Abstract
Recent developments in the legume rhizobium symbiotic interaction particularly those related to the emergence of novel strains of bacteria that nodulate and fix nitrogen in legumes is gaining momentum. These novel strains of bacteria were mostly isolated from the root nodules of indigenous and invasive legumes belonging to the sub families Papilionoideae and Mimosoideae in South Africa, South America and South East China. These rhizobia are phylogenetically and taxonomically different from the traditional ‘alpha rhizobia’ and are termed ‘β-rhizobia’ as they belong to the β-sub class of Proteobacteria. There are also new reports of novel species of root nodulating bacteria from the α-Proteobacteria, not known for several decades since the discovery of rhizobia. However, in this review focus is given to the emerging β-rhizobia isolated from the indigenous Papilionoid legumes in the Cape Floristic regions in South Africa and the indigenous and invasive Mimosoid legumes in South America and South East Asia respectively. The nodulation of the indigenous South African Papilionoid legumes including that of Aspalathus linearis (rooibos) is discussed in a bit detail. Previous reports indicated that A. linearis is very specific in its rhizobium requirement and was reported to be nodulated by the slow growing Bradyrhizobium spp. This review however summarizes that the bacteria associated with the root nodules of A. linearis belong to members of both the alpha (α) Proteobacteria that include Mesorhizobium, Rhizobium and Bradyrhizobium spp. and the beta (β) Proteobacteria represented by the genus Burkholderia (now reclassified as Paraburkholderia). In addition, the occurrence of Paraburkholderia as the newly emerging root nodule symbionts of various other legumes has been discussed. In doing so, the review highlights that nodulation is no longer restricted to the traditional ‘rhizobia’ group following the emergence of the new beta rhizobia as potential nodulators of various indigenous legumes. It thus provides some insights on the status of the legume–rhizobium host specificity concept and the loss of this specificity in several symbiotic associations that puts the long held dogma of host specificity of the legume rhizobium symbiosis in a dilemma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
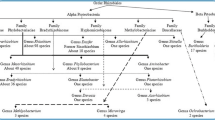
Ever since Frank (1889) had first described the bacterial genus Rhizobium, the legume rhizobium symbiotic interaction has been studied for almost 130 years now owing to its huge significance in sustainable agricultural productivity, soil fertility and ecological benefits. Until 27 years ago, the bacteria that are involved in nodulating the majority of legume species were all classified under a single genus Rhizobium (Gyaneshwar et al. 2011). A breakthrough in bacterial phylogenies based on sequences of the conserved small sub unit of the 16S ribosomal RNA changed the ‘one genus Rhizobium’ concept (Young and Haukka 1996). This resulted in the division of the rhizobia into the well-known genera Rhizobium, Bradyrhizobium, Ensifer (Syn: Sinorhizobium), Mesorhizobium, Allorhizobium and Azorhizobium all belonging to the alpha subclass of Proteobacteria (alpha-proteobacteria) (Graham 2008; Willems 2006; Velazquez et al. 2010). Due to the ecological significance and economic importance, the rhizobia-legume symbiosis has since been studied extensively and currently, the genera that contains the root nodule forming bacteria has risen to 14 (Deng et al. 2011). The new inclusions are the genera Methylobacterium (Sy et al. 2001), Devosia (Rivas et al. 2002), Ochrobactrum (Ngom et al. 2004), and Shinella (Li et al. 2008) that belong to the Alphaproteobacteria and Burkholderia (Moulin et al. 2001) and Cupriavidus (Chen et al. 2001) from the Betaproteobacteria (Fig. 1). The newly discovered rhizobia particularly those belonging to the beta-subclass of Proteobacteria are the subject of this review that will briefly discuss some recent studies, which disclosed the emergence of new group of bacteria as root nodulating symbionts of certain indigenous and exotic legumes.
Phylogenetic tree of Proteobacteria constructed from the 16S rRNA gene sequences. The genera containing rhizobia are indicated in bold (adapted from Barreda and Fikri-Benbrahim 2014 with permission)
Overview on the rhizobium-legume host specificity
Host specificity refers to the phenomenon whereby a given rhizobium species or strain nodulates (i.e. its ability to form root nodules) only a specific or limited number of legume species. Few famous examples of such high host specificity include the symbiosis between Rhizobium leguminosarum (bv. trifolii) and Trifolium spp. and the symbiosis between Rhizobium etli (bv. phaseoli) and Phaseolus vulgaris (Jiao et al. 2015). Other examples of host specificity include Mesorhizobium muleine CCBAU83963 for Cicer arietinum (Zhang et al. 2012) and Sinorhizobium meliloti for only three genera: Medicago, Melilotus and Trigonella (Roche et al. 1996; Albrecht et al. 1999). The mutualistic interaction between the two symbiotic partners results in the formation of a new plant organ, the root nodule. The rhizobia within nodules (which now become the bacteroides) convert atmospheric nitrogen into ammonia, a form that can be directly used by the legume plant (Wang et al. 2012). However, prior to the formation of nodules and the subsequent biological process that converts atmospheric nitrogen into a usable form, the legume and its micro symbiont need to establish a successful interaction. This interaction requires a signal recognition between the two symbiotic partners, which signifies that symbiotic specificity evolves because of the interaction of both the rhizobia and the host legume genes (Wang et al. 2012, 2018).
The first step in the legume rhizobium symbiosis is a ‘molecular dialogue’ between the two partners which starts after the legume secretes special molecules called flavonoids that immediately diffuse across the bacterial membrane. Moments after the bacteria perceives the flavonoid signals, it results in the activation of the bacterial nodulation (nod) genes, which encode the enzyme needed for the synthesis of the rhizobial Nod factors. These are lipochito-oligosaccharides with four or five 1,4-linked N-acetyl-glucosamine residues that carry a fatty acyl chain of varying length attached to the C-2 position of the non-reducing end and various species-specific chemical decorations at both the reducing and non-reducing ends (Wang et al. 2018). Generally, Nod factors are the signal molecules essential for initiating symbiotic development in the legumes and are required for root hair curling, induction of nodulation and entry of the rhizobia into the roots (Pueppke and Broughton 1999; Oldroyd et al. 2011). Only a particular type of Nod factor allows a given rhizobial strain to nodulate a certain legume host, giving rise to the first rhizobial determinant of host specificity (Fauvart and Michiels 2008). A typical example could be the variation in the Nod factors between Sinorhizobium meliloti, a root nodulating nitrogen fixing symbiont of Medicago spp. and Rhizobium leguminosarum bv. viciae a nitrogen fixing symbiont of Vicia faba (Fig. 2). These two rhizobia spp. have different legume host preference for nodulation mainly due to the variation in the nod factors, which differ at the reducing terminal sugar unit and by the structure of the acyl chain. S. meliloti Nod factors are characterized by four glucosamine units and an acyl chain of 16 C-atoms in length with two unsaturated bonds, an acetyl group at the non-reducing end and a sulfate group at the reducing terminal end in the nod factor. On the other hand, the R. leguminosarum nod factor consists of a mixture of factors that contain several compounds, and the length of the glucosamine backbone is four or five units that carry an acyl chain of 18 C-atoms with either one or four unsaturated bonds (Albrecht et al. 1999).
Nod factor variation in Sinorhizobium meliloti (a) and Rhizobium leguminosarum bv viciae (b) that determine host specificity for nodulation. The two Nod factors differ at the reducing terminal sugar unit and by the structure of the acyl chain. The S. meliloti Nod factor contains four glucosamine units, an acyl chain of 16 C-atoms in length with two unsaturated bonds, an acetyl group at the non-reducing end and a sulfate group at the reducing terminal sugar residue. In contrast, R. leguminosarum bv Vicia Nod factor contains glucosamine backbone of four or five units carrying an acyl chain of 18 C-atoms with either one or four unsaturated bonds which could be O-acetylated at the non-reducing terminal sugar residue (adapted from Albrecht, the EMBO Journal with permission)
The idea of Nod factors being the major determinants in the rhizobium-legume host specificity is still in debate. In a report published by Perret et al. (2000), the molecular basis of host specificity and symbiotic promiscuity has been dealt with in detail. The report, supported with few evidences, states that the Nod factors produced by rhizobia and the legumes they nodulate have no strict correlation. For instance, although the Nod factors produced by Rhizobium etli and Rhizobium loti (now Mesorhizobium loti) are identical, the two rhizobia have distinct host ranges in which M. loti nodulates Lotus spp. whereas R. etli is specific to Phaseolus spp. (Cardenas et al. 1995). In yet another example, R. leguminosarum bv. viciae produce Nod factors which are the same as that of R. leguminosarum bv. trifolii, but the two rhizobium biovars nodulate different legume hosts (Orgambide et al. 1995; Spaink et al. 1995).
On the other hand, several recent investigations back up the idea that Nod factors are still the major determinants in legume host specificity where each rhizobium produces a particular mix of Nod factors characterized by chemical substitutions at certain positions that give specificity to the signalling (Wang et al. 2012, 2018; Clua et al. 2018). In a decade long study since 1990, a concerted research by scientists that involved microbiologists, geneticists, plant cell biologists, biochemists and analytical chemists from known universities globally resulted in the isolation and characterization of Nod factors from several species of Rhizobium (Long 2001). The outcome of this work showed that diverse Rhizobium species produce Nod factors with similar chito-oligo backbones, but different side chains that include sugars, acetyl or carbamyl residues and modified lipids. This difference in the side chains of the Nod factors provides host specificity in the legume nodulation.
Nodulation of legumes by both alpha and beta rhizobia
First report of legume nodulation by β-rhizobia
For almost more than a century, it was believed that all legumes are nodulated only by Rhizobium and its relatives in the alpha sub class of Proteobacteria such as Bradyrhizobium, Ensifer (Sinorhizobium) species. However, over the last 20 years, sufficient evidence has accumulated that indicate the emergence of new members of root nodulating bacteria belonging to the genera, Methylobacterium, Devosia, Ochrobactrum, Shinella, Burkholderia and Cupriavidus. The latter two genera fall into the sub class of β-Proteobacteria and are collectively termed as ‘beta-rhizobia’ (Moulin et al. 2001; Gyaneshwar et al. 2011). In many parts worldwide, unlike the food legumes soybean (Glycine max L.), cowpea (Vigna unguiculata (L.) Walp, common beans (Phaseolus vulgaris L.) to mention a few, the symbiotic properties of several other indigenous legumes not directly used for human consumption have not been investigated thoroughly. A few examples include the indigenous South African legumes such as Aspalathus, Cyclopia, Lotononis, Desmodium as well as the medicinally important Crotalaria species, which still warrant such studies to elaborate on their symbiotic properties. Although recent reports on the nodulation properties of Cyclopia and Aspalathus are gaining momentum recently, there is still a big gap in the elucidation of the diversity of the rhizobia and their taxonomic and phylogenetic relatedness, which warrants more work in this regard. For instance, members of the genus Aspalathus including the popular A. linearis have been considered to be nodulated by the slow growing Bradyrhizobium species until recently. It was repeatedly reported that members of the genus Aspalathus are specific in their Bradyrhizobium requirements (Staphorst and Strijdom 1975; Deschodt and Strijdom 1976). These reports were however based solely on cultural and growth properties of the bacteria without the availability of any molecular data (Hassen et al. 2012).
Moulin et al. (2001) was the first to disclose the occurrence of nodulation of legumes by members of the β-subclass of Proteobacteria. In this pioneer discovery, the researchers showed that the ability of rhizobia to establish a symbiotic interaction with legumes is more widespread than has been anticipated to date. The study revealed that two bacteria isolated from the nodules of Aspalathus (strain STM678) and Machaerium (strain STM815) were taxonomically found to be very distant from known rhizobia. For quite a long time before this discovery, strain STM678 originally isolated from the South African Aspalathus carnosa was thought to belong to the slow growing Bradyrhizobium species (Boone et al. 1999; Dakora 1998). Nevertheless, the study by Moulin et al. (2001) using phylogenetic analysis of the gene sequences of the 16S ribosomal RNA showed that the strains do not belong to any of the major branches of rhizobia described and not even to the α-subclass of Proteobacteria, but rather belong to the β-subclass of Proteobacteria. Further phylogenetic analysis of partial sequence of the 23S rRNA gene and the dnaK gene encoding the chaperon heat shock protein were consistent with the 16S rRNA analysis that unambiguously positioned these strains in the Burkholderia genus within the β-subdivision of Proteobacteria.
The discovery of nodulating bacteria other than the traditional rhizobia belonging to the Alphaproteobacteria prompted extensive work on the phylogeny and taxonomy of the newly nodulating strains of β-Proteobacteria. This resulted in the description of the two new nodulating strains as Burkholderia tuberum STM678 and Burkholderia phymatum STM815 (Vandamme et al. 2002). Later, Chen et al. (2003) reported that legume nodulation and nitrogen fixation by the β-Proteobacteria is widespread in nature. Following the pioneer study in 2001, research in the past two decades has accumulated ample evidences worldwide about the nodulation and nitrogen fixation of native and exotic legumes by the newly emerging ‘β-rhizobia’ group.
Nodulation of Papilionoid legumes by both the α and β-Proteobacteria
Nodulation of Aspalathus linearis burm. f.
Several works are available that report about the nodulation of legumes belonging to the sub family Papilionoideae. However, there was very little information regarding the taxonomy of rhizobia that nodulate Aspalathus species until the work by (Moulin et al. 2001; Hassen et al. 2012) who first reported the nodulation of Aspalathus carnosa and A. linearis by members of the β-sub class of Proteobacteria respectively. A few years later after the discovery of legume nodulation by the beta group of rhizobia, the nodulation properties of the South African indigenous legume, A. linearis burm f. and their associated symbionts was studied. A. linearis burm. f. commonly referred to as ‘rooibos’, derived from the Afrikaans roy boss (= red bush), is an indigenous South African legume normally found in the winter rainfall area of the Cederberg region of the Western Cape province. It is a legume in the family Leguminosae, tribe Crotalaria of the sub family Papilionoideae. In South Africa and many other countries, A. linearis is used to make rooibos tea, a popular herbal tea that has several health benefits (Joubert and de Beer 2011; Hassen et al. 2012).
Like several other legumes, A. linearis forms a symbiotic relationship with certain group of Rhizobia that form root nodules and fix well over 100 kg nitrogen ha−1 annually (Sprent et al. 2009; Hassen et al. 2012). A. linearis is generally characterized by the formation of indeterminate nodules; elongated nodules with persistent meristem that continuously give rise to new nodule cells similar to that of Medicago sativa or Pisum sativum (Gage 2004) (Fig. 3). These are different from determinate nodules, which are circular in shape and without meristem that contain homogenous population of symbiotic cells (Maroti and Kondorosi 2014) such as those in soybeans and cowpea. The study by Hassen et al. (2012) reported for the first time that this indigenous Papilionoid legume is nodulated by members of both the α- and β-sub class of Proteobacteria. These workers initially isolated the bacteria using soil trap technique in the glasshouse from soil samples collected from the rhizosphere of wild growing A. linearis at various sites in the Cederberg mountain areas in the Western Cape. The study evaluated the ability of the isolated bacteria to nodulate the original legume host. Their results indicated that the majority of the trapped isolates were able to nodulate their original legume host.
a Indeterminate, elongated pink nodules of Aspalathus linearis.b Different nodule zones in the mature longitudinal root nodule section formed in S. meliloti, M. truncatula symbiosis showing the meristem (i), infection zone (ii), nitrogen fixation zone (iii) and senescence zone (iv). c Symbiotic cells containing the differentiating endosymbionts stained with green fluorescence and the host cytoplasm fully packed with long nitrogen-fixing bacteroides (c). Adapted from (a) Hassen et al. (2012); (b, c) Maroti and Kondorosi (2014), Frontiers in Microbiology, with permission
Unlike the previous thoughts and reports that A. linearis have a specific Bradyrhizobium requirement for their nodulation (Boone et al. 1999; Dakora 1998), the root nodule isolates from this indigenous legume displayed variations in growth rate, colony size, colour and cultural morphology (Fig. 4). Analysis of the taxonomic positions and phylogeny using nucleotide sequence of the 16S ribosomal RNA revealed that the majority of the nodule isolates from A. linearis belong to three genera (Mesorhizobium, Rhizobium, Burkholderia) (Fig. 5). The finding of this work outlines that contrary to the long-time belief that A. linearis are specifically nodulated Bradyrhizobium species, only 2% of the nodule isolates were closely related to Bradyrhizobium spp. In a previous work by Muofhe (1997), the absence of appropriate bacterial strains that nodulate and fix atmospheric nitrogen within the Cape region and which could constraint nodulation and growth of A. linearis was reported. This is contrary to what was found out by Hassen et al. (2012) in which they reported the formation of an average of 3–12 nodules per plant by rhizobia. More than one species of rhizobia including those belonging to both the alpha and beta rhizobia nodulated A. linearis (Fig. 6). The rhizobia also resulted in statistically significant increase in plant biomass (dry weight and fresh weight) indicating that the Cape floristic region is not poor in its microbial diversity that effectively colonize and form nodules in Aspalathus species.
Morphologically different strains of rhizobia isolated from the root nodules of Aspalathus linearis (rooibos): aRhizobium sp., bBurkholderia and Mesorhizobium spp. and cBradyrhizobium and Herbasprillum spp. adapted from Hassen et al. (2012)
Neighbour Joining 16S rRNA phylogenetic tree of rhizobial isolates that nodulated Aspalathus linearis. The tree shows the taxonomic placements of the characterized isolates that contain representative species from both the alpha and beta Proteobacteria. Adapted from Hassen et al. (2012)
Aspalathus linearis is nodulated by different species of bacteria belonging to Mesorhizobium (a), Rhizobium (b, c) and Burkholderia species (d). In all cases, the nodules are indeterminate and pink. Adapted from Hassen et al. (2012)
The isolation of Mesorhizobium spp. (alpha rhizobia) and Burkholderia spp. (beta rhizobia) which both nodulated A. linearis can be attributed to many factors, one of which is that both genera occur in similar acidic and very poor nutrient soils (Lamaire et al. 2015). However, the vast majority of the nodule symbionts detected in A. linearis, like in many other legumes, are the alpha-rhizobia, in this particular case Mesorhizobium followed by Rhizobium spp. (Fig. 5). One possible reason, amongst several other hypotheses, could be that shortly after the transfer of the nod genes from beta to alpha-rhizobia, there was a rapid expansion of legumes into environments that favoured alpha-rhizobia more. For instance, Burkholderia strains mainly occur as symbionts in acidic and very poor soil, but they could be outcompeted by other symbionts (e.g. Mesorhizobium spp.) when the concentration of soil nutrients in particular nitrogen increases (Sprent et al. 2017).
Nodulation of Lebeckia spp.
Another South African indigenous legume for which extensive study on the symbiotic performance and the rhizobium associated with it has been conducted was Lebeckia spp., a legume of the Leguminosae family and the tribe Crotalarieae mainly indigenous to the Southern and Western Cape regions of South Africa (Phalane 2008). This genus falls under the sub family Papilionoideae and consists of close to 35 species that are mainly indigenous to the Western and Southern regions of South Africa. In the study to determine the diversity and taxonomy different Lebeckia spp. (Phalane 2008), used various species of the indigenous Lebeckia collected from nine localities in the western and northern Cape regions. These workers isolated several morphologically different rhizobia which were then characterized to infer the phylogenetic positions at molecular level by PCR amplification using Nif gene derived primer RPO1 (Richardson et al. 1995) and the 16S rRNA primers 16f 27 and 16r 1485 (Lane 1991) followed by sequencing. The results from this analysis indicated that the various species in the genus Lebeckia are nodulated by both the alpha (Mesorhizobium, Bradyrhizobium and Sinorhizobium spp.) and rhizobia of the beta Proteobacteria group. This work presented the first report on the taxonomic identities of the rhizobia associated with the root nodules of this unique legume indigenous to South Africa. The essential finding of this work was the nodulation of all the needle shaped leaf Lebeckia spp. by the newly emerging beta sub group of rhizobia (Burkholderia) and the promiscuous nodulation of Lebeckia sericea by both Mesorhizobium and Bradyrhizobium species.
Despite the report by Phalane (2008) that the alpha (α)-Proteobacteria including Mesorhizobium, Rhizobium and Bradyrhizobium spp. are reported as the predominant symbionts of Lebeckia species, later studies indicated that the indigenous Lebeckia spp. are mainly nodulated by Burkholderia species (De Meyer et al. 2013, 2014). The study on the symbiotic properties of South African fynbos legumes spanned from 2002 until recently with an aim to domesticate herbaceous South African legumes such as Rhynchosia and Lebeckia as perennial pasture legumes in Australian agricultural systems exposed to drying climate (Howieson et al. 2013). Several novel Paraburkholderia species have since been isolated and characterized from different Lebeckia spp. of which Lebeckia ambigua, a perennial suffrutescent legume of the fynbos in South Africa, has been extensively studied. One of such studies investigated the symbiotic properties and taxonomic positions of 23 bacterial strains isolated from this legume (Howieson et al. 2013). According to the 16S rRNA phylogeny study, most of the rhizobia isolates belonged to the genus Paraburkholderia having high sequence similarity with P. graminis, P. caledonia and P. tuberum. These findings prompted for more detailed studies using polyphasic approaches including 16S rDNA, gyrB, recA gene phylogenies, DNA-DNA hybridizations, MALDI-TOF MS analysis as well as physiological and biochemical tests. These studies helped to describe new species of Paraburkholderia such as P. sprentiae sp. nov. (De Meyer et al., 2013), P. dilworthii sp. nov. (De Meyer et al. 2014) and P. fynbosensis sp. nov. (De Meyer et al. 2018) from the previously characterized 23 strains isolated from Lebeckia ambigua (Fig. 7). Unlike most of the previously studied Paraburkholderia mainly in South East China and the Americans, Paraburkholderia strains isolated within South Africa are primarily associated with Papilionoid legumes rather than the Mimosa species (De Meyer et al. 2015).
Phylogenetic tree showing the evolutionary relationship and taxonomic positions of Paraburkholderia species including the newly described novel isolates from Lebeckia ambigua nodules. The tree construction was based on analysis of the 16S ribosomal RNA sequences. Adapted from De Meyer et al. (2018), with permission
Nodulation of other Papilionoideae legumes
Cyclopia spp. are the other endemic Papilionoid legumes that represent part of the fynbos biome of the South African Cape region. Two of the species C. intermedia and C. subternata are predominantly used in the production of a local beverage drink known as honey bush tea with several health benefits as well as economic value on a foreign market. The diversity of the root nodule rhizobia associated with several Cyclopia species has been studied in South Africa using molecular characterization techniques that involved the 16S–23S IGS-RFLP analysis (Kock 2003). Both alpha and beta rhizobia were detected from the nodules as nitrogen fixing symbionts, but the majority belonged to members of the beta rhizobia (Burkholderia) most of which were phylogenetically related to Burkholderia tuberum. Then, Elliott et al. (2007) demonstrated the two Burkholderia tuberum strains STM678 and DUS833 described earlier can nodulate Cyclopia species and other Papilionoid legumes from wildly different tribes. Nodules formed by B. tuberum ST678 or DUS833 on Cyclopia genistoides were typically intermediate with invasion and nitrogen fixing zones. Beukes et al. (2013) also confirmed that the majority of these Burkholderia isolates from Cyclopia are conspicuous to B. tuberum that could expand the host range of Cyclopia species for this species of Burkholderia.
Parallel with the disclosure of the nodulation of the Papilionoid legumes Aspalathus and Cyclopia spp. by both the alpha and beta-sub group of Proteobacteria (Moulin et al. 2001; Kock 2003; Hassen et al. 2012), several new β-rhizobia that nodulate other Papilionoid legumes have been described. In an effort to gather more evidences that South African Papilionoid legumes are nodulated by Paraburkholderia strains different from those nodulating Mimosa spp. in China and the Americans, Garau et al. (2009) conducted a similar study on the indigenous herbaceous legume Rhynchosia ferulifolia. Two root nodule strains Burkholderia sp. WSM3937 and WSM3930 isolated from this legume were studied for their symbiotic performance using the same host in the glasshouse and confirmed that both nodulated Rhynchosia effectively. Using analysis of the 16S rRNA gene as well as intragenic sequences of nodA and nifH the nodule isolates from R. ferulifolia, belong to the genus Burkholderia with substantial similarity with Burkholderia tuberum STM678T, the β-rhizobium from South Africa, but have been found to have a distant relationship with South American Mimosa-nodulating b-rhizobia. This study by Garau et al. (2009) was of course the first to report N-fixation between β-rhizobia and an herbaceous, Papilionoid legume from which the strains were originally isolated.
Later, Beukes et al. (2013), reported the occurrence of diverse Burkholderia spp. in the nodules of the endemic Papilionoid legumes of the tribe Hypocalypteae and Podalyrieae from Cape Floristic Region in South Africa. Based on the analysis of molecular techniques that involved 16S rRNA and recA housekeeping gene, as well as analysis of the genes that encode for nifH and nodA, there exists a horizontal gene transfer that determines their symbiotic performance and their evolutionary origins were by far distinct from Burkholderia from other parts of the world. Similar reports of nodulation of legumes by the newly emerging beta rhizobia that mainly include Paraburkholderia (formerly Burkholderia) is gaining momentum in the past decades. Very recently, Beukes et al. (2019a) investigated and reported the nodulation of an indigenous legume tree Vachellia karroo (formerly Acacia karroo), by both members of the alpha and beta Proteobacteria. The study, which used a 16S rRNA sequence analysis as well as recA-based phylogenetic analyses, identified nodule isolates from V. karroo down to species level many of which belonged to Paraburkholderia spp. In a separate study, the taxonomic status of rhizobia that nodulate the indigenous South African fynbos legume Hypocalyptus sophoroides was elucidated using a genealogical concordance analysis that include six loci (16S rRNA, atpD, recA, rpoB, lepA and gltB) as well as comparison of average nucleotide identity. The result of this study disclosed that these nodulating strains are novel strains of Paraburkholderia which were divergent from their closest relatives and were classified as Paraburkholderia strydomina sp. nov. and Paraburkholderia steynii sp. nov. (Beukes et al. 2019b). Recently, Paraburkholderia species that nodulate soybean have been identified by means of multilocus sequence typing analysis in Venezuela (Ramírez et al. 2019). This is despite the fact that there is very little evidence that demonstrates the occurrence of Burkholderia and Paraburkholderia as the predominant soybean rhizobia in agricultural fields. The authors argue that such occurrence of soybean nodulating Paraburkholderia in Venezuela soils located in different climatic and topographical regions could have resulted through horizontal gene transfer.
Nodulation of Mimosoideae legumes by β-rhizobia
The genus Mimosa is one of the largest genera of legumes (500 species) belonging to the family Leguminosae, sub family Mimosoideae most of which form a symbiotic association with rhizobia (Sprent et al. 2009; Elliott et al. 2007). The principal centre of diversification for Mimosa are the Americans including Central Brazil, Mexico and Sub tropical South America. However, three main invasive Mimosa spp. (M. diplotricha, M. pigra and M. pudica) have recently become widespread in tropical and subtropical areas in South East Asia (Liu et al. 2012). In terms their symbiotic properties, several investigations revealed that the β-rhizobia are the common nodulating symbionts of the tribe Mimosa (Chen et al. 2003, 2005; Platero et al. 2016; Liu et al. 2012). The two major beta-rhizobia that are the preferred symbionts of Mimosa spp. are Burkholderia (now Paraburkholderia) and Cupriavidus spp. Chen et al. (2001) were the first to isolate bacteria from Mimosa nodules, which were taxonomically different from the conventional ‘Rhizobia’ belonging to α-Proteobacteria. The nodule isolates obtained from M. diplotricha and M. pudica were very similar to the genus Ralstonia (syn = Wautersia) in the β-Proteobacteria group and were later confirmed to belong to a new species Ralstonia taiwanensis (Chen et al. 2001) which was later re-described as Cupriavidus taiwanensis (Vandamme and Coenye 2004). C. taiwanensis strain LMG19424 has been reported to be the most effective nitrogen fixing symbiont of the invasive Mimosa shrubs in Taiwan. Taxonomic characterization of several symbiotic isolates from the nodules of M. pudica and M. diplotricha throughout the entire island in Taiwan revealed that 94% were β-rhizobia of which 93% belong to C. taiwanensis, which suggests that C. taiwanensis may be the preferred symbionts of these Mimosa species (Chen et al. 2005). However, subsequent investigations in later years confirmed that invasive Mimosa spp. have colonized large areas of southern China and Burkholderia and Cupriavidus spp. were reported to be the dominant symbionts of the three invasive Mimosa spp. (Liu et al. 2012). According to this report, a more accurate representation of the origins of the symbiotic relationship established by rhizobia with both the native and exotic legumes was obtained by examining the symbiosis related genes such as the N2-fixation (nifH) and nodulation (nodA) genes. Both Burkholderia and Cupriavidus spp. nodulating the invasive Mimosa legumes harbour these symbioses related genes which are thought to be acquired through lateral gene transfer from rhizobia and/or one of the relatives in the α-Proteobacteria (Chen et al. 2005).
Prior to the studies on the nodulation properties of invasive Mimosa spp. mainly in China and Taiwan, and before the discovery of the β-rhizobia as their predominant symbionts, little was known about the symbionts of Mimosa species in their native South American environments. Although several strains of β-rhizobia have been isolated since the first report of Moulin et al. (2001) on Burkholderia strains STM815 and STM678, the majority of them were isolated from Mimosa spp. in Asia, with only very few in the Americas (Barrett and Parker 2005). The works of Chen et al. (2005) provided a very conclusive proof for the nodulation of native South American Mimosa spp. by novel strains of Burkholderia spp. In this study, 16S rRNA sequence analysis and RFLP of amplified 16S rRNA genes of several Mimosa nodulating strains collected from Brazil and Venezuela revealed that Burkholderia strains form effective symbiosis with native Mimosa spp. in South America.
More recent works have since been reported on the nodulation of the native South American Mimosa spp. by the β-rhizobia with Burkholderia (Paraburkholderia) being the preferred major symbionts. The first demonstration of biological nitrogen fixation of Mimosa by β-rhizobial symbionts was held under field conditions in two of the major centres of diversity of Mimosa spp. in Brazil, Cerrado and Caatinga (dos Reis Jr et al. 2010). The major finding of this study was that Burkholderia spp. were the predominant symbionts in these native soils, and no single Cupriavidus sp. was isolated suggesting that the native South American Cerrado and Caatinga Mimosa spp. prefer Burkholderia as their major symbiont. However, in contrast to the reports for the major centres of diversification of the genus Mimosa (Brazil and Mexico) as being nodulated mainly by Burkholderia spp., the native Uruguayan Mimosa spp. are exclusively nodulated by β-rhizobia belonging to the genus Cupriavidus (Platero et al. 2016). An interesting part of this investigation is that all the five tested Uruguayan native species of Mimosa effectively nodulated in the field with rhizobia belonging to the genus Cupriavidus but none of them belonged C. taiwanensis.
Later, Lammel et al. (2015) conducted the symbiotic properties of native woody Mimosa spp. in Brazil using Mimosa scabrella and Mimosa bimucronata as trap legumes, previously tested and verified to be nodulated and fix nitrogen with Burkholderia spp. (Chen et al. 2006, 2007). More recently, Agnol et al. (2017) conducted similar work in the Atlantic Forest (Mata Atlantica) which is the centre of Mimosa diversity and that represents one of the largest and richest biomass in Brazil. In this study, rhizobia were trapped from several soil samples collected at different locations of the Atlantic Forest using Mimosa pudica as the trap legume. The genetic diversity of the trapped isolates in the nodules of M. pudica from the soils in the different sites was determined using a concatenated 16S rRNA-recA phylogeny and it was found that the majority of the isolates trapped in the nodules belonged to the Paraburkholderia species. These findings of course supported the results of previous and initial work by Bontemps et al. (2010) who characterized nodule isolates from 47 Mimosa spp. based on concatenated 16S rDNA and recA sequences. The concatenation revealed that these nodulating isolates were related to the new β-rhizobia species including Burkholderia nodosa and Burkholderia tuberum, but were in several distinct clades that indicated they could probably be new Paraburkholderia species. It is noteworthy that the root nodulating beta-rhizobia species described as ‘Burkholderia’ throughout this review are reclassified as the genus Paraburkholderia based on analysis of their phylogenetic positions and beneficial traits to separate them from plant and/or animal pathogenic Burkholderia strains (Dobritsa and Samadpour 2016).
Concluding summary
Reports of novel strains of root nodulating β-Proteobacteria, mainly that of Paraburkholderia which nodulate and fix nitrogen with several indigenous legumes adapted to acidic and nutrient poor soils is increasing in the past few years (Moulin et al. 2001; Chen et al. 2003; Garau et al. 2009; Elliott et al. 2007). These successive findings of the nodulation of legumes by the β-rhizobia has changed the long held dogma that only rhizobia, bacteria belonging to the Alphaproteobacteria, are able to nodulate legumes. The phenomenon that a single legume plant is nodulated by more than one species of rhizobia has been a point of debate with regard to the legume–rhizobium host specificity. A good example of such phenomena could be the fact that Mesorhizobium and Burkholderia spp. are both capable of nodulating A. linearis (Hassen et al. 2012) and probably other related legumes. One possible argument is that horizontal gene transfer of nod genes from one species to another could have resulted in the loss of specificity of many rhizobia to their legume host (Donate-Correa et al. 2007). This loss of specificity and the emergence of new group of root nodulating rhizobia (both α- and β-Proteobacteria) can also be attributed to the plasticity of the nodulation traits. For instance, in Rhizobium and Sinorhizobium, the nodulation traits are easily transferable plasmid borne genes whereas in the genus Mesorhizobium, there is a mobile symbiosis island in the chromosome (Gyaneshwar et al. 2011). Another possibility of the observed promiscuity (being nodulated by more than one species of rhizobia) in A. linearis or any other legume that nodulates promiscuously could be associated with the presence of more than one type of flavonoids, nod gene inducers, released by the same legume which warrants future investigation.
Coupled with the aforementioned traits and genomic features of the rhizobia, the spread of crop plants beyond the range of their native ancestors might also have resulted in the selection of a less stringent symbiotic specificity (Mutch and Young 2004). It is also argued that the nodulation by the newly emerging β-rhizobia belonging to the genus Burkholderia therefore seem to be associated more with a physical niche, such as acid, sandy, infertile soils, than with a particular legume phylogeny, hence not abiding by the principles of the legume host specificity for nodulation. The driving force for legume host specificity in the nodulation process is still debatable since, in the past two decades, several novel species of rhizobia that nodulate more than one legume host were detected. Moreover, several indigenous legumes, for example the South African indigenous Papilionoid legume A. linearis, are nodulated by more than one rhizobium species including both alpha and beta rhizobia. Therefore, whether Nod factors are generally the decisive molecular signals for legume–rhizobium host specificity is yet to be fully understood to further elucidate the mechanisms behind long held legume–rhizobium host specificity dogma. Rhizo-biologists have now a concern (Howieson et al. 2013) that the emergence of new alpha rhizobia and the discovery of several novel species of Paraburkholderia (β-rhizobia) from root nodules could be the microbiological challenge facing the domestication of legumes to promote sustainable agriculture in the future.
References
Agnol RFD, Bournaud C, Faria SM, Bena G, Moulin M, Hungria M (2017) Genetic diversity of symbiotic Paraburkholderia species isolated from nodules of Mimosa pudica (L.) and Phaseolus vulgaris (L.) grown in soils of Brazilian Atlantic Forest (Mata Atlântica). FEMS Microbiol Ecol 93:1–15
Albrecht C, Guerts R, Bisseling T (1999) Legume nodulation and mycorrhizae formation; two extremes in host specificity meet. EMBO J 18:281–288
Barreda H, Fikri-Benbrahim K (2014) Taxonomy of rhizobia: current perspectives. Br Microbiol Res J 4:616–639
Barrett CF, Parker MA (2005) Prevalence of Burkholderia sp. nodule symbionts on four mimosoid legumes from Barro Colorado Island, Panama. Syst Appl Microbiol 28(1):57–65
Beukes CW, Venter SN, Law IJ, Phalane FL, Steenkamp ET (2013) South African Papilionoid legumes are nodulated by diverse Burkholderia with unique nodulation and nitrogen-fixation loci. PLoS ONE 8:1–13
Beukes CW, Boshoff FS, Phalane FL, Hassen AI, le Roux MM, Stepkowski T, Venter SN, Steenkamp ET (2019a) Both alpha and beta-rhizobia occupy the root nodules of Vachellia karro in South Africa. Front Microbiol 10:1195. https://doi.org/10.3389/fmicb.2019.01195
Beukes CW, Steenkamp ET, van Zyl E, Avontuur J, Chan WY, Hassen AI, Palmer M, Mthombeni LS, Phalane FL, Sereme TK, Venter SN (2019b) Paraburkholderia strydomiana sp. nov. and Paraburkholderia steynii sp. nov.: rhizobial symbionts of the fynbos legume Hypocalyptus sophoroides. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01269-5
Bontemps C, Elliott GN, Simon MF, dos Reis FB, Jr GE, Lawton RC, Neto NE, Loureiro MF, de Faria SM, Sprent JI, James EK, Young JPW (2010) Burkholderia species are ancient symbionts of legumes. Mol Ecol 19:44–52
Boone CM, Olsthroon MMA, Dakpra FD, Spaink HP, Thomas-Oats JE (1999) Structural characterization of lipo-chitin oligosaccharides isolated from Bradyrhizobium aspalati, microsymbionts of commercially important South African legumes. Carbohydr Res 317:155–163
Cárdenas L, Domínguez J, Quinto C, López-Lara IM, Lugtenberg BJ, Spaink HP, Rademaker GJ, Haverkamp J, Thomas-Oates JE (1995) Isolation, chemical structures and biological activity of the lipo-chitin oligosaccharide nodulation signals from Rhizobium etli. Plant Mol Biol 29(3):453–464
Chen WM, Laevens S, Lee TM, Coeney T, de Vos P, Mergeay M, Vandamme P (2001) Ralstonia taiwanensis sp. nov. isolated from root nodules of Mimosa species and spatum of a cyctic fibrosis patient. Int J Syst Evol Microbiol 51:1729–1735
Chen W-M, Mouline L, Bontemps C, Vandamme P, Bena G, Bovine-Masson C (2003) Legume symbiotic nitrogen fixation by β-Proteobacteria is widespread in nature. J Bacteriol 185:7266–7272
Chen W-M, James EK, Chou J-H, Sheu SY, Yang SZ, Sprent JI (2005) β-Rhizobia from Mimosa pigra, a newly discovered invasive plant in Taiwan. New Phytol 168:661–675
Chen WM, James EK, Coennye T, Chou J-H, Barrios E, de Faria SM, Elliott GN, Sheu S-Y, Sprent JI, Vandamme P (2006) Burkholderia mimosarum sp. nov., isolated from root nodules of Mimosa spp. from Taiwan and South America. Int J Syst Evol Microbiol 56:1847–1851
Chen WM, Faria SM, James EK, Elliott GN, Kuan-Yin Lin K-Y, Jui-Hsing Chou J-H, Shih-Yi Sheu S-Y, Cnockaert M, Sprent JI, Vandamme P (2007) Burkholderia nodusa sp. no., isolated from root nodules of the woody Brazillian legumes Mimosa bimucronata and Mimosa scabrella. J Syst Evol Microbiol 57:1055–1059
Clua J, Roda C, Zaneti ME, Blanco FA (2018) Compatibility between legumes and rhizobia for the establishment of a successful nitrogen fixing symbiosis. Genes 9:125
Dakora FD (1998) Nodulation specificity of Aspalathus linearis subsp. linearis, a shrub tea legume indigenous to the Western Cape. In: Elmerich C, Kondorosi A, Newton WE (eds) Biological nitrogen fixation for the 21st century. Kluwer, Dordrecht, pp 671–672
De Meyer S, Cnockaert M, Ardley JK, Maker J, Yates R, Howieson JG, Vandamme P (2013) Burkholderia sprentiae sp. no., isolated from Lebeckia ambigua root nodules. Int J Syst Evol Microbiol 63:3950–3957
De Meyer S, Cnockaert M, Ardley JK, Van Wyk B-E, Vandamme PA, Howieson JG (2014) Burkholderia dilworthii sp. nov., isolated from Lebeckia ambigua root nodules. Int J Syst Evol Microbiol 64:1090–1095
De Meyer SE, Tian R, Seshadri R, Reddy TBK, Markowitz V, Ivanova N, Pati A, Woyke T, Kyrpides N, Ron Yates R, Howieson J, Reeve W (2015) High-quality permanent draft genome sequence of the Lebeckia ambigua-nodulating Burkholderia sp. strain WSM4176. Stand Genomic Sci 10:79
De Meyer SE, Cnockaert M, Moulin L, Howieson JG, Vandamme P (2018) Symbiotic and non-symbiotic Paraburkholderia isolated from South African Lebeckia ambigua root nodules and the description of Paraburkholderia fynbosensis sp. nov. Int J Syst Evol Microbiol 68:2607–2614
Deng ZS, Zhao FL, Kong ZY, Yang WQ, Lindstrom K, Wang ET, Wei GH (2011) Diversity of endophytic bacteria within nodules of the Spherophysa salsula in different regions of Loess Plateau in China. FEMS Microbiol Ecol 76:463–475
Deschodt CC, Strijdom BW (1976) Effective nodulation of Aspalathus linearis sub. spp. linearis by rhizobia from other Aspalathus species. Phytophylactica 8:103–104
Dobritsa AP, Samadpour M (2016) Transfer of eleven species of the genus Burkholderia to the genus Paraburkholderia and proposal of Caballeronia gen. nov. to accommodate twelve species of the genera Burkholderia and Paraburkholderia. Int J Syst Evol Microbiol 66:2836–2846
Donate-Correa J, Leon-Barrios M, Hernandez M, Perez-Galdona R, del Arco-Aguilar M (2007) Different Mesorhizobium species sharing the same symbiotic gene nodulate the shrub legume Angaris latifolia. Syst Appl Microbiol 30:615–623
dos Reis Jr FB, Simon MF, Gross E, Boodey RM, Elliott GN, Netto NE, de Loureiro MF, de Querzo LP, Scotti MR, Chen W-M, Noren A, Rubio MC, de Faria SM, Bontemps C, Goi SR, Young JPW, Sprent JI, James EK (2010) Nodulation and nitrogen fixation by Mimosa spp. in the Cerrado and Caatinga biomes of Brazil. New Phytol 186:934–946
Elliott GN, Chen WM, Bontemps C, Chou JH, Young JPW, Sprent JI, James EK (2007) Nodulation of Cyclopia spp. (Leguminosae, Papilionoideae) by Burkholderia tuberum. Ann Bot 100:1403–1411
Fauvart M, Michiels J (2008) Rhizobial secreted proteins as determinants of host specificity in the rhizobium-legume symbiosis. FEMS Microbiol Lett 285:1–9
Frank B (1889) Über die Pilzsymbiose der Leguminosen. Ber Dtsch Bot Ges 7:332–346
Gage DJ (2004) Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol Mol Biol Rev 68:280–300
Garau G, Yates RJ, Deiana P, Howieson JG (2009) Novel strains of Burkholderia have a role in nitrogen fixation with papilionoid herbaceous legumes adapted to acid, infertile soils. Soil Biol Biochem 41:125–134
Graham PH (2008) Ecology of the root nodule bacteria of legumes. In: Dilworth MJ, James EK, Sprent JI, Newton WE (eds) Nitrogen fixing legume symbioses. Springer, Dordrecht, pp 23–58
Gyaneshwar P, Hirsh AM, Moulin L, Chen W-M, Elliott GN, Bontemps C, Estrada-de Los Santos P, Gross E, dos Reis B, Jr F, Sprent JI, Young JPW, James EK (2011) Legume nodulating Betaproteobacteria: diversity, host range, and future prospects. Mol Plant Microbe Interact 24:1276–1288
Hassen AI, Bopape FL, Habig J, Lamprecht SC (2012) Nodulation of rooibos (Aspalathus linearis Burm. f), an indigenous South African legume by members of both the α-Proteobacteria and β-Proteobacteria. Biol Fert Soils 48:295–303
Howieson JG, De Meyer SE, Vivas-Marfisi A, Ratnayake S, Ardley JK, Yates RJ (2013) Novel Burkholderia bacteria isolated from Lebeckia ambigua – A perennial suffrutescent legume of the fynbos. Soil Biol Biochem 60:55–64
Jiao YS, Li YH, Yan H, Wang ET, Tian CF, Chen WX, Guo BL, Chen WF (2015) Rhizobial diversity and nodulation characteristics of the extremely promiscuous legume Sophora flavescens. Mol Plant Microbe Interact 28:1338–1352
Kock MM (2003) Diversity of root nodulating bacteria associated with Cyclopia species. PhD Thesis, University of Pretoria, Pretoria
Joubert E, de Beer D (2011) Rooibos (Aspalathus linearis) beyond the farm gate: from herbal tea to potential phytopharmaceutical. S Afr J Botany 77(4):869–886
Lamaire B, Dlodlo O, Chimpango S, Stirton C, Schrire B, Boarwright JS, Honny O, Smets E, Sprent J, James EK, Muasya AM (2015) Symbiotic diversity, specificity and distribution of rhizobia in native legumes of the core Cape subregion (South Africa). FEMS Microbiol Ecol 91:1–17
Lammel DR, Cruz LM, Mescolotti D, Sturmer SL, Cardoso EJBN (2015) Woody Mimosa species are nodulated by Burkholderia in ombrophylous forest soils and their symbiosis are enhanced by arbuscular mycorrhizal fungi (AMF). Plant Soil 393:123–135
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–175
Li HJ, Wang FT, Chen WF, Chen WX (2008) Genetic diversity and potential for promotion of plant growth detected in nodule endophytic bacteria of soybean grown in Heilongjiang province of China. Soil Biol Biochem 40:238–246
Liu XY, Shuang W, Fang W, James EK, Gua XY, Zagar C, Xia LG, Dong X, Yi PW (2012) Burkholderia and Cupriavidus spp. are the preferred symbionts of Mimosa spp. in Southern China. FEMS Microbiol Ecol 80:417–426
Long SM (2001) Genes and signals in the Rhizobium-legume symbiosis. Plant Physiol 75:69–72
Maroti G, Kondorosi E (2014) Nitrogen-fixing Rhizobium-legume symbiosis: are polyploidy and host peptide-governed symbiont differentiation general principles of endosymbiosis? Front Microbiol 5:326. https://doi.org/10.3389/fmicb.2014.00326
Moulin L, Munive A, Dreyfus B, Boivin-Masson C (2001) Nodulation of legumes by members of the β-subclass of Proteobacteria. Lett Nat 411:948–950
Muofhe ML (1997) N2 fixation and rhizosphere ecology of Aspalathus linearis subsp. linearis (Rooibos tea). MSc Thesis, Cape University of Peninsula
Mutch LA, Young JP (2004) Diversity and specificity of Rhizobium leguminosarum biovar viciae on wild and cultivated legumes. Mol Ecol 13:2435–2444
Ngom A, Nakagawa Y, Sawada H (2004) A novel symbiotic nitrogen fixing member of te Ochrobactrum clade isolated from root nodules of Acacia mangium. J Gen Appl Microbiol 50:17–27
Oldroyd GE, Murray JD, Poole PS, Downie JA (2011) The rules of engagement in the legume rhizobial symbiosis. Annu Rev Genet 45:119–144
Orgambide GG, Lee JI, Holingsworth RI, Dazo FB (1995) Structurally diverse chitolipooligosaccharide Nod factors accumulate primarily in membranes of wild type Rhizobium leguminosarum biovar trifolii. Biochemistry 34:3832–3840
Perret X, Staehelin BWJ, Broughton WJ (2000) Molecular basis of symbiotic specificity. Microbiol Mol Biol Rev 64:180–201
Phalane FL (2008) The diversity of root nodule bacteria associated with Lebeckia species in South Africa. MSc Thesis, University of Pretoria
Platero R, James KE, Rios C, Iriarte A, Sandes L, Zabaleta M, Battistoni F, Fabiano E (2016) Novel Cupriavidus strains isolated from root nodules of native Uruguayan species. Appl Environ Microbiol 82:3150–3164
Pueppke SG, Broughton WJ (1999) Rhizobium sp. strain NGR234 and R fredii USDA257 share exceptionally broad, nested host range. Mol Plant Microbe Interact 12:293–318
Ramírez MDA, España M, Aguirre C, Kojima K, Ohkama-Ohtsu N, Sekimoto H, Yokoyama T (2019) Burkholderia and Paraburkholderia are predominant soybean Rhizobial genera in Venezuelan soils in different climatic and topographical regions. Microbes Environ 34:43–58
Richardson AE, Viccars LA, Watson JM, Gibson AH (1995) Differentiation of Rhizobium strains using the polymerase chain reaction with random and directed primers. Soil Biol Biochem 27:515–524
Rivas R, Velazquez E, Willems A, Vizcaino M, Subba-Rao NS, Mateos PF, Gills M, Dazzo FB, Martinez Molina EA (2002) New species of Devosia that froms a unique nitrogen fixing root nodule symbiosis with the aquatic legume Neputina natans (L. f) Druce. Appl Environ Microbiol 68:5217–5222
Roche P, Fabienne M, Claire P, Debele F, Ferro M, Georges T, Prome J-C, Denarie J (1996) The common nodABC genes of Rhizobium meliloti are host range determinants. Proc Natl Acad Sci USA 93:15305–15310
Spaink HP, Wijfjes AHM, Lugtenberg BJJ (1995) Rhizobium NodI and NodJ protiens play a role in the efficiency of secretion of lipochitin oligosaccharides. J Bacteriol 177:6276–6281
Sprent JI, Odee DW, Dakora FD (2009) African legume: a vital but underutilized resource. J Exp Bot 25:1–9. https://doi.org/10.1093/jxb/erp342
Sprent JI, Ardley J, James EK (2017) Biogeography of nodulated legumes and their nitrogen fixing symbionts. New Phytol 215:40–56
Staphorst JL, Strijdom BW (1975) Specificity in the Rhizobium symbiosis of Aspalathus linearis (Burm (f.)) Dahlgr. spp. linearis. Phytophylactica 7:95–96
Sy A, Giraud E, Jourand P, Garcia N, Willems A, de Lajudie P, Prim Y, Neyra M, Gills M, Bovin-Dryefus B (2001) Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. J Bacteriol 183:214–220
Vandamme P, Coenye T (2004) Taxonomy of the genus Cupriavidus: a tale of lost and found. Int J Syst Evol Microbiol 54:2285–2289
Vandamme P, Goris J, Chen W-M, de Vos P, Willems A (2002) Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov. nodulate the roots of tropical legumes. System Appl Microbiol 25:507–512
Velazquez E, Garcia-Fraile P, Ramirez-Bahena MH, Peix A, Rivas R (2010) Proteobacteria forming nitrogen-fixing symbiosis with higher plants. In: Sezena ML (ed) Proteobacteria: phylogeny, metabolic diversity and ecological effects. Nova Science, New York
Wang D, Shengming Y, Fang T, Hongyan Z (2012) Symbiosis specificity in the legume-rhizobial mutualism. Cell Microbiol 14:334–342
Wang Q, Liu J, Zhu H (2018) Genetic and molecular mechanisms underlying symbiotic specificity in legume-rhizobium interactions. Front Plant Sci 9:313. https://doi.org/10.3389/fpls.2018.00313
Willems A (2006) The taxonomy of rhizobia: an overview. Plant Soil 287:3–14
Young JPW, Haukka K (1996) Diversity and phylogeny of rhizobia. New Phytol 133:87–94
Zhang JJ, Liu TY, Chen WF, Wang ET, Sui XH, Zhang XX, Li Y, Li Y, Chen WX (2012) Mesorhizobium muleiense sp. nov., nodulating with Cicer arietinum L. Int J Syst Evol Microbiol 62(11):2737–2742
Acknowledgements
Due acknowledgement goes to the Agricultural Research Council—Plant Health and Protection for the material and moral assistance rendered in the publication of this work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hassen, A.I., Lamprecht, S.C. & Bopape, F.L. Emergence of β-rhizobia as new root nodulating bacteria in legumes and current status of the legume–rhizobium host specificity dogma. World J Microbiol Biotechnol 36, 40 (2020). https://doi.org/10.1007/s11274-020-2811-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-020-2811-x