Abstract
Bacterial genomes contain a huge amount of different genes. These genes are spatiotemporally expressed to accomplish some required functions within the organism. Inside the cell, any step of gene expression may be modulated at four possible places such as transcription initiation, translation regulation, mRNA stability and protein stability. To achieve this, there is a necessity of strong regulators either natural or synthetic which can fine-tune gene expression regarding the required function. In recent years, riboswitches as metabolite responsive control elements residing in the untranslated regions of certain messenger RNAs, have been known to control gene expression at transcription or translation level. Importantly, these control elements do not prescribe the involvement of protein factors for metabolite binding. However, they own their particular properties to sense intramolecular metabolites (ligands). Herein, we highlighted current important bacterial riboswitches, their applications to support genetic control, ligand-binding domain mechanisms and current progress in synthetic riboswitches.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
More developments in gene regulation reveal that natural and synthetic RNA-based elements have been demonstrated as key components applied in the diverse genetic control (Henkin 2008). Among them, riboswitches have received attention as attractive sensor–actuator hybrids that can control gene expression in response to intracellular metabolites concentration. Such regulatory RNA sequences are typically found in the 5′-untranslated regions of messenger RNAs, where they control gene expression by binding to small specific metabolites normally called ‘effectors’ (Berens and Suess 2015; Wang et al. 2015). Comparatively to other regulatory RNA elements, riboswitches directly interact with their effectors without requisition of intermediate factors which suggest them to be one of the oldest regulatory systems (Alexey et al. 2003).
Riboswitches regulate several metabolic pathways including the biosynthesis of vitamins (e.g. riboflavin, thiamin and cobalamin) and the metabolism of methionine, lysine and purines (Barrick and Breaker 2007). Results obtained using a riboswitch sensor to examine coenzyme B12 metabolism and transport in Escherichia coli indicated that riboswitch based sensors can provide valuable information on intracellular small molecule concentrations that can be employed in the study of related cellular processes (Fowler et al. 2010). From these results, potential research on ligand concentrations has been carried out and it was reported that high ligand concentrations are needed to effectively regulate gene expression (Garst et al. 2011). However, additional research on this query revealed that in the most cases an exaggerated ligand concentration can cause the closures in one way or another of the expression (Khani et al. 2018). Even though most known riboswitches occur in bacteria, functional TPP riboswitch have been discovered in plants, some fungi and have also been predicted in archaea (Sudarsan 2003). Additionally, monitoring conditional gene expression by a tetracycline-riboswitch showed an influence on translation initiation in archaea (Demolli et al. 2014). Nowadays, metabolite-binding mRNAs are possibly involved in eukaryotic gene regulation and some riboswitches might be representatives of an ancient form of genetic control. This explains why recent researchers are in battle to intensify the transfer of riboswitch regulatory systems to mammalian cell culture and preferably to transgenic animals and plants (Berens and Suess 2015; Sudarsan 2003). Again, an intriguing research on cis-acting regulatory elements of genes has demonstrated that riboswitches are not only exclusively found in non-pathogenic bacteria, but also found in pathogenic species such as Mycobacterium tuberculosis, Staphylococcus aureus, Streptococcus pneumoniae, Vibrio cholera, E. coli and Pseudomonas aeruginosa (Miotto et al. 2012; Ramesh 2015; Wang et al. 2017).
Mandal et al. (2003) reported that most of riboswitches found in Bacillus subtilis sense and respond to small target molecules which play a crucial role in central metabolic pathways of some organisms. These findings emphasized the influence of riboswitches in diverse living systems especially in B. subtilis. Whether artificial or natural, the function of riboswitches to switch on or off the gene expression remains a big challenge. The persistent challenge yet observed in gene regulation has deeply encouraged biologist engineers to worry about the existence of cis-acting regulatory elements affecting regulation in the genome and how they can be synthetically engineered to respond to the endogenous ligands. This further suggests that riboswitches can be used for metabolic control, especially in novel and synthetic cell factory for biofuels and chemicals production. Several researchers in this field explored about riboswitches mechanisms and their effect on downstream coding sequence in host cells. However, up to now, a query of understanding sophisticated phenomena of sensing and responding on intra or extracellular molecules known as ligands still contributes a limiting factor on gene expression.
In this manuscript, we focused on mentioning important riboswitches currently found in bacteria, their related mechanisms as well as their diversified applications. We also briefly describe other riboswitches found in other microorganisms and end up with some model perspectives in the future.
General mechanisms of discovered riboswitches
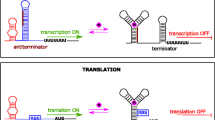
Among proposed riboswitches mechanisms, transcription termination, translation initiation, and RNA cleavage are the common mechanisms frequently found in bacterial species (Fig. 1).
Established or proposed mechanisms of riboswitch-mediated gene regulation. a Translation activation in the presence of ligand. The ribosome binding site (RBS) is sequestered in the absence of ligand. Upon ligand binding, the RNA undergoes a conformational shift, revealing the RBS and enabling translation. b Transcription continues in the presence of ligand. In the absence of ligand, an intrinsic terminator stops transcription. Ligand binding induces a conformational shift that forms an anti-terminator, enabling expression of the downstream genes. c RNA cleavage in the presence of ligand slows down gene expression. The glmS riboswitch functions by a ligand-dependent splicing mechanism, whereby protein expression is diminished in the presence of the ligand
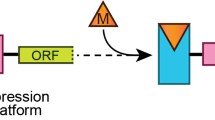
Riboswitches are often conceptually divided into two parts: an aptamer domain and an expression platform. The receptor (aptamer) domain directly binds the small target molecule (effector), and the expression platform or regulatory domain undergoes structural changes in response to the changes in the aptamer and communicates this response to other gene expression components of the cell (Garst et al. 2011). Like engineered aptamers, each natural aptamer serves as a molecular sensor embedded within the riboswitch, where it selectively senses and recognizes its corresponding target molecule within the complex sea of other metabolites (Bastet et al. 2011; Mandal and Breaker 2004). The expression platform is what regulates gene expression by typically shut down gene expression in response to the small molecule, but some turn it on (Garst et al. 2011) (Fig. 2).
The general organization of riboswitch RNAs in bacteria mRNAs. Binding of the target ligand to the aptamer domain destabilizes a stable conformation of the expression platform that results in changing gene expression for the downstream gene(s). The gene is expressed in a normal way in the absence of ligand
In this context, the conformational changes in the riboswitch aptamer domain are used to couple ligand binding to the folding of a downstream structural switch in the expression platform that in turn instructs the transcriptional initiation and elongation of mRNA, translational initiation, and presumably even the stability and splicing of mRNA transcript machinery (Berens et al. 2015; Mandal and Breaker 2004; Serganov and Nudler 2013). Transcription interference or possibly antisense action, dual transcription and translation control, and ligand-dependent self-cleaving ribozyme action were reported but they may be prevalent nonetheless (Breaker 2012). Previous study has demonstrated that binding alone does not determine the effectiveness of a compound to regulate the activity of a riboswitch. Therefore, different aspects of an interaction of the effector with the RNA must be preserved for a chemical analog to be able to also regulate the riboswitch (Trausch and Batey 2014). Reviewed in “single-molecule studies of riboswitch folding”, the author showed that magnesium seems to promote folding intermediates and/or fully folded states that are competent to bind ligand (Savinov et al. 2014). Recently discovered riboswitch classes have surprisingly complex mechanisms for regulating gene expression and new high-resolution structural models of these RNAs provide insight into the molecular details of metabolite recognition by natural RNA aptamers (Berens et al. 2015). Besides, structural studies have revealed that riboswitches, which bind a variety of small molecule metabolites, including purine bases, S-adenosylmethionine, amino acids and cofactors, may contain a complicated tertiary architecture that facilitates their function. Hence, these structures guide the messenger RNA to adopt one of two mutually exclusive forms, dictating the outcome of transcription or translation. Another highly structured messenger RNA element known as the viral internal ribosomal entry site, able to manipulate the ribosome and take over from the function of initiation factors by promoting gene expression has been identified (Batey 2006).
Among the discovered riboswitches, guanine was the first riboswitch to show the possibility of being engineered through point mutation which tends its molecular specificity to bind to adenine (Kim et al. 2007). Basing on metabolite-binding mechanisms of natural riboswitches which exist in different bacteria, more than six elements (glmS, gcvT, ydaO_yuaA, ykkC_yxkD, ykoK, and yybP_ykoY) within the B. subtilis genome that exhibit characteristics of riboswitch function have been identified and added on what are currently present with the well-defined functions. Further researches revealed that these motifs are not only existed in B. subtilis but also exhibit an extensive sequence and secondary-structure conservation among many other bacterial species and occur upstream of related gene (Barrick et al. 2004). In previous years, research studies confirmed glmS (Watson and Fedor 2011) and ydaO motives as glmS and ATP riboswitches (Watson and Fedor 2012) respectively. Moreover, coenzyme-B12, thiamin pyrophosphate, S-adenosyl methionine, flavin mononucleotide, purine, lysine, glycine, and preq1 riboswitches are considered as potential riboswitches often found in bacteria as well as in other microorganisms. These riboswitches will be described in detail below.
Coenzyme-B12 riboswitch structure and gene-control function
Cobalamin riboswitch (B12-element), is a natural RNA device which binds adenosylcobalamin to regulate cobalamin biosynthesis and transport of cobalamin and similar metabolites, and other genes (Nahvi et al. 2004).
Similar to other riboswitches, it is comprised of a conserved aptamer domain and an expression platform which broadly changes with evolution. Also, this riboswitch controls gene expression by transcription termination and translation control mechanisms. At the transcriptional level, the binding of a ligand (B12) to the aptamer domain triggers the formation of an anti-terminator stem or an intrinsic terminator stem which favors RNA polymerase to block transcription before the formation of the coding portion of mRNA. Translation control is conveyed by ligand binding to the aptamer domain which influences the formation of an anti-anti-RBS (ribosome-binding site) stem and an anti-RBS stem, resulting in an allosteric change along the expression platform and stop translation initiation (Mandal and Breaker 2004; Vitreschak 2003).
Recently, through genome mining applied in Klebsiella pneumonia, a sequence was identified by bioinformatics and proposed to be a B12 riboswitch regulated by a coenzyme B12. Further, in-line probing experiments were used to explore the structural rearrangement along the RNA sequence while Isothermal Titration Calorimetry (ITC) confirmed the thermodynamic parameters of the interaction between the riboswitch and its metabolite. Consequently, the interaction of coenzyme B12 with the butB riboswitch of K. pneumoniae found to be the exothermic process with a 1:1 binding stoichiometry and binding affinities ranging from log KA = 6.73 ± 0.02 at 15 °C to log KA = 6.00 ± 0.09 at 30 °C (Palou-Mir et al. 2016).
Moreover, research revealed that btuB riboswitch from E. coli, once binds to its ligand, coenzyme B12 undergoes a conformational change and takes part in the gene regulation of an outer membrane B12-transport protein in E. coli.
This RNA is characterized by its selective high-affinity binding to coenzyme B12 and by the structural rearrangement it undergoes upon this interaction (Gallo and Sigel 2018). Very recently, the distribution analysis of different riboswitches families in cyanobacteria genomes was achieved. Significantly, the observation analysis highlighted four classes of riboswitch plentiful in cyanobacteria where B12 (Cob)/AdoCbl/AdoCbl-variant being the most abundant. This research also confirmed a large number of genes regulated by riboswitches which may assist in the elaborating diversity among cyanobacteria species (Singh et al. 2018). To the best of our knowledge on this type of riboswitch, further studies, which take B12 riboswitch mechanisms into account, will need to be undertaken in other B12 producing bacteria. Additional study was carried out in other cyanobacteria to know the effect of 5′-UTR sequences on genes involved in methionine metabolism, which thus led to the identification and validation of a cobalamin riboswitch, with potential transcriptional regulation, in the 5′-UTR of the metE gene from Synechococcus sp. strain PCC 73109. Crucially, this discovery provided a novel genetic tool for controlling gene expression responsible for de novo methionine biosynthesis in this cyanobacterium (Perez et al. 2016).
Thiamin pyrophosphate riboswitches
TPP is the active form of thiamine (vitamin B1), an essential coenzyme synthesized by coupling of pyrimidine and thiazole moieties in bacteria (Hallera et al. 2013). With latest technological advances in gene regulatory networks, scientists demonstrated that thiamin biosynthesis and transport, as well as transport of similar metabolites are governed by TPP riboswitches which bind thiamin pyrophosphate (TPP) in its aptamer domain and alter conformation in expression platform of 5′-UTR region of corresponding gene (Winkler and Breaker 2003). However, this is particularly interesting given the fact that it is the only riboswitch found so far in eukaryotes and nowadays can be found within all three kingdoms (McRose et al. 2014; Moldovan et al. 2018; Zhang et al. 2017). To date, this riboswitch is known as a highly conserved RNA secondary structure and researchers consider its domain as a direct contributor to the mechanism of regulating genes by modulating transcription termination, translation initiation, or even (in eukaryotes) alternative RNA splicing mediated translational regulation (Henkin 2008; Savinov et al. 2014). Moreover, this regulatory RNA sequence serves as a riboswitch that binds directly to thiamine pyrophosphate (TPP) to regulate gene expression through a variety of mechanisms in archaea, bacteria and eukaryotes (McRose et al. 2014; Moldovan et al. 2018; Zhang et al. 2017). Furthermore, in a more recent advance, Singh et al. (2018) showed that TPP riboswitches are distributed across 290 cyanobacterial genomes. This research stressed with a clear evidence that TPP riboswitches reside in upstream of thiC, which encodes an essential thiamine biosynthesis enzyme, regulate mainly thiC gene in cyanobacteria. Many hypothetical genes were also mentioned as riboswitch regulated for this family. Apart from thiC gene often regulated by TPP riboswitch in terrestrial cyanobacteria, aquatic marine and aquatic flesh water cyanobacteria showed the presence of TPP riboswitch in the upstream of their various genes (Singh et al. 2018). These discoveries may have a potential promising application and offer a crucial evidence for future exploitations of more regulatory RNA sequences governing various metabolic pathways in cyanobacteria.
SAM riboswitch (S-box)
Other class of riboswitches, “S-box (SAM-I)” has been primarily found in low-G + C gram-positive bacteria where they are involved in the regulation of sulfur metabolism. These riboswitches, once bound to S-adenosyl methionine (SAM), they undergo a regulation of methionine, SAM biosynthesis and transport as well (Huang et al. 2012). Until now, three kinds of these riboswitches are known: (1) SAM-I which is widespread located in bacteria especially in B. subtilis, (2) SAM-II uniquely found in alpha-, beta- and a few γ-proteobacteria and (3) SMK box which only found in the order lactobacillales where it regulates gene expression at the level of translation initiation (Hickey and Hammond 2014; Wilson et al. 2011). The metX SAM-II riboswitch, firstly identified in the Sargasso Sea metagenome, is thought to regulate gene expression at the translational level by sequestering the SD sequence of its associated gene in an H-type pseudoknot, with pseudoknot formation being promoted by ligand binding (Savinov et al. 2014). To date, there are no obvious similarities among these three varieties of the riboswitch regarding to the sequence or structure. A fourth variety, SAM-IV and SAM-I riboswitches, seems to have a similar ligand-binding core, except in riboswitch scaffold. SAM-SAH riboswitches bind both SAM and SAH with similar affinities. Since they are still found in a position to regulate genes encoding methionine adenosyltransferase, it was proposed that only their binding to SAM is physiologically relevant (Mandal and Breaker 2004).
Tomsic et al. (2008) demonstrated that in case SAM binds to S-box leader RNA, there is a creation of an allosteric change which immediately induces the premature termination of transcription at the S-box leader region terminators. However, methionine, as one of the most valuable amino acids known, its metabolism regulation ought to be driven by S-box riboswitch which often regulates genes expression in s-box gene family. Similar to other techniques applied to the 5′-UTR to regulate downstream sequence, Lu et al. (2010) introduced mutations in the SAM-binding region resulting in loss of SAM binding and constitutive termination. Further, regarding the crucial role they play in medical pathogens to regulate the expression of survival and/or virulence essential genes, SAM riboswitches might be important targets for the development of new antimicrobial agents (Trausch et al. 2014). SAM-II riboswitch which is usually found in proteobacteria, is solely found in five cyanobacterial species namely Fischerella PCC 9605, Mastigocoleus testarum BC008, Obscuribacter phosphatis Mle1 12, ThermoSynechococcus sp. NK55a and Scytonema millei VB511283, while SAM-I/IV-variant riboswitch was observed in only two cyanobacterial species namely Gastranaerophilaceae (Zag_1) and Gastranaerophilaceae (Zag_111). Recent findings regarding SAM riboswitches revealed that a gene belongs to the CBS domain family protein which is under the regulation of SAM-I/IV riboswitch is anticipated to play a major role fermentation (Singh et al. 2018). Taken together, these evidential results suggest that further screening and applications of this riboswitch for valuable compounds production in cyanobacteria could be advantage.
Flavin mononucleotide riboswitch
Flavin mononucleotide riboswitch is a highly conserved RNA element frequently found in the 5′-UTR of prokaryotic mRNAs that encode for flavin mononucleotide (FMN) biosynthesis, transport proteins as well as transport of riboflavin, the nonphosphorylated precursor of FMN (Sklyarova and Mironov 2014). Similarly, it was well established that the phosphorylated form of roseoflavin can be converted into a roseoflavin mononucleotide which in turn competes with FMN for riboswitch binding and consequently causes FMN downregulation which subsequently results in cell growth challenges (Lee et al. 2014; Ott et al. 2014). It is well known that ligand binding properties (kinetics and affinity) of aptamer domain are the most important parameters to be considered when engineering riboswitches. In this way, a kinetic analyses of binding reaction between flavin mononucleotide and several natural mutations in aptamer domains of FMN-specific riboswitches were performed. To well execute this, a rational modification in the tuning regions was used to generate variants of synthetic riboswitches which subsequently provides further evidence that the gene expression can be controlled by rational adjustment of tuning regions (Rode et al. 2015). The experiments conducted by Pedrolli et al. (2012) demonstrated that ribFMN riboswitch of S. davawensis is the only promised model confided to discriminate between the closely similar flavins FMN and RoFMN and finally they showed an opposite response to these ligands. In B. subtilis, this riboswitch controls gene expression by causing premature transcription termination within the 5′-UTR of the ribDEAHT operon and precluding access to the ribosome-binding site of ypaA mRNA (Winkler and Breaker 2003). Moreover, an additional study conducted in B. subtilis, revealed that a ypaA gene which encodes riboflavin transporter presents in its 5′-UTR sequence a FMN riboswitch which binds flavin mononucleotide, then resulting in translation inhibition due to the sequestration of SD-sequence (Sklyarova and Mironov 2014). In cyanobacteria, FMN riboswitch plays a great role on genes belonging to PIN domain superfamily, where it regulates tryptophanyl-tRNA synthetase and tRNA 2-selenouridine synthase (Singh et al. 2018).
Purine riboswitches
Purine riboswitches are normally known as adenine and guanine riboswitches which control gene expression (Kim et al. 2009), purine metabolism and transport in the response to their purine ligand molecule (Delfosse et al. 2010). The expression platform consists of an anti-terminator hairpin that shares nucleotides with the alternatively-formed P1 hairpin of the aptamer. Guanine binding promotes folding of the aptamer domain over the anti-terminator, leading to transcription termination via an intrinsic terminator hairpin (Savinov et al. 2014). Different forms of purine riboswitches bind guanine, adenine or 2′-deoxyguanosine (2′-dG). Homologous types of purine riboswitches bind deoxyguanosine, but have more significant differences than a single nucleotide mutation (Marcano-Velazquez and Batey 2015). Research carried out in B. subtilis, revealed that the pbuE adenine riboswitch (A riboswitch) acts as a genetic switch which turns on transcription. In contrast, xpt–pbuX guanine riboswitch (G-riboswitch) found in B. subtilis appears to control more than five transcription units and acts as a genetic switch that turns ‘off’ transcription (Delfosse et al. 2010; Mandal and Breaker 2004).
Moreover, structure evaluation of purine riboswitches aptamer by X-ray, showed that these riboswitches are characterized by a compact fold in where the ligand forms a Watson–Crick base pair with residue 65 (Delfosse et al. 2010). Nowadays, Singh et al. (2018) affirmed the presence of this riboswitch in cyanobacteria; their results demonstrated that purine riboswitches control relevant genes for purine biosynthesis and salvage. These genes with upstream purine riboswitch are present in the pathway of purine de novo biosynthesis.
Lysine riboswitch (L-box or LYS-element)
Lysine riboswitch also called L-box, is found in the leader sequence of bacterial mRNAs coding for proteins related to the regulation of biosynthesis, catabolism, and transport of lysine (Doerks et al. 2002; Wang et al. 2015). Similar to other riboswitches, its aptamer domain encapsulates the ligand and undergoes a conformational change hence regulating the enzyme activity upon interacting with lysine (Yang et al. 2013).
Lysine may exert its control by inducing premature transcription termination by an unknown mechanism (Kochhar and Paulus 1996). This proposition was emphasized by Rodionov et al. (2003) who demonstrated that mechanism of the lysine-specific riboswitch is similar to the other metabolite-specific riboswitches found in B. subtilis and involves either transcriptional or translational attenuation in various groups of bacteria. Moreover, the lysC leader RNA not only uses some common mechanisms, like shape complementarity and metal ion assistance for lys binding and recognition, but also makes nucleotide-specific interactions for ligand identification. These interactions differ from the mechanism used by the glycine riboswitch to recognize similar amino acid features. For this riboswitch, the unique arrangement of contacts for ligand recognition and gene regulation are frequently used (Wilson-Mitchell et al. 2012).
Mutations applied within the lysine-responsive riboswitch play great importance in the acquisition of resistance to antimicrobial lysine analogs. This indicates that lysine induces only limited and local conformational changes upon ligand binding (Garst et al. 2008). However, the knowledge about genes driven by this riboswitch activity has remained undiscovered for many years. Mukherjee et al. (2017) analyzed about 2785 bacterial gene sequences of which 468 were natural lysine riboswitches that regulate almost all biosynthetic and transporter genes in Firmicutes and Gammaproteobacteria. This research confirmed that lysine riboswitches are relatively rare in all other prokaryotic phyla where if it is present they are primarily found upstream to operons containing many lysine biosynthesis genes. In recent years, research has demonstrated that the regulation of biosynthesis and transport of amino acid in cyanobacteria is controlled by lysine riboswitch. For instance, d-hydantoinase, Holliday junction DNA helicase subunit (RuvA) and an amino acid carrier protein showed in their upstream sequence the presence of lysine riboswitches (Singh et al. 2018). Even though these discoveries brought many benefits in cyanobacteria gene regulation, further investigations based on molecular evolution are also needed on this riboswitch to better understand its effect on in vivo expression of these genes.
glmS riboswitch
The glmS is a unique riboswitch not only known as gene regulator, but also as nucleic acid catalyst (Fei et al. 2014), and this regulation is monitored through a self-cleavage activity (Davis et al. 2011). Here, the reaction requires the hydrated Mg2+ which is simultaneously cleaved in the presence of glucosamine-6-phosphate (GlcN6P) (Barrick et al. 2004; Winkler et al. 2004). Alternatively, the glmS ribozyme adopts a compact double pseudoknot tertiary structure, with two strictly packed helical stacks (Cochrane et al. 2007).
It has been demonstrated that genetic switches may have functioned as metabolite sensors in primitive organisms, and other researchers have suggested that modern cells retain some of these ancient hereditary control systems (Barrick et al. 2004). In this case, glmS riboswitch shows a unique mechanism compared to other riboswitch classes studied which utilize ligand-induced changes in expression platform sequences to regulate numerous gene expression processes. Further, this research has emphasized that ribozyme acts as a metabolite responsive genetic switch that cleaves itself and represses the glmS gene whose activity depends on binding in the active site of the ribozyme and sufficient concentration of glucosamine-6-phosphate. This makes the amine to function as a general acid and electrostatic catalyst (Barrick and Breaker 2007; Ferre-D’Amare 2011).
Apart from GlcN6P, other researches revealed that glmS riboswitch can respond to multiple metabolites but not triggered by a conformational change. To provide evidence on this, the comparison of inhibitor apo, and activator which bound this ribozyme structures were further carried out and revealed that the active site is preorganized even in the absence of the effector (Butler et al. 2011; Cochrane et al. 2007; Watson and Fedor 2011).
A 246-nt sequence found in 5′-UTR of the glmS gene in B. subtilis explains the cleavage at a specific site in the RNA (Barrick and Breaker 2007; Barrick et al. 2004). Further, the ribozyme is pre-folded but inactive in the absence of GlcN6P, meaning that it has evolved strict dependence on the small exogenous molecule. To elucidate this, research carried out on the use of coenzyme by the glmS ribozyme–riboswitch demonstrated that replacement of the ribozyme by catalytically compromised mutant results in abrogation of sporulation by B. subtilis. In this research, the in vitro study on self-cleaving domain of the glmS mRNA further demonstrated that a ribozyme can be designed to function as a multiple-turnover catalyst (Ferre-D’Amare 2011). Recent findings regarding reporter gene experiments in B. subtilis confirmed that this riboswitch integrates positive and negative chemical signals in its natural biological context (Watson and Fedor 2011). The current discovery demonstrates that glmS riboswitch resides in cyanobacteria with a function of controlling amidohydrolase, thioredoxin and cytochrome c biogenesis protein (CcdA) expression (Singh et al. 2018).
Glycine riboswitch
Glycine riboswitch binds glycine to regulate glycine metabolism genes, including the use of glycine as an energy source (Butler et al. 2011). This natural regulator belongs among the earliest known riboswitches and has been known in various bacteria including Streptomyces griseus (Mandal et al. 2004; Tezuka and Ohnishi 2014). So far, glycine-sensing riboswitch found in B. subtilis works as a rare genetic on switch for the gcvT gene, which codes for proteins that form the glycine cleavage system. For this riboswitch, a leader sequence was recognized but the linker interaction was not well characterized. Mostly, these riboswitches combine two ligand-binding domains which function cooperatively to more closely approximate a two-state genetic switch which is important to ensure the excess glycine. Here, there are used efficiently to provide carbon flux by means of the citric acid cycle even to maintain adequate amounts of the amino acid for protein synthesis (Mandal et al. 2004). In addition, an effect on gene expression regulation by a putative glycine riboswitch located in the 5′-UTR of a sodium: alanine symporter family (SAF) protein gene residing in A Streptococcus pyogenes strain 591 (serotype M49) was evaluated. Hence, the evaluation of the presence or absence of glycine pointed out that high glycine concentrations downregulate gene expression whereas less concentration of glycine led to the production of a full-length transcript. Finally, this research concluded that putative glycine riboswitch in S. pyogenes strain 591 (serotype M49) represses expression of the SAF protein gene and the downstream putative cation efflux protein gene in the presence of high glycine concentrations (Khani et al. 2018). Similarly, within this year, seven cyanobacterial genomes namely Leptolyngbya PCC 7375, Mastigocoleus testarum BC008, Halothece PCC 7418, Myxosarcina sp. GI1, Prochlorococcus sp. scB243_498P3, Scytonema millei VB511283 and Tolypothrix campylonemoides VB511288 have been mined and confirmed to have glycine riboswitch where aspartate racemase, aspartyl/asparaginyl beta-hydroxylase encoding genes are glycine riboswitch regulated (Singh et al. 2018).
PreQ1 riboswitches
7-Aminomethyl-7-deazaguanine (PreQ1) have been known as the smallest riboswitches which sense mRNA domains. This class of riboswitch binds to pyrrolopyrimidine to regulate genes involved in the biosynthesis or transport of precursor queuosine, universally found in four RNA tRNAs (Kang et al. 2014; McCown et al. 2014; Meyer et al. 2008). Two entirely distinct classes of PreQ1 riboswitches are known: PreQ1-I and PreQ1-II (Rieder et al. 2010). The binding domain of PreQ1-I riboswitches are unusually small among naturally occurring riboswitches and, divalent cations are required for its high-affinity binding. It was found that the P4 helix in PreQ1 class I and flanking adenine residues play crucial and unexpected roles to control pseudoknot formation and, in turn, to sequester the Shine–Dalgarno sequence (Kang et al. 2014). PreQ1-II riboswitches mainly found in the genera Streptococcus and Lactococcus, can also be found in B. subtilis (Petrone et al. 2011), but their response mechanism remains undetermined at the molecular and biophysical level (Rieder et al. 2010). A study conducted by comparing the free-state 3D structure of the T. tengcongensis and B. subtilis PreQ1 riboswitch, revealed that T. tengcongensis PreQ1 riboswitch remains the same in its bound state whereas B. subtilis PreQ1 riboswitch showed a remarkable change.
Further discussions and evaluation on some regulation functions for these riboswitches and the unfolding of their aptamers by all-atom molecular dynamic simulation indicated that they have similar unfolding or folding pathways and ligand-binding processes. Beyond this, only what has been noticed about their difference is the folding intermediate states. Preferably, the results obtained from this evaluation were suggested to be used to understand the regulation mechanism of different riboswitches with free-state 3D structures similar to their bound states (Gong et al. 2012). Moreover, to deepen elegantly way, the study concerned ligand-free conformation mechanisms of PreQ1 aptamer domains for B. subtilis has been conducted. As results, an overall 1.5 µs all-atom molecular dynamics simulations were used and showed a state stability of ligand-free aptamers supported by a folded P1-L3 with an open binding pocket (Gong et al. 2014). In the last 2 years, rational re-engineering of a transcriptional silencing PreQ1 riboswitch was then introduced in B. subtilis resulting in the identification of an orthogonal riboswitch-ligand pairing that effectively repressed the transcription of selected genes in B. subtilis. Among these candidate riboswitches, the resulting artificial riboswitch, M1 (C17U), responded to the diamino analog of PreQ0 (DPQ0) and effectively repressed transcription of mreB, a gene for cell morphology in B. subtilis, upon addition of 2 mM DPQ0 (Robinson et al. 2016; Wu et al. 2015).
Furthermore, apart from B. subtilis, research carried out on mycobacteria revealed that these organisms do not have preQ1 biosynthetic genes, this absence pushed researchers to study whether preQ1 could be used as an exogenous non-metabolite ligand to control riboswitches in mycobacteria. Hence, PreQ1 riboswitches were assayed and successfully drove PreQ1-dependent repression of a green fluorescent protein reporter in Mycobacterium smegmatis. Above all, researchers, by practicing engineering approaches on naturally occurring PreQ1 riboswitches, they have not only extended the tools available for inducible gene regulation in mycobacteria but also uncovered new behavior of these riboswitches (Van Vlack et al. 2017). In very recent years, the curiosity of exploring the importance of riboswitch on cyanobacteria genes regulation accelerated scientists to conduct experiments on this riboswitch and results led to the confirmation that PreQ1-I riboswitches in cyanobacteria are thinly dispersed with only 14 genomes showing their presence. This research has further showed that the largest number of riboswitch regulated genes are present in Microcystis aeruginosa NIES 843 and Trichodesmium erythraeum IMS 101 while only single gene was found to be regulated under PreQ1-II (pre-queuosine) riboswitch in Leptolyngbya sp. The PreQ1-II riboswitch also showed its present upstream of soluble [2Fe-2S] ferredoxin gene in Leptolyngbya sp. (Singh et al. 2018).
Based on chronological discoveries of regulatory RNA elements involved in gene regulation, it has been confirmed that highly structured domains residing within non-coding regions of certain bacterial mRNAs serve as metabolite-responsive genetic switches. Yet, early 2004, around seven classes of riboswitch and related mechanisms have been experimentally demonstrated. A few years later until now, with considerable progress made in this field, different classes of natural and synthetic riboswitches together with their related application were discovered. In the above text, we reviewed nine studied natural riboswitches in detail (Table 1). However, there are still some other riboswitches especially the synthetic ones showing potential application in gene regulation (Table 2). In the next section, we will discuss progresses in synthetic riboswitches.
Progress in synthetic riboswitches
Artificial devices such as synthetic riboswitches which own similar properties as natural riboswitches, have shown potential in introducing unnatural phenotypic perturbation due to their synthetic traits which are distinct from that of innate metabolism (Lee and Oh 2015). Heretofore, two remarkable limitations should be carried away to allow researchers to build a beneficial riboswitch. The first one is the lack of a large number of alternative aptamers as ligand-binding modules and the second is the large discrepancy between the in vitro affinities of aptamers for their ligands and some cases the high concentrations of ligand needed in vivo to flip the corresponding switches (Berens et al. 2015). More elegant solution to these problems is that different synthetic riboswitches that display deficient background levels of gene expression in the absence of the desired ligand and robust increases in expression when it is present have been suggested (Lynch et al. 2007). Contrariwise, the construction of synthetic regulators of transcription and translation has failed repeatedly. Importantly, over the past few years, engineered riboswitches most often targeting translation initiation, either by controlling access to the ribosomal binding site through helix slippage or by sequestering its sequence were reported in bacteria (Suess et al. 2004). However, a more in-depth, accessible and understandable design is quite imperative to discover and combat the inefficacy of riboswitches towards regulation of metabolic systems (Fig. 3).
Development procedure of new riboswitch. (a) Bacterial chromosome showing the location of natural riboswitch in 5′-UTR between two endogenous genes. (b) Scheme of synthetic riboswitch library construction from 5′-UTR, natural riboswitch (aptamer and expression platform) or simulated sequences. (c) In silico riboswitch sequence prediction. (d) Sequence engineering, plasmid construction, library screening and (e) dilution of culture and set up ligand incubations followed by fluorescence measurement and expression level quantitation. (f) Results analysis and selection of the strongest riboswitch. (g) In vivo essay: ON or OFF switch response in accordance with internal or external ligand effect
A continuous incentive on this query underlined the possibility of performing selections for novel synthetic riboswitches which function similarly both in vitro and in vivo (Mishler and Gallivan 2014), and different previous researches revealed that riboswitches for translational control could be designed successfully. Furthermore, an in-silico pipeline for the rational design of synthetic riboswitches that regulate gene expression at the transcriptional level have been reported (Wachsmuth et al. 2013; Wittmann and Suess 2012). Here, a theophylline aptamer was used as a sensor to design an actuator part as RNA sequences that can fold into functional intrinsic terminator structures. Based on biochemical characterization observed during an in silico pipeline for the rational design of synthetic riboswitches, several of the designed constructs demonstrated ligand-dependent control of gene expression in E. coli, promising that it is possible to engineer riboswitches for both translational and transcriptional regulation.
De novo design of a synthetic riboswitch based on a connection of functional building blocks has been achieved. This riboswitch candidate should be evaluated by folding simulations to further tune the transcription termination in organisms (Wachsmuth et al. 2013).
Recently, different techniques based on screening or rational design methods to construct artificial riboswitches that function in either bacterial or eukaryotic translational systems were described (Ogawa 2014). Urgent studies on riboswitches have been conducted and results are now assured to be used in biosensor design. For instance, the combination of different permutations topology of the ligand sensing domain of natural riboswitches with in vitro selected fluorogenic aptamers proved to be a promising technology to enhance fluorescence turn-on and ligand binding affinity compared to the non-permuted topology. In the same way, SAM-I riboswitch circular permutation of the riboswitch ligand sensing domain also gives functional biosensors (Truong et al. 2018). Some azaaromatic molecules are bound by the RNA with nanomolar dissociation constants, and a subset of these ligands activate riboswitch-mediated gene expression in the cells. Based on this supposition, Lubelski et al. (2006) used a dual strategy of formulating ligand hypotheses based on rare gene associations and testing a variety of diverse chemical compounds for possible binding by yjdF motif RNAs. Results obtained demonstrated that yjdF motif RNAs are riboswitches that can regulate gene expression by binding to an unusually large diversity of polycyclic aromatic nitrogen-containing heterocycles, which are sometimes called PANHs or azaaromatics. From these findings, a profound research is needed to confirm the hypothesis raised up that these “azaaromatic riboswitches” might have evolved to bind natural member classes of large, planar, and hydrophobic compounds to activate production of the YjdF protein, perhaps as a mechanism for detoxification. If true, azaaromatic riboswitches would be analogous to protein receptors that bind diverse polycyclic aromatic hydrocarbon (PAH) compounds and activate gene expression to regulate natural PAH production or to overcome PAH toxicity as seen in some bacterial species (Huillet et al. 2006). Few examples of protein sensor-regulators for flavonoids have been shown (Siedler et al. 2014; Wenzel et al. 2012). More recent research (Jang et al. 2017) develops artificial riboswitches that activate gene expression in response to naringenin. The results showed a high response to naringenin and activated gene expression up to 2.91 fold. The reported naringenin riboswitches will be valuable tools in metabolic engineering of microorganisms for the production of flavonoids.
In the last few years, researches in synthetic biology and biotechnology have evolved rapidly, and their application for bioengineering are nowadays remarkably achieved to meet human needs. CRISPR technology is among the current useful biotechnology tool used in prokaryotic and eukaryotic organisms for gene editing intention even though they are some challenges remained to alleviate. Very recently, a combination of riboswitch and CRISPR development technology has been launched with the aim to functionalize synthetic sgRNA designs to enable inducible and spatiotemporal regulation of CRISPR-based genetic editors in response to cellular or extracellular stimuli. Interestingly, some riboswitches can be developed and adapted to control sgRNA activity by adding corresponding sequences to its 5′end to repress the guide sequence in the unbound confirmation and reconstitute the active confirmation after ligand binding. In this sense, current study on genome editing and transcriptional activation in mammalian cells was able to incorporate ligand-responsive self-cleaving catalytic RNAs (aptazymes) into guide RNAs resulting a developed set of aptazyme-embedded guide RNAs that enable small molecule-controlled nuclease-mediated genome editing and small molecule-controlled base editing, as well as small molecule-dependent transcriptional activation in mammalian cells (Tang et al. 2017). Also, the advantage of using sgRNA-based inducible systems for synthetic biology applications has been recently showcased in a study demonstrating the ability to rewire cellular pathways by CRISPR-TR with modified sgRNAs containing ligand-responsive riboswitches. These sgRNA ‘signal conductors’ employ a strand-displacement mechanism to transition between OFF and ON states and can be coupled to a variety of inducers and dCas9 effectors (Liu et al. 2016). Furthermore, aptamer-controlled hammerhead ribozymes (so-called aptazymes) have been shown to be a versatile platform for the engineering of novel gene regulators. However, the generation of these novel aptazymes requires a functional aptamer-ribozyme connection, which can be difficult to engineer. Interestingly, Wieland et al. (2012), defined and elaborated new methods and protocols for FACS-mediated aptazyme identification in bacteria and for engineering an aptazyme-based gene control system in mammalian cells.
Roßmanith and Narberhaus (2016) established three riboswitch-RNAT systems conferring dual regulation of transcription and translation depending on the two triggers ligand binding and temperature. This concept relied on designing thermoswitches by integration of a thermosensor into riboswitches from different classes (TPP and lysine) as well as synthetic riboswitches (theophylline) in modular ways to gain functional regulatory elements. These thermoswitches respond to the cognate ligand at low temperatures and are turned into a continuous on-state by a temperature upshift. These findings might help scientists to deplore riboswitches and RNATs for engineering synthetic RNA regulators due to their modular behavior.
Most recently, engineered drug-inducible catalytically inactive Cpf1 nuclease has been used to effectively construct transcriptional repressors in bacteria and plants. Besides, when it is fused to transcriptional activation domains, the construct enabled to tune the expression of endogenous genes in human cells. It is in this sense that, a programmable ligand-controlled dAsCpf1 systems either by coupling crRNAs with engineered riboswitches or by fusing dAsCpf1 proteins with G protein-coupled receptors has been constructed. These approaches using tunable CRISPR–Cpf1-based transcription factors allow the regulation of the transcription of endogenous genes in response to diverse classes of ligands, thus constructing artificial signaling pathways with rewired cellular input–output behaviors (Liu et al. 2017; Tak et al. 2017). Referring to a crucial impact of environmental pH on metabolism and behavior of living cells, Pham et al. (2017) engineered a set of riboswitch-based pH-sensing genetic devices to allow the control of gene expression. A digital pH sensing system designed can use the analog-sensing characteristic of these devices for high-resolution recording of host cell exposure to the external pH levels. This valuable innovation could be used in multiple engineering aspects of host cell for improved tolerance to a narrow range of organic acids, bioremediation and a precious phenotype for metabolic engineering.
Concluding remarks and future perspectives
Riboswitches, regulatory RNA elements which control a wide set of metabolic pathways in bacteria are nowadays used as molecular tools for the precise control of gene expression in many genetic studies. They only consist of RNA molecules and are composed of aptamer domain (high affinity and specific binding of metabolites) and expression platform (regulation of gene expression). Their output effect can intervene in controlling transcription termination (kinetically controlled), translation initiation, RNA stability and protein stability. Natural riboswitches are considered as an ideal target for constructing synthetic regulatory systems for gene expression and some synthetic riboswitches have been achieved so far. Basing on their simple use without any involvement of intermediate factors, riboswitches could not only be manipulated for biosynthetic regulation and transport of cognate ligand, but also in various domains such as microbial food detection, antimicrobial drug targets, antibiotic resistance, boolean logic gates as well as bacterial cell behavior as shown in Table 2. Interestingly, riboswitches demonstrated good functionality associated with both transcription initiation and translation initiation in gram positive bacteria especially in B. subtilis as well as in other microorganisms. However, riboswitches remain the rarest genetic parts hitherto less known but promising a greater importance in the regulation of chromosomal genes. Despite their insufficiency in the genome, the design of synthetic variants could be an interesting direction for future research.
In addition to their essential role in molecular engineering as well as in biochemical molecules production, their sequences may also be a requisite tool to be deeply explored in order to better understand drug resistance challenges, genetic-associated syndromes, cancerous genes regulation, designing of synthetic aptamers required for the detection of harmful substances in food microorganisms as well as the discovery of epigenetic related diseases. Moreover, the implementation to alleviate the above challenges, would require much concerted efforts from synthetic biologists and metabolic pathway engineers to deploy synthetic biology and genetic engineering techniques at the forefront. When these requisites are filled in, it will facilitate the use of these RNA molecules and understand their effect in different areas of life. It would be an essential matter to explore these regulatory tools and discover new intracellular ligands which could likewise be used as metabolites ligand sensing. Similarly, in-depth studies on the response of 5′-UTR to metabolites should be an essential focus in diversified organisms. Subjected to our critical analysis, the results outlined in this review point out the urgent need for multiple comprehensive studies using mathematical modelling and computational simulation as well as experimental practices to demonstrate the effect of RNA molecules during the expression of endogenous genes. Whether it is an artificial or natural, the function of riboswitches to regulate genes involved in metabolic pathways abide a big challenge. Researchers in this field have intensively delved into riboswitches mechanisms and their effect on downstream coding sequence in host cells, nevertheless, there is still a serious quest of understanding the sophisticated phenomena of sensing and responding on intra or extracellular molecules known as ligands which encompass limiting factors on gene expression.
References
Alexey GV, Dimitry AR, Andrey AM, Mikhail SG (2003) Riboswitches: the oldest mechanism for the regulation of gene expression? Trends Genet 20:44–50
Babitzke P, Gollnick P (2001) Posttranscription initiation control of tryptophan metabolism in Bacillus subtilis by the trp RNA-binding attenuation protein (TRAP), anti-TRAP, and RNA structure. J Bacteriol 183:5795–5802. https://doi.org/10.1128/JB.183.20.5795-5802.2001
Barrick JE, Breaker RR (2007) The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol 8:R239. https://doi.org/10.1186/gb-2007-8-11-r239
Barrick JE et al (2004) New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci USA 101:6421–6426. https://doi.org/10.1073/pnas.0308014101
Bastet L, Dube A, Masse E, Lafontaine DA (2011) New insights into riboswitch regulation mechanisms. Mol Microbiol 80:1148–1154. https://doi.org/10.1111/j.1365-2958.2011.07654.x
Batey TR (2006) Structures of regulatory elements in mRNAs. Curr Opin Struct Biol 16:299–306
Berens C, Suess B (2015) Riboswitch engineering—making the all-important second and third steps. Curr Opin Biotechnol 31:10–15. https://doi.org/10.1016/j.copbio.2014.07.014
Berens C, Groher F, Suess B (2015) RNA aptamers as genetic control devices: the potential of riboswitches as synthetic elements for regulating gene expression. Biotechnol J 10:246–257
Blount KF, Wang JX, Lim J, Sudarsan N, Breaker RR (2007) Antibacterial lysine analogs that target lysine riboswitches. Nat Chem Biol 3:44–49. https://doi.org/10.1038/nchembio842
Bocobza S, Adato A, Mandel T, Shapira M, Nudler E, Aharoni A (2007) Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Genes Dev 21:2874–2879. https://doi.org/10.1101/gad.443907
Breaker RR (2012) Riboswitches and the RNA world. Cold Spring Harb Perspect Biol 4:a003566. https://doi.org/10.1101/cshperspect.a003566
Butler EB, Xiong Y, Wang J, Strobel SA (2011) Structural basis of cooperative ligand binding by the glycine riboswitch. Chem Biol 18:293–298. https://doi.org/10.1016/j.chembiol.2011.01.013
Cochrane JC, Lipchock SV, Strobel SA (2007) Structural investigation of the glmS ribozyme bound to its catalytic cofactor. Chem Biol 14:97–105. https://doi.org/10.1016/j.chembiol.2006.12.005
Davis JH, Dunican BF, Strobel SA (2011) glmS riboswitch binding to the glucosamine-6-phosphate alpha-anomer shifts the pKa toward neutrality. Biochemistry 50:7236–7242. https://doi.org/10.1021/bi200471c
Delfosse V, Bouchard P, Bonneau E, Dagenais P, Lemay JF, Lafontaine DA, Legault P (2010) Riboswitch structure: an internal residue mimicking the purine ligand. Nucleic Acids Res 38:2057–2068. https://doi.org/10.1093/nar/gkp1080
Demolli S, Geist MM, Weigand JE, Matschiavelli N, Suess B, Rother M (2014) Development of beta-lactamase as a tool for monitoring conditional gene expression by a tetracycline-riboswitch in Methanosarcina acetivorans. Archaea 2014:725610. https://doi.org/10.1155/2014/725610
Doerks T, Copley RR, Schultz J, Ponting CP, Bork P (2002) Systematic identification of novel protein domain families associated with nuclear functions. Genome Res 12:47–56
Fei X et al (2014) Phosphatase-inert glucosamine 6-phosphate mimics serve as actuators of the glmS riboswitch. ACS Chem Biol 9:2875–2882
Ferre-D’Amare AR (2011) Use of a coenzyme by the glmS ribozyme–riboswitch suggests primordial expansion of RNA chemistry by small molecules. Philos Trans R Soc Lond B Biol Sci 366:2942–2948. https://doi.org/10.1098/rstb.2011.0131
Fowler CC, Brown ED, Li Y (2010) Using a riboswitch sensor to examine coenzyme B(12) metabolism and transport in E. coli. Chem Biol 17:756–765. https://doi.org/10.1016/j.chembiol.2010.05.025
Gallo S, Sigel RKO (2018) Covalent and non-covalent binding of platinated vitamin B12-derivatives to a B12 responsive riboswitch. Inorg Chim Acta 472:214–220. https://doi.org/10.1016/j.ica.2017.09.018
Garst AD, Heroux A, Rambo RP, Batey RT (2008) Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem 283:22347–22351. https://doi.org/10.1074/jbc.C800120200
Garst AD, Edwards AL, Batey RT (2011) Riboswitches: structures and mechanisms. Cold Spring Harb Perspect Biol 3: a003533. https://doi.org/10.1101/cshperspect.a003533
Gong Z, Zhao Y, Chen C, Xiao Y (2012) Computational study of unfolding and regulation mechanism of preQ1 riboswitches. PLoS ONE 7:e45239. https://doi.org/10.1371/journal.pone.0045239
Gong Z, Zhao Y, Chen C, Duan Y, Xiao Y (2014) Insights into ligand binding to PreQ1 riboswitch aptamer from molecular dynamics simulations. PLoS ONE 9:e92247. https://doi.org/10.1371/journal.pone.0092247
Hallera A, Altmanb RB, Soulièrea MF, Blanchardb SC, Micuraa R (2013) Folding and ligand recognition of the TPP riboswitch aptamer at single-molecule resolution. Proc Natl Acad Sci USA 10:4188–4193
Henkin TM (2008) Riboswitch RNAs: using RNA to sense cellular metabolism. Genes Dev 22:3383–3390. https://doi.org/10.1101/gad.1747308
Hickey SF, Hammond MC (2014) Structure-guided design of fluorescent S-adenosylmethionine analogs for a high-throughput screen to target SAM-I riboswitch RNAs. Chem Biol 21:345–356. https://doi.org/10.1016/j.chembiol.2014.01.004
Huang W, Joohyun K, Shantenu J, Aboul-ela F (2012) Conformational heterogeneity of the SAM-I riboswitch transcriptional ON state: a chaperone-like role for S-adenosyl methionine. J Mol Biol 418:331–349. https://doi.org/10.1016/j.jmb.2012.02.019
Huillet E, Velge P, Vallaeys T, Pardon P (2006) LadR, a new PadR-related transcriptional regulator from Listeria monocytogenes, negatively regulates the expression of the multidrug efflux pump MdrL. FEMS Microbiol Lett 254:87–94. https://doi.org/10.1111/j.1574-6968.2005.00014.x
Jang S, Jung GY (2018) Systematic optimization of l-tryptophan riboswitches for efficient monitoring of the metabolite in Escherichia coli. Biotechnol Bioeng 115:266–271. https://doi.org/10.1002/bit.26448%5D
Jang S, Jang S, Xiu Y, Kang TJ, Lee SH, Koffas MAG, Jung GY (2017) Development of artificial riboswitches for monitoring of naringenin in vivo. ACS Synth Biol 6:2077–2085. https://doi.org/10.1021/acssynbio.7b00128
Jin Y, Watt RM, Danchin A, Huang JD (2009) Use of a riboswitch-controlled conditional hypomorphic mutation to uncover a role for the essential csrA gene in bacterial autoaggregation. J Biol Chem 284:28738–28745. https://doi.org/10.1074/jbc.M109.028076
Kang M, Eichhorn CD, Feigon J (2014) Structural determinants for ligand capture by a class II preQ1 riboswitch. Proc Natl Acad Sci USA 111:E663–E671. https://doi.org/10.1073/pnas.1400126111
Ketzer P et al (2014) Artificial riboswitches for gene expression and replication control of DNA and RNA viruses. Proc Natl Acad Sci USA 111:E554–E562. https://doi.org/10.1073/pnas.1318563111
Khani A, Popp N, Kreikemeyer B, Patenge N (2018) A glycine riboswitch in Streptococcus pyogenes controls expression of a sodium:alanine symporter family protein gene. Front Microbiol 9:200. https://doi.org/10.3389/fmicb.2018.00200
Kim JN, Roth A, Breaker RR (2007) Guanine riboswitch variants from Mesoplasma florum selectively recognize 2′-deoxyguanosine. Proc Natl Acad Sci USA 104:16092–16097. https://doi.org/10.1073/pnas.0705884104
Kim JN, Blount KF, Puskarz I, Lim J, Link KH, Breaker RR (2009) Design and antimicrobial action of purine analogs that bind guanine riboswitches. ACS Chem Biol 4:915–927
Klein DJ, Ferré-D’Amaré AR (2009) Crystallization of the glmS ribozyme–riboswitch. Methods Mol Biol 540:129–139
Kochhar S, Paulus H (1996) Lysine-induced premature transcription termination in the lysC operon of Bacillus subtilis. Microbiology 142:1635–1639
Lee SW, Oh MK (2015) A synthetic suicide riboswitch for the high-throughput screening of metabolite production in Saccharomyces cerevisiae. Metab Eng 28:143–150. https://doi.org/10.1016/j.ymben.2015.01.004
Lee ER, Blount KF, Breaker RR (2009) Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol 6:187–194. https://doi.org/10.4161/rna.6.2.7727
Lee ER, Blount KF, Breaker RR (2014) Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol 6:187–194. https://doi.org/10.4161/rna.6.2.7727
Liu Y et al (2016) Directing cellular information flow via CRISPR signal conductors. Nat Methods 13:938–944. https://doi.org/10.1038/nmeth.3994
Liu Y, Han J, Chen Z, Wu H, Dong H, Nie G (2017) Engineering cell signaling using tunable CRISPR–Cpf1-based transcription factors. Nat Commun 8:2095. https://doi.org/10.1038/s41467-017-02265-x
Lu C et al (2010) SAM recognition and conformational switching mechanism in the Bacillus subtilis yitJ S box/SAM-I riboswitch. J Mol Biol 404:803–818. https://doi.org/10.1016/j.jmb.2010.09.059
Lubelski J, de Jong A, van Merkerk R, Agustiandari H, Kuipers OP, Kok J, Driessen AJ (2006) LmrCD is a major multidrug resistance transporter in Lactococcus lactis. Mol Microbiol 61:771–781. https://doi.org/10.1111/j.1365-2958.2006.05267.x
Lunse CE, Schuller A, Mayer G (2014) The promise of riboswitches as potential antibacterial drug targets. Int J Med Microbiol 304:79–92. https://doi.org/10.1016/j.ijmm.2013.09.002
Lynch SA, Desai SK, Sajj HK, Gallivan JP (2007) A high throughput screen for synthetic riboswitches reveals mechanistic insights into their function. Chem Biol 14:173–184
Mandal M, Breaker RR (2004) Gene regulation by riboswitches. Nat Rev Mol Cell Biol 5:451–463. https://doi.org/10.1038/nrm1403
Mandal M, Barrick JE, Breaker RR, Boese B, Winkler WC (2003) Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113:577–586
Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR (2004) A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science 306:1477
Marcano-Velazquez JG, Batey RT (2015) Structure-guided mutational analysis of gene regulation by the Bacillus subtilis pbuE adenine-responsive riboswitch in a cellular context. J Biol Chem 290:4464–4475. https://doi.org/10.1074/jbc.M114.613497
Martini L, Mansy SS (2011) Cell-like systems with riboswitch controlled gene expression. Chem Commun 47:10734–10736. https://doi.org/10.1039/c1cc13930d
McCown PJ, Liang JJ, Weinberg Z, Breaker RR (2014) Structural, functional, and taxonomic diversity of three preQ1 riboswitch classes. Chem Biol 21:880–889. https://doi.org/10.1016/j.chembiol.2014.05.015
McRose D et al (2014) Alternatives to vitamin B1 uptake revealed with discovery of riboswitches in multiple marine eukaryotic lineages. ISME J 8:2517–2529. https://doi.org/10.1038/ismej.2014.146
Meyer MM, Roth A, Chervin SM, Garcia GA, Breaker RR (2008) Confirmation of a second natural preQ1 aptamer class in Streptococcaceae bacteria. RNA 14:685–695. https://doi.org/10.1261/rna.937308
Miotto P et al (2012) Genome-wide discovery of small RNAs in Mycobacterium tuberculosis. PLoS ONE 7:e51950. https://doi.org/10.1371/journal.pone.0051950
Mishler DM, Gallivan JP (2014) A family of synthetic riboswitches adopts a kinetic trapping mechanism. Nucleic Acids Res 42:6753–6761. https://doi.org/10.1093/nar/gku262
Moldovan MA, Petrova SA, Gelfand MS (2018) Comparative genomic analysis of fungal TPP-riboswitches. Fungal Genet Biol 114:34–41. https://doi.org/10.1016/j.fgb.2018.03.004
Moore SJ, Mayer MJ, Deery RB, Warren E MJ (2014) Towards a cell factory for vitamin B12 production in Bacillus megaterium: bypassing of the cobalamin riboswitch control elements. N Biotechnol 31:553–556
Mukherjee S, Barash D, Sengupta S (2017) Comparative genomics and phylogenomic analyses of lysine riboswitch distributions in bacteria. PLoS ONE 12:e0184314. https://doi.org/10.1371/journal.pone.0184314
Nahvi A, Barrick JE, Breaker RR (2004) Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res 32:143–150
Ogawa A (2014) Artificial riboswitches: methods and protocols. Humana Press, New York
Ott E, Stolz J, Mack M (2014) The RFN riboswitch of Bacillus subtilis is a target for the antibiotic roseoflavin produced by Streptomyces davawensis. RNA Biol 6:276–280. https://doi.org/10.4161/rna.6.3.8342
Palou-Mir J, Musiari A, Sigel RK, Barcelo-Oliver M (2016) Characterization of the full-length btuB riboswitch from Klebsiella pneumoniae. J Inorg Biochem 160:106–113. https://doi.org/10.1016/j.jinorgbio.2015.12.012
Pedrolli DB, Matern A, Wang J, Ester M, Siedler K, Breaker R, Mack M (2012) A highly specialized flavin mononucleotide riboswitch responds differently to similar ligands and confers roseoflavin resistance to Streptomyces davawensis. Nucleic Acids Res 40:8662–8673. https://doi.org/10.1093/nar/gks616
Perez AA, Liu Z, Rodionov DA, Li Z, Bryant DA (2016) Complementation of cobalamin auxotrophy in Synechococcus sp. strain PCC 7002 and validation of a putative cobalamin riboswitch in vivo. J Bacteriol 198:2743–2752. https://doi.org/10.1128/JB.00475-16
Petrone PM, Dewhurst J, Tommasi R, Whitehead L, Pomerantz AK (2011) Atomic-scale characterization of conformational changes in the preQ1 riboswitch aptamer upon ligand binding. J Mol Graph Model 30:179–185. https://doi.org/10.1016/j.jmgm.2011.07.006
Pham HL, Wong A, Chua N, Teo WS, Yew WS, Chang MW (2017) Engineering a riboswitch-based genetic platform for the self-directed evolution of acid-tolerant phenotypes. Nat Commun 8:411. https://doi.org/10.1038/s41467-017-00511-w
Ramesh A (2015) Second messenger—sensing riboswitches in bacteria. Semin Cell Dev Biol 47–48:3–8. https://doi.org/10.1016/j.semcdb.2015.10.019
Ramesh A, Winkler WC (2014) Magnesium-sensing riboswitches in bacteria. RNA Biol 7:77–83. https://doi.org/10.4161/rna.7.1.10490
Rieder U, Kreutz C, Micura R (2010) Folding of a transcriptionally acting preQ1 riboswitch. Proc Natl Acad Sci USA 107:10804–10809. https://doi.org/10.1073/pnas.0914925107
Robinson CJ, Medina-Stacey D, Wu MC, Vincent HA, Micklefield J (2016) Rewiring riboswitches to create new genetic circuits in bacteria. Methods Enzymol 575:319–348. https://doi.org/10.1016/bs.mie.2016.02.022
Rode AB, Endoh T, Sugimoto N (2015) Tuning riboswitch-mediated gene regulation by rational control of aptamer ligand binding properties. Angew Chem Int Ed Engl 54:905–909. https://doi.org/10.1002/anie.201407385
Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS (2003) Regulation of lysine biosynthesis and transport genes in bacteria: yet another RNA riboswitch? Nucleic Acids Res 31:6748–6757
Roßmanith J, Narberhaus F (2016) Exploring the modular nature of riboswitches and RNA thermometers. Nucleic Acids Res 44:5410–5423. https://doi.org/10.1093/nar/gkw232
Savinov A, Perez CF, Block SM (2014) Single-molecule studies of riboswitch folding. Biochim Biophys Acta 1839:1030–1045. https://doi.org/10.1016/j.bbagrm.2014.04.005
Serganov A, Nudler E (2013) A decade of riboswitches. Cell 152:17–24. https://doi.org/10.1016/j.cell.2012.12.024
Shin JH, Wakeman CA, Goodson JR, Rodionov DA, Freedman BG, Senger RS, Winkler WC (2014) Transport of magnesium by a bacterial Nramp-related gene. PLoS Genet 10:e1004429. https://doi.org/10.1371/journal.pgen.1004429
Siedler S, Stahlhut SG, Malla S, Maury J, Neves AR (2014) Novel biosensors based on flavonoid-responsive transcriptional regulators introduced into Escherichia coli. Metab Eng 21:2–8. https://doi.org/10.1016/j.ymben.2013.10.011
Singh P, Kumar N, Jethva M, Yadav S, Kumari P, Thakur A, Kushwaha HR (2018) Riboswitch regulation in cyanobacteria is independent of their habitat adaptations. Physiol Mol Biol Plants 24:315–324. https://doi.org/10.1007/s12298-018-0504-9
Sklyarova SA, Mironov AS (2014) Bacillus subtilis ypaA gene regulation mechanism by FMN riboswitch. Russ J Genet 50:319–322
Sudarsan N (2003) Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA 9:644–647. https://doi.org/10.1261/rna.5090103
Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker RR (2005) Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem Biol 12:1325–1335. https://doi.org/10.1016/j.chembiol.2005.10.007
Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR (2006) Tandem riboswitch architectures exhibit complex gene control functions. Science 314:300–304. https://doi.org/10.1126/science.1130716
Suddala KC et al (2013) Single transcriptional and translational preQ1 riboswitches adopt similar pre-folded ensembles that follow distinct folding pathways into the same ligand-bound structure. Nucleic Acids Res 41:10462–10475. https://doi.org/10.1093/nar/gkt798
Suess B, Fink B, Berens C, Stentz R, Hillen W (2004) A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res 32:1610–1614
Tak YE et al (2017) Inducible and multiplex gene regulation using CRISPR–Cpf1-based transcription factors. Nat Methods 14:1163–1166. https://doi.org/10.1038/nmeth.4483
Tang W, Hu JH, Liu DR (2017) Aptazyme-embedded guide RNAs enable ligand-responsive genome editing and transcriptional activation. Nat Commun 8:15939. https://doi.org/10.1038/ncomms15939
Tezuka T, Ohnishi Y (2014) Two glycine riboswitches activate the glycine cleavage system essential for glycine detoxification in Streptomyces griseus. J Bacteriol 196:1369–1376
Tomsic J, McDaniel BA, Grundy FJ, Henkin TM (2008) Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in Bacillus subtilis exhibit differential sensitivity to SAM In vivo and in vitro. J Bacteriol 190:823–833. https://doi.org/10.1128/JB.01034-07
Topp S, Gallivan JP (2007) Guiding bacteria with small molecules and RNA. J Am Chem Soc 129:6807–6811. https://doi.org/10.1021/ja0692480
Trausch JJ, Batey RT (2014) A disconnect between high-affinity binding and efficient regulation by antifolates and purines in the tetrahydrofolate riboswitch. Chem Biol 21:205–216. https://doi.org/10.1016/j.chembiol.2013.11.012
Trausch JJ, Xu Z, Edwards AL, Reyes FE, Ross PE, Knight R, Batey RT (2014) Structural basis for diversity in the SAM clan of riboswitches. Proc Natl Acad Sci 11:6624–6629
Truong J, Hsieh YF, Truong L, Jia G, Hammond MC (2018) Designing fluorescent biosensors using circular permutations of riboswitches. Methods 143:102–109. https://doi.org/10.1016/j.ymeth.2018.02.014
Van Vlack ER, Topp S, Seeliger JC (2017) Characterization of engineered PreQ1 riboswitches for inducible gene regulation in Mycobacteria. J Bacteriol. https://doi.org/10.1128/JB.00656-16
Vicens Q, Mondragon E, Batey RT (2011) Molecular sensing by the aptamer domain of the FMN riboswitch: a general model for ligand binding by conformational selection. Nucleic Acids Res 39:8586–8598. https://doi.org/10.1093/nar/gkr565
Vitreschak AG (2003) Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. RNA 9:1084–1097. https://doi.org/10.1261/rna.5710303
Wachsmuth M, Findeiss S, Weissheimer N, Stadler PF, Morl M (2013) De novo design of a synthetic riboswitch that regulates transcription termination. Nucleic Acids Res 41:2541–2551. https://doi.org/10.1093/nar/gks1330
Wang J, Gao D, Yu X, Li W, Qi Q (2015) Evolution of a chimeric aspartate kinase for l-lysine production using a synthetic RNA device. Appl Microbiol Biotechnol 99:8527–8536. https://doi.org/10.1007/s00253-015-6615-0
Wang H et al (2017) Dual-targeting small-molecule inhibitors of the Staphylococcus aureus FMN riboswitch disrupt riboflavin homeostasis in an infectious setting. Cell Chem Biol 24:576–588 e576. https://doi.org/10.1016/j.chembiol.2017.03.014
Watson PY, Fedor MJ (2011) The glmS riboswitch integrates signals from activating and inhibitory metabolites in vivo. Nat Struct Mol Biol 18:359–363. https://doi.org/10.1038/nsmb.1989
Watson PY, Fedor MJ (2012) The ydaO motif is an ATP-sensing riboswitch in Bacillus subtilis. Nat Chem Biol 8:963–965. https://doi.org/10.1038/nchembio.1095
Welz R, Breaker RR (2007) Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA 13:573–582. https://doi.org/10.1261/rna.407707
Wenzel M et al (2012) Characterization of the flavonoid-responsive regulator FrrA and its binding sites. J Bacteriol 194:2363–2370. https://doi.org/10.1128/JB.06567-11
Wieland M, Ausländer D, Fussenegger M (2012) Engineering of ribozyme-based riboswitches for mammalian cells. Methods 56:351–357. https://doi.org/10.1016/j.ymeth.2012.01.005
Wilson RC, Smith AM, Fuchs RT, Kleckner IR, Henkin TM, Foster MP (2011) Tuning riboswitch regulation through conformational selection. J Mol Biol 405:926–938. https://doi.org/10.1016/j.jmb.2010.10.056
Wilson-Mitchell SN, Grundy FJ, Henkin TM (2012) Analysis of lysine recognition and specificity of the Bacillus subtilis L. box riboswitch. Nucleic Acids Res 40:5706–5717. https://doi.org/10.1093/nar/gks212
Winkler WC, Breaker RR (2003) Genetic control by metabolite-binding riboswitches. Chembiochem 4:1024–1032. https://doi.org/10.1002/cbic.200300685
Winkler WC, Adam Roth AN, Collins JA, Breaker RR (2004) Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428:281–286
Wittmann A, Suess B (2012) Engineered riboswitches: expanding researchers’ toolbox with synthetic RNA regulators. FEBS Lett 586:2076–2083. https://doi.org/10.1016/j.febslet.2012.02.038
Wu MC, Lowe PT, Robinson CJ, Vincent HA, Dixon N, Leigh J, Micklefield J (2015) Rational re-engineering of a transcriptional silencing PreQ1 riboswitch. J Am Chem Soc 137:9015–9021. https://doi.org/10.1021/jacs.5b03405
Yang J, Seo SW, Jang S, Shin SI, Lim CH, Roh TY, Jung GY (2013) Synthetic RNA devices to expedite the evolution of metabolite-producing microbes. Nat Commun 4:1413
Zhang Y-Y, Cheng H, Sun Y, Wang J-E, Wu Z-Y, Pei R-J (2017) Engineering of thiamine pyrophosphate fluorescent biosensors based on ribozyme switches in mammalian cells. Chin J Anal Chem 45:157–162. https://doi.org/10.1016/s1872-2040(16)60992-1
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sinumvayo, J.P., Zhao, C. & Tuyishime, P. Recent advances and future trends of riboswitches: attractive regulatory tools. World J Microbiol Biotechnol 34, 171 (2018). https://doi.org/10.1007/s11274-018-2554-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-018-2554-0