Abstract
Scorpion long-chain insect selective neurotoxin AaIT has the potential to be used against agricultural insect pests. However, there is still a lack of a heterologous gene expression system that can express AaIT efficiently. Here, using X33 as the host strain and pPICZαA as the expression vector, one transformant had the highest expression of recombinant AaIT (rAaIT) was obtained, and secreted as high as 240 mg/l rAaIT in fed-batch fermentation. Secretory rAaIT was purified by Ni2+-nitriloacetic affinity and CM chromatography, and 8 mg of high purity rAaIT were purified from 200 ml fed-batch fermentation cultures. Injecting silkworm (Bombyx mori Linnaeus) and Galleria mellonella larvae with rAaIT resulted in obvious neurotoxin symptoms and led to death. These results demonstrate that a large amount of anti-insect active rAaIT could be prepared efficiently.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The extensive use of synthetic pesticides to kill pests will also kill insect-eating birds, which provide many ecosystem benefits, thereby impairing ecosystem function (Zhong et al. 2016; Chaudhary et al. 2017). This problem has generated widespread concern; thus, it is desirable to search for natural, green insecticidal substances from animals, plants, and microorganisms (Michiels et al. 2010; Kawachi et al. 2013; Olombrada et al. 2017). Insectivorous animals are an important source of these insect specific toxins. Among insect toxins, many low-molecular-weight proteins that act on the insect nervous system have the most practical value because of their high toxicity and specificity. Scorpions often use poison to capture their prey, often via neurotoxins that are highly toxic to insects. Scorpion venom comprises low-molecular-weight proteins with highly selective activities against the insect nervous system (Pelhate et al. 1998; Zlotkin et al. 2000; Gurevitz et al. 2007). These highly efficient neurotoxins, known as anti-insect toxins, include excitatory toxins and depressant toxins. Scorpion neurotoxins, which have high affinities for sodium channel, are composed of 60–70 amino acids with several pairs of cross-linked disulfide bonds (Zlotkin et al. 1991, 1993). As the first purified insect-selective excitatory and a depressant insect toxin, AaIT, a 70-amino-acid neurotoxin, was isolated from the venom of the Androctonus australis Hector scorpion (Zlotkin et al. 1971). Because of its strict selectivity and high bioactivity, AaIT has great potential value as a biological pesticide (Regev et al. 2006; Pava-Ripol et al. 2008; Liu et al. 2016; Nalcacioglu et al. 2016). Previously, AaIT was used to enhance the insecticidal activity of entomopathogenic fungi, for example, AaIT improved the activity of microbial insecticides from Metarhizium acridum against locusts (Locusta migratoria manilensis) (Wang and Leger 2006; Pava-Ripol et al. 2008). However, a lack of purified active recombinant AaIT (rAaIT) is still the major obstacle preventing analyses of its insecticidal activity against agricultural pests.
Both prokaryotic and eukaryotic systems are used commonly for heterologous protein expression, and these systems have their own advantages and disadvantages (Forstner et al. 2007; Valero 2012). Although many heterologous expression systems have successfully reported the expression of scorpion long chain insect neurotoxins, the expression of recombinant toxins in the systems were low or it was difficult to prepare a large amount of purified toxins (Ji et al. 2002). P. pastoris is a well-known eukaryotic expression system efficiently used to overexpress proteins especially with low molecular weight. However, the previous studies expressed AaIT at a very low level in P. pastoris strain KM71 using the expression plasmid pPIC9K and it was difficult to obtain high-purity rAaIT (Li and Xia 2009). Changing the host strains of P. pastoris or expression vectors may effectively improve the expression level of recombinant proteins. There were not established matching expression vectors and host strains for efficient secretory expression of scorpion long-chain neurotoxins. In the study, we report a suitable method for selection of high level secretory expression transformants of rAaIT from P. pastoris strain X33 using the expression plasmid pPICZɑA. There is also no report on the preparation of large amount of scorpion long chain insect neurotoxins by high density fermentation using P. pastoris. Therefore, high level fed-batch expression and preparation of active rAaIT will help to develop the similar insecticidal proteins to AaIT as new biocontrol agents.
Materials and methods
Materials
All analytical grade reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA) and Sangon Biotech (Shanghai, China). P. pastoris strain X-33 and the expression vector pPICZαA were obtained from Invitrogen (Carlsbad, CA, USA). Low salt Luria–Bertani agar plate containing 40 µg/ml zeocin, yeast extract-peptone-dextrose (YPD) plates containing 100, 200, 500 and 1000 µg/ml zeocin, BMGY growth medium, and BMMY induction medium were prepared as recommended (Invitrogen 2008). Ni2+-nitriloacetic acid (NTA) resin was purchased from Qiagen (Valencia, CA, USA). Dialysis bags and filter devices were purchased form Millipore (Guangzhou, China, molecular weight cut-off: 3 kDa). Rabbit anti-six-histidine IgG and goat anti-rabbit horseradish peroxidase-conjugated (HRP) IgG were purchased from CST (Guangzhou, China).
Screening transformants
AaIT cDNA was amplified by polymerase chain reaction (PCR) from our previous reported plasmid pPIC9K/aait using forward (5′-GCCTCGAGaaaagaAAGAAG AACGGTTACGCTG-3′) and reverse (5′-GCTCTAGAtcaATGATGATGATG ATGATGGTTGATGATAGTGGTGTCGC-3′) primers (Li and Xia 2009). To obtain natural N- terminal rAaIT, a Kex2 signal cleavage site (aaaaga) was fused upstream of the Aait. DNA fragments of amplified AaIT with a stop codon and a 6 × His tag (reverse, 5′-tcaATGATGATGATGATGATG-3′) were digested with XhoI (CTCGAG) and XbaI (TCTAGA) and fragments were inserted into the pPICZαA vector. The recombinant vector was confirmed by sequencing. The expression vector was linearized by digestion with SacI and transformed into P. pastoris X-33 using LiCl as recommended (Invitrogen 2008). P. pastoris transformants were grown on YPD (1% yeast extract, 2% peptone, 2% glucose and 1.8% agar, pH 6.0) plates containing 100 µg/ml of zeocin. Resulting colonies were transferred to YPD plates containing 200, 500 and 1000 µg/ml of zeocin, respectively. After incubation at 28 °C for 3 days, large colonies from YPD plates containing 1000 µg/ml of zeocin were picked and subjected to a PCR analysis. As previously described, small-scale cultivation and expression was performed by using 96-deep-well plates (Bel-Art Scienceware, NJ, USA) (Weis et al. 2004; Barnard et al. 2010). To assay the expression of rAaIT, the induced supernatant samples were transformed into 96-well plates and analyzed by an enzyme-linked immunosorbent assay (ELISA) using rabbit anti-six-histidine IgG and goat anti-rabbit horseradish peroxidase-conjugated IgG according to previously published methods. Finally, 0.02% o-phenylenediamine was used as the substrate in a chromogenic reaction. After 10 min of incubation at room temperature, the plate wells were read on an ELISA plate reader at OD495. The transformants corresponding to the wells with the greater light absorption at 495 nm were considered had the higher rAaIT expression level. The supernatants of the higher light absorption cultures of the selected transformants that were induced by methanol collected from 96-deep-well plates were further verified by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE).
Small-scale fermentation
The highest secretory colony selected by the ELISA and SDS was grown in BMGY [1% yeast extract, 2% peptone, 2% glycerol, 100 mM potassium phosphate (pH 6.0), 1.34% yeast nitrogen broth and 0.4 mg/l biotin] medium with shaking (250 rpm) at 28 °C until the OD600 was between 10 and 12. Then, the cells were collected by centrifugation at room temperature and 1500×g for 10 min and resuspended in 40 ml of BMMY medium [1% yeast extract, 2% peptone, 1% methanol, 100 mM potassium phosphate (pH 6.0), 1.34% yeast nitrogen broth and 0.4 mg/l biotin], followed by incubation at 28 °C with shaking at 250 rpm. Each day, 0.4 ml of 100% methanol was added to the culture. One-milliliter samples were collected at 0, 24, 48, 72, and 96 h after induction with methanol. The protein concentrations of the induced supernatants were assayed by the Bradford method. Then, the supernatants (0.2 ml) were mixed with 0.6 ml of ice cold acetone. After centrifugation, precipitates were dissolved in 20 µl of 1 × SDS loading buffer and heated at 95 °C for 5 min, 10 µl denatured samples were subjected to SDS–PAGE. The amount of secretory rAaIT was assayed by gel scanning density analysis method with software Quantity One (Biorad, USA) and 10 µg bovine serum albumin (BSA) was used as the protein concentration standard. The rAaIT protein was further confirmed by Western blotting with the anti-six-histidine antibody.
Fed-batch fermentation
A single colony of the most highly expressing X33 transformant was inoculated into 5 ml YPG media and grown for 20 h at 28 °C, and inoculated to 300 ml fed-batch basal salts medium with 2% (w/v) glycerol and shaken at 28 °C and 250 rpm to OD600 at about 10. Fed-batch fermentation was in a 5-L bioreactor (NCBIO, China) with 2.8 l basal salts medium. Compressed air was maintained at 5 l/min, dissolved oxygen (DO) was maintained higher than 30% saturation by stirring at 200–1000 rpm at 28 °C. Fermentation was started by adding 300 ml fed-batch basal salts seed culture into the basal salts medium, with pH maintained at 5.5 with 30% ammonium hydroxide. To maintain glycerol concentration in the glycerol fed-batch phase, feed solution was added automatically according to DO > 30% saturation by series regulation of feeding and DO to the wet cell weight at 400 g/l. After the feeding glycerol was consumed and starvation for 30 min, induction solution was added for 84 h, with medium pH maintained at 5.5 with 30% ammonium hydroxide. For methanol feeding, the feeding rate of methanol was controlled at 3 ml/h for 24 h, 6 ml/h for 48 h, and 3 ml/h for 24 h. Cell cultures (10 ml) were collected at 0, 12, 24, 36, 48, 60, 72, 84 and 96 h after methanol induction. For each time point, 0.9 ml supernatants of collected cultures were mixed with 100 µl 100% TCA and incubated at − 20 °C for 4 h. After centrifugation at 15,000×g for 30 min at 4 °C, precipitates were resuspended in 90 µl 1 × SDS loading buffer with 8 M urea and 5 µl samples were subjected to SDS–PAGE. The amount of secretory AaIT was assayed by gel scanning and BSA was used as the protein concentration standard. Wet weights of cultured cells were obtained by centrifugation and removal of supernatant. Total protein concentration in supernatants was estimated using Bradford assays.
Optimization of affinity purification conditions
Supernatants from bioreactor (100 ml) were adjusted to pH 7.6 with NaOH, after centrifugation at 15,000×g for 60 min at 4 °C all supernatants were loaded onto a 30-ml column containing 3 ml Ni+-NTA resin pre-equilibrated with buffer A (40 mM Na2HPO4, pH 7.6, 200 mM NaCl). The column was washed with buffer B (40 mM Na2HPO4, pH 7.6, 200 mM NaCl, 20 mM imidazole), and then eluted with buffer B containing increasing concentrations of imidazole from 50 to 300 mM. The eluted protein from affinity column was analyzed by SDS–PAGE.
Up-scale purification and molecular weight determination of rAaIT
Two hundred ml supernatants from bioreactor we adjusted to pH 7.6 with NaOH, after centrifugation at 15,000×g for 60 min at 4 °C all supernatants were loaded onto a 30-ml column containing 4 ml Ni+-NTA resin pre-equilibrated with buffer A by using a peristaltic pump sampling to 1 ml per minute. Then, the column was washed with 200 ml of buffer A containing 50 mM imidazole to remove contaminated proteins. rAaIT was eluted with 200 imidazole in buffer A. The collected solution was dialyzed against buffer C (20 mM NaH2PO4, pH 5.8) at 4 °C overnight. After centrifugation at 15,000×g for 30 min at 4 °C, the supernatant was collected and loaded onto a 1-ml HiTrap CM FF column that was pre-equilibrated with buffer C. After washing with buffer C containing 40 mM NaCl, rAaIT was eluted with buffer D (20 mM Na2HPO4, pH 7.4, 100 mM NaCl). The final solution was collected and analyzed by SDS–PAGE under the condition of using or not using DTT reduction. The molecular weight of purified and concentrated rAaIT was determined by matrix-assisted laser desorption/ionization tandem time-of-flight MS (MALDI-TOF) using a Voyager-DE MALDI TOF mass spectrometer (Framingham,MA/Applied Biosystems, Foster City, USA) equipped with a 337 nm nitrogen laser.
Bioactivity assays
To evaluate the injection bioactivity of purified rAaIT, in the final instar larval stage, Galleria mellonella (G. mellonella) larvae (weighed 0.25 ± 0.05 g) and fifth instar silkworm larvae (Bombyx mori Linnaeus) (weighed 4 ± 0.2 g) were used. G. mellonella and silkworms were incubated on ice prior to use. Recombinant AaIT proteins were injected at the junction of the third and fourth abdominal segments using 10-µl sharp Hamilton syringes (Sigma-Aldrich, St. Louis, MO, USA). Each larva was injected with 1, 5, 10, 20 and 50 µg/g body weight of purified rAaIT (2 mg/ml) or the corresponding volume of PBS as a control. Post-injection toxicity was monitored with a lethality test at 24 and 48 h, and dead larvae were removed. For each treatment, 30 larvae were tested.
Results
Screening transformants
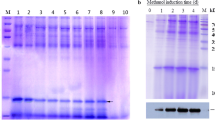
The AaIT gene was cloned into pPICZαA under the control of the oxidase I promoter as shown in Fig. 1a. Transformants that grew on YPD plates containing 1000 µg/ml zeocin were picked and subjected to PCR amplification. As seen in Fig. 1b, the amplified fragment was approximately 200 bp, as was predicted, indicating that the AaIT gene was introduced successfully into the P. pastoris genome. In an ELISA analysis of 45 P. pastoris transformants from the 1000 µg/ml zeocin YPD plates, five transformants had higher light absorption at 495 nm. As shown in Fig. 2, SDS–PAGE results showed that one transformant, named A1, had the highest expression level, a less than 10 kDa secreted protein, the same molecular weight as predicted for rAaIT was presented.
Construction of expression vector and determination of transformants. a Map of expression vector construction. b PCR amplification of AaIT from P. pastoris transformants. Four AaIT P. Pastoris transformants from YPD plate (1 g/l zeocin) and one colony of strain X33 of P. pastoris were tested by PCR using the specific primers. Lanes 1–4, transformants of pPICZaA/AaIT; Lane 5, negative control with one colony of the strain X33; lane M, DNA marker
Detection of high level secreted rAaIT protein transformants by SDS–PAGE. Lanes A1-A5, supernatans induced by methanol from the ELISA selected highest light absorption transformants; lanes B1–B3, supernatans induced by methanol from 3 transformants growth on 100 µg/ml zeocin YPD plates; lanes C1–C3, supernatants from 3 empty pPICZaA X33 transformants; lane M, protein markers; rAaIT was marked with an arrow
Small-scale fermentation in flasks
As shown in Fig. 3, a less than 10 kDa secreted protein was detected by Coomassie blue R250 staining at 24 h post-induction with methanol, and it gradually reached a maximal level after 3 days of induction. Subsequently, the band decreased because of substantial degradation, probably by yeast proteases that were secreted into the culture medium. The size of the observed recombinant protein was similar to the estimated molecular weight of rAaIT (theoretical molecular weight of 8654.7 Da, as determined using the ExPASy Compute pI/MW tool, http://www.expasy.ch/cgi-bin/pi_tool) based on the predicted amino acid sequence of KKNGYAVDSSGKAPECLLSNYCNNECTKVHYADKGYCCLLSCYCFGLNDDK KVLEISDTRKSYCDTTIINHHHHHH. The highest protein concentration was approximately 163 mg/l at day 4, but the rAaIT concentration decreased subsequently, as determined by a gel-staining analysis. SDS–PAGE analysis showed that the transformant screened by ELISA expressed and secreted rAaIT at a high level. The gel scanning assays by software Quantity One (Biorad, USA) showed post 3 days methanol induction, the band of rAaIT was about 8 µg which meant that the target protein amounts to 80 mg/l in the media. Western blotting results with rabbit anti-six-histidine IgG confirmed that the recombinant protein was indeed rAaIT, and that the greatest concentration of rAaIT was obtained after 3 days of methanol induction (Fig. 3).
Fed-batch fermentation
The results of gel scanning for rAaIT expression were as follows (Fig. 4a). Secretory rAaIT could be detected post 12 h methanol induction, the secreted rAaIT was increased rapidly post 12–72 h induction. Secreted rAaIT was increased to a peak at 72 h induction with methanol. However, the induction time of methanol was prolonged, the expression of rAaIT decreased as a result of degradation. The result of gel scanning determined that the proportion of target protein was as high as 37% of total protein amounted to approximately 240 mg/l rAaIT in supernatant after 72 h of methanol induction (Fig. 4b), which was about three times higher than that in shake flasks.
Analysis of rAaIT expressed in fed-batch fermentation. a SDS–PAGE identified the time course of rAaIT expression in 5-L bioreactor. Culture supernatants were collected at the indicated time after methanol induction. b Detection the amounts of rAaIT by gel scanning. Culture supernatants were collected at the indicated time after methanol induction
Optimization of affinity purification conditions
After removing unbound proteins with buffer B from the column, the elution of the recombinant protein was performed using buffer A with increasing imidazole respectively. As shown in Fig. 5, the optimal imidazole concentration for elution was 50–300 mM, and the predicted protein with a molecular weight lower than 10 kDa was observed. According to the amino acid sequence of rAaIT, the theoretical molecular weight is about 8.6 kDa which was closed to the gel staining results. As the SDS–PAGE results shown, when the imidazole concentration was lower than 50 mM, rAaIT was almost not eluted, and when the imidazole concentration was higher than 200 mM, almost all rAaIT was eluted. Although, washing Ni+-NTA column with buffer B containing 50 mM imidazole could remove most of contaminated proteins, some contaminated proteins with the molecular weights of 10–15 kDa were still not removed. In order to obtain higher purity rAaIT, other purification methods are needed.
Up-scale purification and molecular weight determination of rAaIT
The rAaIT eluted with 200 mM imidazole from the Ni+-NTA column were collected. Approximately 18.6 mg of rAaIT was obtained from 200 ml of culture supernatant. The dialyzed supernatant was loaded onto a CM ion-exchange column and washing with buffer C to remove unbound proteins, eluates with an absorbance at 280 nm greater than 20 mAU were collected (Fig. 6a). The collected samples were analyzed by SDS–PAGE. As shown in Fig. 6b, the purified rAaIT was a single band in the absence of or without a reducing agent. According to the intensity value of gel scanning, approximately 8 mg of high purity rAaIT was obtained by CM ion-exchange chromatography; the purification yield of each step was summarized in Table 1. The precise molecular weight of the purified rAaIT was 8650.4 Da, which was closed to the the theoretical molecular weight of the expressed rAaIT (Fig. 6c). A single protein peak indicated that rAaIT purified by CM chromatography had a high purity.
Up-scale purification and molecular weight determination of rAaIT. a Elution curves of rAaIT from a HiTrap CM FF ion exchange column. The eluents were collected at the value of OD280nm > 20 mAU. The blue curve and the red curve represent the UV absorbance (mAU) and ionic strength (mS/cm) of the eluent respectively. b SDS–PAGE results of concentrated rAaIT under reduced and non reducing conditions. Lane M, protein markers. c The mass of purified rAaIT protein was measured using MALDI TOF mass spectrometer. The mass denotes the Mr of rAaIT is 8650.4. Mass per charge is indicated by use of m/z
Bioactivity assays
G. mellonella and silkworms were injected with rAaIT, and they all showed classic symptoms of neurotoxin poisoning, including immediate contraction paralysis and abdominal contraction, while control larvae that were injected with PBS did not show any such symptoms (Fig. 7). G. mellonella toxicity results are shown in Table 2. When the dose was 1 µg/g body weight, post 24 h injection, almost all of the larvae lose mobility but no death. Increasing the injection dose to 20 µg/g body weight, post 24 h injection, 19 of larvae (63.3%) died post 24 h injection, 27 of larval (90%) were died post 48 h injection. When the dose was 50 µg/g body weight, post 24 h injection, 28 of larvae (93.3%) died post 24 h injection, all larval (100%) were died post 48 h injection. Although 2 of control G. mellonella died after 48 h, the death larvae did not show any symptoms of neurotoxin poisoning.
Silkworm injection toxicity results are shown in Table 3. When the dose was 1 µg/g body weight, almost all of the larvae lose mobility, but no death post 48 h of injection. Upon increasing the dose to 5 µg, 7 of larvae (23.3%) died post 24 h injection and 12 of larval were died post 48 h injection. Increasing the injection dose to 10 µg/g body weight, 16 of larvae (53.3%) were died post 24 h injection and 25 of larval (83.3%) were died post 48 h injection. When the dose was 20 µg/g body weight, post 24 h injection, 26 of larvae (86.7%) died post 24 h injection and all larvae (100%) were died post 48 h injection. When the dose was 50 µg/g body weight, post 24 h injection, all larval were died. Although one of control silkworm died after 48 h, the death larvae did not show any symptoms of neurotoxin poisoning.
Disscussion
Many factors, especially host strains and expression plasmids affect the expression of heterologous proteins in P. pastoris (Rabert et al. 2013; Looser et al. 2015). Thus, the Invitrogen Company provides several host strains such as X33, KM71 and SMD1168, and several secretory expression vectors containing pPIC9k, pPIC3.5 k, and pPICZαA. Therefore, choosing the proper expression plasmid and host strain is critical for achieving high-level protein expression. Using pPIC9k as the expression plasmid, X33 and SMD1168 as the host strains, we did not select the transformants which were significantly higher than KM71 (data not shown). Similarly, KM71 was used as host strain and pPIC3.5 k and pPICZαA as the expression vectors, the transformant which significantly higher than the previous reports was not obtained (data not shown). In the case of identical gene sequences and signal peptides (different cleavage sites), using pPICZαA as the expression vector and strain X33 as the host strain, we selected several high level secretory expression rAaIT transformants and the highest one which was about eightfold greater that that achieved in our previous studies using pPIC9K as the expression vector and strain KM71 as the host strain. Therefore, matching expression plasmid with host strain is one of the key factors to obtain high-level secreted expression transformants. Under the condition of fed batch fermentation, the expression level of rAaIT was over three times higher than that of shake flask. Thus, fed batch fermentation could enhance the expression level and facilitate the preparation of rAaIT. To our knowledge, this is by far the highest secretory expression of scorpion insecticidal neurotoxin in P. Pastoris.
Gene copy number is another important factor affecting the expression level of recombinant proteins, the most common strategy for selecting P. pastoris transformants with high heterologous protein expression levels is to focus on screening multicopy integrations of an expression cassette (Liu et al. 2007; Yang et al. 2016; Cámara et al. 2017). In this study, initially, we could not obtain high-level secretory expression of rAaIT using standard selection protocols from low concentration zeocin plates and we had to select additional transformants by using a much higher level of zeocin on the YPD plates. Using real-time PCR method, we assayed the copy number of AaIT gene from 1 g/l zeocin YPD plates, and cassettes of these 45 transformants was between 6.3 and 8.8 (6–9 expression cassettes). Five transformant that grew on YPD plate containing 1 g/l zeocin exhibited a high level of expression of rAaIT, their average cassettes were 8.3 which had the most expression cassettes (data not shown). Therefore, the copy number of the AaIT gene was another critical factor determining the level of rAaIT expression in P. Pastoris. Generally, the more expression cassettes, the easier it is to obtain high levels of secretory expression transformants. By screening for transformants with more expression cassettes, it is possible to obtain the transformants secreted rAaIT at higher levels. However, SDS–PAGE analysis of the supernatants of 5 ELISA selected transformants showed that one transformant named A1 had the highest expressed. In fact, another two transformants had higher light absorption than A1, suggesting that many interfering agents may be induced during the induction of expression. Because of the specificity of antibody and the complexity of antigen, the transformants cannot be screened simply by ELISA. In general, the successful screening of high-level expression P. pastoris transformants often involves the use of multiple methods.
Compared with the shaking flasks fermentation in BMMY medium, the fed-batch culture of P. pastoris produced a significantly higher rAaIT. The NCBIO system could maintain 30% DO before the methanol induction by stirring up to 1000 rpm, but the DO could not always maintained at 30% during the induction course at the maximal agitation, especially at the moment when methanol was added, the DO was reduced to as low as 10%. Therefore, pure oxygen instead of compressed air could improve the productivity of rAaIT. The feeding rate of methanol is also a crucial factor to the expression of rAaIT. In our study, the methanol feeding strategy was crude and the methanol concentration in the culture was unknown during the induction period. The real-time online monitoring of methanol concentration and automatic control of methanol feeding may achieve a higher level of rAaIT secretion.
Most protein purification protocols require multiple steps to achieve highly purified products. An ideal purification protocol results in high yield and purity with a minimum number of steps. Affinity chromatography is an effective way of separating proteins, often with only one step being needed to separate a protein of interest from a complex protein mixture. In addition, the target protein is concentrated during the purification process, thereby shortening the purification time, which is especially beneficial for the purification of unstable proteins, as well as maintaining protein activity. Ni2+-affinity chromatography is one of the simplest and most effective methods for protein purification, and it is often used to purify histidine-tagged proteins (Lin et al. 2015; Terpe 2003). In this study, a six-histidine tag was added to the carboxyl-terminus of rAaIT, which greatly simplified the purification steps. With a simple, one-step purification via the Ni+-NTA column, we obtained rAaIT that was up to 85% pure. In previous studies, although the expression of rAaIT was achieved, an effective purification method was not found, and only small amounts of low-purity rAaIT were obtained (Li and Xia 2009). Here, rAaIT was purified efficiently with Ni2+-affinity purification which was convenient for subsequent use of CM chromatography to obtain high purity rAaIT protein.
Soft-bodied silkworms exhibit all kinds of toxic symptoms in response to exposure to various neurotoxins, and they are often used to determine the activity of insect neurotoxins (Figueiredo et al. 1995; Ji et al. 2002). Our data showed that the injection of rAaIT had immediate effects on silkworms, including severe vomiting, mouth movement, abdominal contraction, swelling of the head, and anal exposure (supplement A), which is consistent with the reported symptoms of AaIT. In previous studies, the P. Pastoris was used to express an alpha-neurotoxin named BmalphaTX14 fusion with six-histidine tag at the carboxyl-terminus, the purified rBmalphaTX14 was also bio-active and the selectively did not change (Wang et al. 2006). Therefore, the fusion of the six-histidine tag at the carboxyl-terminus of rAaIT likely did not obvious affect its three-dimensional conformation.
In summary, transformants with high level expression of rAaIT can be efficiently obtained by changing the expression strains, expression plasmids and screening the transformants with multiple expression cassettes. Moreover, high density fermentation can further increase the expression of rAaIT. As a result, we have described a highly efficient method of producing large quantities of functional rAaIT, and this method may facilitate further functional studies of the other similar insect neurotoxins.
References
Barnard GC, Kull AR, Sharkey NS, Shaikh SS, Rittenhour AM, Burnina I, Jiang Y, Li F, Lynaugh H, Mitchell T, Nett JH, Nylen A, Potgieter TI, Prinz B, Rios SE, Zha D, Sethuraman N, Stadheim TA, Bobrowicz P (2010) High-throughput screening and selection of yeast cell lines expressing monoclonal antibodies. J Ind Microbiol Biotechnol 37:961–971
Cámara E, Landes N, Albiol J, Gasser B, Mattanovich D, Ferrer P (2017) Increased dosage of AOX1 promoter-regulated expression cassettes leads to transcription attenuation of the methanol metabolism in Pichia pastoris. Sci Rep 7:44302
Chaudhary S, Kanwar RK, Sehgal A, Cahill DM, Barrow CJ, Sehgal R, Kanwar JR (2017) Progress on Azadirachta indica based biopesticides in replacing synthetic toxic pesticides. Front Plant Sci 8:610
Figueiredo SG, Garcia ME, Valentim AC, Cordeiro MN, Diniz CR, Richardson M (1995) Purification and amino acid sequence of the insecticidal neurotoxin Tx4(6-1) from the venom of the ‘armed’ spider Phoneutria nigriventer (Keys). Toxicon 33:83–93
Forstner M, Leder L, Mayr LM (2007) Optimization of protein expression systems for modern drug discovery. Expert Rev Proteom 4:67–78
Gurevitz M, Karbat I, Cohen L, Ilan N, Kahn R, Turkov M, Stankiewicz M, Stühmer W, Dong K, Gordon D (2007) The insecticidal potential of scorpion beta-toxins. Toxicon 49:473–489
Ji SJ, Liu F, Li EQ, Zhu YX (2002) Recombinant scorpion insectotoxin AaIT kills specifically insect cells but not human cells. Cell Res 12:143–150
Kawachi T, Miyashita M, Nakagawa Y, Miyagawa H (2013) Isolation and characterization of an anti-insect-toxin from the venom of the scorpion Isometrus maculatus. Biosci Biotechnol Biochem 77:205–217
Li HB, Xia YX (2009) High expression and rapid purification of recombinant scorpion anti-insect neurotoxin AaIT. World J Microbiol Biotechnol 25:1251–1257
Lin Z, Zhao Q, Xing L, Zhou B, Wang X (2015) Aggregating tags for column-free protein purification. Biotechnol J 10:1877–1886
Liu Y, Wang Z, Yin Y, Cao Y, Zhao H, Xia Y (2007) Expression, purification, and characterization of recombinant Metarhizium anisopliae acid trehalase in Pichia pastoris. Protein Expr Purif 54:66–72
Liu SM, Li J, Zhu JQ, Wang XW, Wang CS, Liu SS, Chen XX, Li S (2016) Transgenic plants expressing the AaIT/GNA fusion protein show increased resistance and toxicity to both chewing and sucking pests. Insect Sci 23:265–276
Looser V, Bruhlmann B, Bumbak F, Stenger C, Costa M, Camattari A, Fotiadis D, Kovar K (2015) Cultivation strategies to enhance productivity of Pichia pastoris: a review. Biotechnol Adv 33:1177–1193
Michiels K, Van Damme EJ, Smagghe G (2010) Plant-insect interactions: what can we learn from plant lectins? Arch Insect Biochem Physiol 73:193–212
Nalcacioglu R, Muratoglu H. Yesilyurt A, van Oers MM, Vlak JM, Demirbag Z (2016) Enhanced insecticidal activity of Chilo iridescent virus expressing an insect specific neurotoxin. J Invertebr Pathol 138:104–111
Olombrada M, Lázaro-Gorines R, López-Rodríguez JC, Martínez-Del-Pozo Á, Oñaderra M, Maestro-López M, Lacadena J, Gavilanes JG, García-Ortega L (2017) Fungal ribotoxins: a review of potential biotechnological applications. Toxins (Basel) 9:71–91
Pava-Ripol M, Posada FJ, Momen B, Wang C, St Leer RJ (2008) Increased pathogenicity against coffee berry borer, Hypothenemus hampei (Coleoptera: Curculionidae) by Metarhizium anisopliae expressing the scorpion toxin (AaIT) gene. J Invertebr Pathol 99:220–226
Pelhate M, Stankiewicz M, Ben Khalifa R (1998) Anti-insect scorpion toxins: historical account, activities and prospects. C R Seances Soc Biol Fil 192:463–684
Rabert C, Weinacker D, Pessoa Jr A, Farías JG (2013) Recombinants proteins for industrial uses: utilization of Pichia pastoris expression system. Braz J Microbiol 44:351–356
Regev A, Rivkin H, Gurevitz M, Chejanovsky N (2006) New measures of insecticidal efficacy and safety obtained with the 39K promoter of a recombinant baculovirus. FEBS Lett 580:6777–6782
Terpe P (2003) Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol 60:523–533
Valero F (2012) Heterologous expression systems for lipases: a review. Methods Mol Biol 861:161–178
Wang CS, St Leger RJ (2007) A scorpion neurotoxin increases the potency of a fungal insecticide. Nat Biotechnol 25:1455–1456
Wang K, Yin SJ, Lu M, Yi H, Dai C, Xu XJ, Cao ZJ, Wu YL, Li WX (2006) Functional analysis of the alpha-neurotoxin, BmalphaTX14, derived from the Chinese scorpion, Buthus martensii Karsch. Biotechnol Lett 28:1767–1772
Weis R, Luiten R, Skranc W, Schwab H, Wubbolts M, Glieder A (2004) Reliable high-throughput screening with Pichia pastoris by limiting yeast cell death phenomena. FEMS Yeast Res 5:179–189
Yang J, Lu Z, Chen J, Chu P, Cheng Q, Liu J, Ming F, Huang C, Xiao A, Cai H, Zhang L (2016) Effect of cooperation of chaperones and gene dosage on the expression of porcine PGLYRP-1 in Pichia pastoris. Appl Microbiol Biotechnol 100:5453–5465
Zhong G, Cui G, Yi X, Sun R, Zhang J (2016) Insecticide cytotoxicology in China: current status and challenges. Pestic Biochem Physiol 132:3–12
Zlotkin E, Rochat H, Kopeyan C, Miranda F, Lissitzky S (1971) Purification and properties of the insect toxin from the venom of the scorpion Androctonus australis Hector. Biochimie 53:1073–1078
Zlotkin E, Eitan M, Bindokas VP, Adams ME, Moyer M, Burkhart W, Fowler E (1991) Functional duality and structural uniqueness of depressant insect-selective neurotoxins. Biochemistry 30:4814–4821
Zlotkin E, Gurevitz M, Fowler E, Adams ME (1993) Depressant insect selective neurotoxins from scorpion venom: chemistry, action, and gene cloning. Arch Insect Biochem Physiol 22:55–73
Zlotkin E, Fishman Y, Elazar M (2000) AaIT: from neurotoxin to insecticide. Biochimie 82:869–881
Acknowledgements
This work was supported in part by Funds from Project funded by China Postdoctoral Science Foundation (2016M590867), Project funded of Chongqing Postdoctoral Science Foundation (Xm2016075) and Scientific Research Fund of Hunan Provincial Education Department (15A147).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (MOV 25563 KB)
Rights and permissions
About this article
Cite this article
Li, H., Xia, Y. Improving the secretory expression of active recombinant AaIT in Pichia pastoris by changing the expression strain and plasmid. World J Microbiol Biotechnol 34, 104 (2018). https://doi.org/10.1007/s11274-018-2484-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-018-2484-x