
Gene ontology (GO) annotation resulted in the top 5 abundant protein functions: Membrane protein, TetR family transcriptional regulator, Non-specific serine/threonine protein kinase (EC 2.7.11.1), Transcriptional regulator, TetR family, ABC transporter related protein clustered into 10 biological, 10 molecular and 5 cellular branches (Supplementary Figure S2).
The Kyoto Encyclopedia of Genes and Genomes (KEGG) database interpreted that the genes are mainly involved in 27 pathway functions, of which 49 genes are associated with the pathways of metabolism of terpenoids and polyketides responsible for antibiotic synthesis (Fig. 3), namely ansamycins (K00615), enediyne (K20420), Siderosphore (K12240), vancomycin group antibiotics (K16437), along with other potential antibiotics. In addition, 37 genes belong to the pathways of biosynthesis of other secondary metabolites such as novobiocin (K00812), monobactam (K00215), penicillin, cephalosporin (K17836), acarbose, validamycin (K19978), prodigiosin (K00059), streptomycin (K01092), Neomycin, kanamycin and gentamicin (K00845), and carbapenem (K00147) (Supplementary Table S3).
RAST annotation and secondary metabolites prediction
The online antiSMASH database predicted total 24 secondary BGCs regions for NRPS, T1PKS, CDPS, terpene, lanthipeptide class i, iii, iv, RiPP-like, NI-siderophore, LAP, thiopeptide, melanin and ectoine. Region 4 and 6 belongs to terpene and lanthipeptide class-iii, which have shown 100% similarity with isorenieratene and SapB, respectively. Isorenieratene is a carotenoid, and SapB is a peptide used in aerial mycelium formation. Xanthobaccin C was first isolated from Stenotrophomonas sp. strain SB-K88 and is an antifungal drug that shows 50% similarity and is associated with region 16. Neocarzinostatin shows 50% similarity and is an antitumor antibiotic produced by Streptomyces sp. that mediates strand breaking of DNA. There are many drugs that are produced such as Siamycin, mathermycin, duramycin, neocarzilin A/neocarzilin B, jerangolid A/jerangolid D, streptamidine, nonactin/monactin/dinactin/trinactin/tetranactin, azetidomonamide A/azetidomonamide B, 4-hydroxy-3-nitrosobenzamide, grixazone A, ferroverdin/bagremycin, Rosamicin, carrimycin and ectoine (Table 2).
The SEED viewer environment provided the subsystems classification of the encoded proteins, and only 19% comes under subsystem category distribution. As per RAST annotation, 48 genes are involved in stress response, 19 genes in the production of secondary metabolites and 40 genes in virulence diseases and defence mechanisms (Fig. 4).
BAGEL4 identified lanthipeptides class i and iv, SapB, and lactococcin 972. Lactococcin 972 inhibits cell division by preventing the incorporation of septum cell wall precursor and is the only non-lantibiotic that does not target the cytoplasmic membrane (Martínez et al. 2008).
Bioactivity of cell-free extract of N. lucentensis EMB25
The agar-plug assay showed the antibacterial activity against Gram-positive bacteria, namely S. aureus, MRSA, and B. subtilis. However, no activity was observed against Gram-negative P. aeruginosa PseA (Fig. 5A). The organic extract was used for disc diffusion assay and resulted antibacterial activity at 40 and 120 μg/mL against S. aureus and MRSA respectively (Supplementary Figure S3).
Furthermore, the effect of N. lucentensis EMB25 was investigated for eradication of the preformed biofilm and cell viability (log CFU/mL) (Fig. 5B). The extract eradicated the biofilm in a dose-dependent manner and thereby reduced the viable cell count. In order to validate the above observations, confocal microscopy was performed (Fig. 5C). Two dyes SYTO 9 and Propidium iodide (PI), were used to stain the biofilm. SYTO 9 is used to visualize the live cells and gives green color, while PI stains dead cells and appears red in color. The result (Fig. 5C) clearly shows a significant reduction in biofilm and proportionate killing of bacteria colonizing and inhabiting within the biofilm. The N. lucentensis EMB25 secretes potential bioactives that can be used in AMR therapeutics.
Discussion
The N. lucentensis EMB25 was isolated from Salt pan, Pen, Maharashtra, India. It is a rare actinobacteria possessing the capability to produce novel bioactive compounds. We have previously published a study that provides in-depth information regarding its isolation and bioactivity profile (Goel et al. 2023). The preference for carbon source and carbohydrate utilization of the isolate has been studied. It was discovered that monosaccharides were the preferred source for the growth of cell biomass (Supplementary Table S1). However, it is well-documented in the literature that complex carbon sources often stimulate the production of secondary metabolites. Moreover, studies have shown that starch is the most effective substrate for the production of antibiotics (Jonsbu et al. 2002). Hence, the carbon source utilization profile of Nocardiopsis sp. could be instrumental in enhancing the production rate of secondary bioactive compounds.
Our previous study conducted by Goel et al. in 2023 delved into the metabolic profile of N. lucentensis EMB25. We discovered a range of intriguing compounds that have the potential to inhibit and eliminate mature biofilms of P. aeruginosa. The BRIG tool shows that N. lucentensis EMB25 is closely related to Nocardiopsis dassonvillei NOCA502F based on BLAST search, which finds local regions of similarity and can be filtered to minimum percentage identity or E-cut value. The secondary metabolites profile revealed more BGCs in N. lucentensis EMB25 than Nocardiopsis dassonvillei NOCA502F, and three metabolites ectoine, isorenieratene and streptamidine are produced by both Nocardiopsis strains. Streptamidine is an amidine-containing peptide first reported to be produced by Streptomyces albidoflavus J1074. The biological significance of streptamidine remains unknown. However, the presence of streptamidine and streptamidine-like BGCs implies the role in the development and signalling of organisms among various terrestrial actinobacteria (Russell et al. 2021). Here, we have identified the streptamidine BGC presence in rare actinobacteria, Nocardiopsis species.
Microorganisms develop suitable solutes like ectoines to survive in environments with wildly varying osmolarities. Ectoines shield the DNA from ionising radiation, aid in lipid bilayer stabilization, shield protein function from a variety of stresses, and protect the bacterial cell from high temperatures (Goel et al. 2022; Salvador et al. 2018). However, based on the orthoANI values, the query genome is related to N. arvandica DSM4527. Among various overall genome relatedness indices (OGRI) algorithms developed, orthoANI is the most widely used for comparing the genomes and is most accurate. The orthoANI values obtained clearly indicate that N. lucentensis EMB25 has orthoANI values less than 85% of other closely related Nocardiopsis species inferring the difference and how closely related to the existing genome. Both N. lucentensis EMB25 and N. arvandica DSM4527 produces Lanthipeptide class i and LAPs inferring from the results obtained after BAGEL4 analysis (Supplementary Table S3). Lanthipeptides are post-translationally modified peptides which has antimicrobial properties and can be used as preservatives, probiotics, additives in cosmetics, and prophylactics. Class-i lanthipeptides are synthesized by two different enzymes: LanB (dehydratase enzyme) and LanC (cyclase enzyme). N. arvandica DSM4527 contains both domains whereas N. lucentensis EMB25 has only dehydratase domain. Lanthipeptides are post-translationally modified peptides which has antimicrobial properties and can be used as preservatives, probiotics, additives in cosmetics, and prophylactics (Bothwell et al. 2021).
In this study, we aim to uncover the genetic signatures of enzymes in biosynthetic pathways through genome mining. Specifically, our focus is on identifying and predicting the genes responsible for producing bioactive compounds in the marine rare actinobacteria N. lucentencis EMB25. The genetic basis behind production of bioactive compounds opens the door to further discovery and provides an unparalleled opportunity to thoroughly examine the diversity and inner workings of life (Bauman et al. 2021). Furthermore, the Nocardiopsis genus is a valuable natural resource that merits exploration for the production of novel bioactive compounds with diverse structures and significant pharmaceutical applications (Bennur et al. 2015). The rich biosynthetic potential of this strain could be instrumental to predict promising molecules which could not be synthesized in laboratory settings. Additionally, there is a lack of studies on whole genome sequencing for this genus, particularly for this specific species. It is crucial to shift the research community’s attention toward the potential utility of this genus for drug discovery, particularly in the fight against drug-resistant bacteria. This is essential due to the increasing threat of antimicrobial resistance, which is slowly becoming a silent pandemic.
Data Availability
The bacterial genome of N. lucentensis EMB25 has been submitted to NCBI database with accession number: JAMTDN000000000, Bioproject: PRJNA729595 and Biosample: SAMN19135856.
References
Ali R, El-Boubbou K, Boudjelal M (2021) An easy, fast and inexpensive method of preparing a biological specimen for scanning electron microscopy (SEM). Methods X 8:101521
Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA (2011) BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402
Balasubramanian S, Othman EM, Kampik D, Stopper H, Hentschel U, Ziebuhr W, Oelschlaeger TA, Abdelmohsen UR (2017) Marine Sponge-Derived Streptomyces sp. SBT343 extract inhibits staphylococcal biofilm formation. Front Microbiol 8
Bauman KD, Butler KS, Moore BS, Chekan JR (2021) Genome mining methods to discover bioactive natural products. Nat Prod Reps 38:2100–2129
Bennur T, Ravi Kumar A, Zinjarde SS, Javdekar V (2015) Nocardiopsis species: incidence, ecological roles and adaptations. Microbiol Res 174:33–47
Bennur T, Ravi Kumar A, Zinjarde SS, Javdekar V (2016) Nocardiopsis species: a potential source of bioactive compounds. J Appl Microbiol 120:1–16
Billings N, Birjiniuk A, Samad TS, Doyle PS, Ribbeck K (2015) Material properties of biofilms—a review of methods for understanding permeability and mechanics. Rep Prog Phys 78:036601
Blin K, Shaw S, Augustijn HE, Reitz ZL, Biermann F, Alanjary M, Fetter A, Terlouw BR, Metcalf WW, Helfrich EJ, van Wezel GP (2023) antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res 4:gkad344
Bothwell IR, Caetano T, Sarksian R, Mendo S, van der Donk WA (2021) Structural analysis of Class I Lanthipeptides from Pedobacter lusitanus NL19 reveals an unusual Ring Pattern. ACS Chem Biol 16:1019–1029
Chen J, Xu L, Zhou Y, Han B (2021) Natural Products from Actinomycetes Associated with Marine Organisms. Mar Drugs 19:629
Cho JY, Williams PG, Kwon HC, Jensen PR, Fenical W (2007) Lucentamycins A-D, cytotoxic peptides from the marine-derived actinomycete Nocardiopsis lucentensis. J Nat Prod 70:1321–1328
Costerton W, Veeh R, Shirtliff M, Pasmore M, Post C, Ehrlich G (2003) The application of biofilm science to the study and control of chronic bacterial infections. J Clin Investig 112:1466–1477
De Simeis D, Serra S (2021) Actinomycetes: a never-ending source of bioactive compounds—An overview on antibiotics production. Antibiotics 10:483
Eliwa EM, Abdel-Razek AS, Frese M, Wibberg D, Halawa AH, El-Agrody AM, Bedair AH, Kalinowski J, Sewald N, Shaaban M (2017) New bioactive compounds from the marine-derived actinomycete Nocardiopsis lucentensis sp. ASMR2. Z Naturforsch B 72:351–360
Fu P, Liu P, Qu H, Wang Y, Chen D, Wang H, Li J, Zhu W (2011) Α-pyrones and diketopiperazine derivatives from the marine-derived actinomycete Nocardiopsis dassonvillei HR10-5. J Nat Prod 74:2219–2223
Goel N, Singh R, Sood S, Khare SK (2022) Investigation of Streptomyces sp. Strain EMB24 secondary Metabolite Profile has unraveled its extraordinary antibacterial potency against drug-resistant Bacteria. Mar Biotechnol 24:1168–1175
Goel N, Ghosh M, Jain D, Sinha R, Khare SK (2023) Inhibition and eradication of Pseudomonas aeruginosa biofilms by secondary metabolites of Nocardiopsis lucentensis EMB25. RSC Med Chem 14:745–756
Hadj Rabia-Boukhalfa Y, Eveno Y, Karama S, Selama O, Lauga B, Duran R, Hacène H, Eparvier V (2017) Isolation, purification and chemical characterization of a new angucyclinone compound produced by a new halotolerant Nocardiopsis sp. HR-4 strain. World J Microbiol Biotechnol 33:126
Hamed A, Abdel-Razek AS, Frese M, Stammler HG, El-Haddad AF, Ibrahim TMA, Sewald N, Shaaban M (2018) Terretonin N: a New Meroterpenoid from Nocardiopsis sp. Molecules 23(2):299
Ibrahim AH, Desoukey SY, Fouad MA, Kamel MS, Gulder TAM, Abdelmohsen UR (2018) Natural product potential of the Genus Nocardiopsis. Mar Drugs 16:147
Jonsbu E, McIntyre M, Nielsen J (2002) The influence of carbon sources and morphology on nystatin production by Streptomyces noursei. J Biotechnol 95:133–144
Kanehisa M (2000) KEGG: Kyoto Encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30
Kim MC, Kwon OW, Park JS, Kim SY, Kwon HC (2013) Nocapyrones H-J, 3,6-disubstituted α-pyrones from the marine actinomycete Nocardiopsis sp. KMF-001. Chem Pharm Bull 61:511–515
Lahiri D, Dash S, Dutta R, Nag M (2019) Elucidating the effect of anti-biofilm activity of bioactive compounds extracted from plants. J Biosci 44:52
Lee I, Ouk Kim Y, Park SC, Chun J (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103
Li YQ, Li MG, Li W, Zhao JY, Ding ZG, Cui XL, Wen M (2007) Griseusin D, a new pyranonaphthoquinone derivative from a alkaphilic Nocardiopsis sp. J Antibiot 60:757–761
Martínez B, Böttiger T, Schneider T, Rodríguez A, Sahl HG, Wiedemann I (2008) Specific Interaction of the unmodified bacteriocin lactococcin 972 with the Cell Wall Precursor lipid II. Appl Environ Microbiol 74:4666–4670
Ortlieb N, Klenk E, Kulik A, Niedermeyer THJ (2021) Development of an agar-plug cultivation system for bioactivity assays of actinomycete strain collections. PLoS ONE 16:e0258934
Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R (2014) The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–214
Russell AH, Vior NM, Hems ES, Lacret R, Truman AW (2021) Discovery and characterisation of an amidine-containing ribosomally-synthesised peptide that is widely distributed in nature. Chem Sci 12:11769–11778
Salvador M, Argandoña M, Naranjo E, Piubeli F, Nieto JJ, Csonka LN, Vargas C (2018) Quantitative RNA-seq analysis unveils osmotic and thermal adaptation mechanisms relevant for Ectoine Production in Chromohalobacter salexigens. Front Microbiol 9:1845
Samrot AV, Abubakar Mohamed A, Faradjeva E, Si Jie L, Hooi Sze C, Arif A, Chuan Sean T, Norbert Michael E, Yeok Mun C, Xiao Qi N, Ling Mok P, Kumar SS (2021) Mechanisms and impact of Biofilms and Targeting of Biofilms using Bioactive Compounds-A Review. Med (Kaunas Lithuania) 57:839
Schneemann I, Ohlendorf B, Zinecker H, Nagel K, Wiese J, Imhoff JF (2010) Nocapyrones A-D, gamma-pyrones from a Nocardiopsis strain isolated from the marine sponge Halichondria panicea. J Nat Prod 73:1444–1447
Seemann T (2014) Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069
Sharma D, Misba L, Khan AU (2019) Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob Resist Infect Control 8:1–10
Shi T, Wang YF, Wang H, Wang B (2022) Genus Nocardiopsis: a prolific producer of Natural Products. Mar Drugs 20:374
Takahashi A, Hotta K, Saito N, Morioka M, Okami Y, Umezawa H (1986) Production of novel antibiotic, dopsisamine, by a new subspecies of Nocardiopsis mutabilis with multiple antibiotic resistance. J Antibiot 39:175–183
Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624
van Heel AJ, de Jong A, Song C, Viel JH, Kok J, Kuipers OP (2018) BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res 46:W278–W281
Wick RR, Judd LM, Gorrie CL, Holt KE (2017) Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol 13:e1005595
Wu ZC, Li S, Nam SJ, Liu Z, Zhang C (2013) Nocardiamides A and B, two cyclohexapeptides from the marine-derived actinomycete Nocardiopsis sp. CNX037. J Nat Prod 76:694–701
Yu Y, Zhao B, Wang L, Liao L, Song L, Wang X, Liu G (2017) Complete genome of Nocardiopsis dassonvillei strain NOCA502F isolated from sediment of the Arctic Ocean. Mar Genom 34:27–29
Acknowledgements
The first author (NG) would like to acknowledge the financial assistance provided by the Ministry of Human Resource Development (MHRD), Govt. of India, and IIT Delhi. SZ is grateful to CSIR for providing the Senior research fellowship.
Author information
Authors and Affiliations
Contributions
Nikky Goel: Conceptualization, Methodology, Formal analysis, writing original draft; Saniya Zaidi: Methodology, Formal analysis, writing; Sunil K. Khare: Conceptualization, Formal analysis, Supervision, Review MS.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Goel, N., Zaidi, S. & Khare, S.K. Whole genome sequencing and functional analysis of a novel biofilm-eradicating strain Nocardiopsis lucentensis EMB25. World J Microbiol Biotechnol 39, 292 (2023). https://doi.org/10.1007/s11274-023-03738-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-023-03738-6