Abstract
Crude extract from a culture of a soil Streptomyces sp. strain ZDB showed toxicity towards Artemia salina and antimicrobial activity against Escherichia coli, Bacillus subtilis, Staphylococcus aureus, Chlorella vulgaris, and Chlorella sorokiniana. Large scale fermentation of the strain led to the isolation of the macrolide antibiotics, bafilomycins A1 (1), B1 (2), and D (3) together with nonactic acid (4) and bostrycoidin-9-methyl ether (5). Structures of the antibiotics were determined based on spectral data analysis. We describe the isolation of the compounds and characterization of the producing strain.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Currently, there is an urgent need for effective antibiotics with low toxicity and a minor environmental impact (Strobel and Daisy 2003). Studies show that there is a rise in antibiotic resistance worldwide as pathogens continue to develop resistance mechanisms (Hancock 2007; Cowen 2008). Furthermore, ‘superbugs’ and new infectious agents’ emergence have further driven the need for novel and effective antibiotics (Katz et al. 2006; Strobel et al. 2004). Streptomyces have been a rich source of biologically active secondary metabolites for decades. Recently, Jang et al. (2013) have isolated a potent anthrax antibiotic named anthracimycin from a Streptomyces species recovered from shore marine sediments found near Santa Barbara. Further study on anthracimycin by Hensler et al. (2014) showed that it effectively inhibited methicillin-resistant (MRSA) and vancomycin-resistant strains of Staphylococcus aureus both in vitro and in vivo. Similarly, researchers from Germany and USA have discovered a new antibiotic, teixobactin, which kills pathogen without detectable resistance from the culture of a bacterium species (Ling et al. 2015).
In the course of our search for bioactive compounds from microorganisms, we isolated a strain of Streptomyces from soil samples collected from nearshore sediment of lake Koka, Ethiopia. We investigated the strain for metabolic competence and biological activities against larvae of A. salina and other test organisms. From the fermentation broth of the strain, five bioactive compounds were isolated and characterized. In this study, we describe the isolation of the active compounds together with characterization of the producing strain.
Materials and methods
Characterizations of the Streptomyces sp. strain ZDB
Streptomyces sp. Strain ZDB was isolated from soil samples collected from nearshore sediment of Lake Koka (8°26′N 39°02′E), Ethiopia according to a previously described method (Ruanpanun et al. 2010).
The morphology and cultural characteristics of the strain were examined following the guidelines of the International Streptomyces Project (ISP) (Shirling and Gottlieb 1966). The cultural aspects of the pure isolate were observed on various ISP media following incubation at 28 °C for 14 days. Colors of aerial and substrate mycelia were determined and recorded using the National Bureau of Standards (NBS) Color Name Charts (KL 1958). Growth range for temperature and pH were examined on ISP2 medium and through culturing for 7 days. Cell wall diaminopimelic acid (DAP) isomers and sugar composition were examined using a TLC according to procedures described by (Hasegawa et al. 1983). Physiological characteristics were determined as recommended by (Williams et al. 1983).
Phylogenetic analyses
The phylogenetic position of Streptomyces sp. strain ZDB was determined by 16S rRNA gene sequence analysis. Genomic DNA was extracted from biomass of the strain and PCR-mediated amplification and 16S rRNA gene sequence of the purified product achieved, as described by Ruanpanun et al. (2011). The resultant, almost complete 16S rRNA gene sequence (1467 nucleotides), was submitted to the EzTaxon server (http://eztaxon-e.ezbiocloud.net; Kim et al. 2012) and aligned with 16S rRNA gene sequences of closely related Streptomyces species using MUSCLE version 3.8 software (Edgar 2004). Phylogenetic tree was generated using neighbor-joining (Saitou and Nei 1987) tree-making algorithms drawn from MEGA 7 (Kumar et al. 2016); an evolutionary distance matrix for the neighbor joining analysis was prepared using the Jukes and Cantor (1969) model. The stability of the clades in the trees was evaluated after 1000 bootstrap replicates of the neighbour-joining data (Felsenstein 1985). The root position in the neighbour-joining tree was inferred by using S. longisporus ISP 5166T (accession number AJ399475).
Artemia salina microwell cytotoxicity assay
Larvae of A. salina (SERA Artemia Salinenkrebseier, SERA Heinsberg—Salinenkrebsfutter: micro cell DOHSE Aquaristik KG Bonn) were transferred to a deep-well microtiter plate filled with 0.2 mL of salt water. Dead larvae were counted (N). The crude extract (100 µg/mL) was added to the plate and kept at room temperature for 24 h. A number of the dead larvae (A) was recorded under a microscope. The still living larvae were killed by addition of ca. 0.5 mL methanol so that subsequently the total number (G) of larvae could be determined. The mortality rate M was calculated in %. The test was accompanied by a blind sample with pure DMSO (B) and a control sample with 1 µg/test actinomycin D. The mortality rate M was calculated using the following formula:
Antimicrobial activity assay
Antimicrobial activity was measured by the disc diffusion method on nutrient agar (bacteria; culture at 37 °C for 24 h) and water agar [algae; culture at room temperature (25 ± 3 °C) for 7–14 days]. Test organisms were E. coli, B. subtilis, S. aureus, C. vulgaris, C. sorokiniana. The strain ZDB was grown in ISP2 medium broth on a rotatory shaker at 130 rpm and 28 °C for 7 days (Ruanpanun et al. 2011). The culture broth was extracted with ethyl acetate and the organic phase evaporated to dryness. The crude extract was dissolved in 0.5% DMSO at a concentration of 10 mg/mL; 40 μL of this solution was absorbed on paper discs (9 mm diameter, Schleicher & Schüll No. 321 261).
Fermentation and isolation of the bioactive compounds
Five litres culture broth of the terrestrial Streptomyces sp. strain ZDB was used to inoculate 25 L of ISP2 medium in a 50 L fermenter Biostat U (B. Braun Dissel Biotech GmbH) consisted of a 70 L metallic container (50 L working volume), propeller stirrer, and culture container covered with thermostat for autoclaving, cooling and thermostating (Braun Melsungen, Germany). The culture was maintained at 28 °C for 7 days. The resulting brown broth was separated into biomass and water phase using a method previously reported (Dame et al. 2015). The biomass was extracted with ethyl acetate and acetone, respectively. The water phase was adsorbed on Amberlite XAD-16 (Rohm and Haas Frankfurt) column and eluted with methanol. The methanolic extract was concentrated and further extracted with ethyl acetate. Organic phases from both the biomass and water phase extracts were combined and evaporated under vacuum at 38 °C to afford 5 g of crude extract.
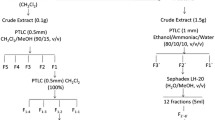
The crude extract was fractionated on silica gel column using CH2Cl2/MeOH with a stepwise gradient of increasing polarity. The fractions were monitored by TLC (DC-Folien Polygram SIL G/UV254 (Macherey-Nagel & Co.). A total of three fractions were obtained (Fig. 1) from which the first fraction (Fraction I) was found to be fat and was not further analyzed. The second fraction (Fraction II) contains UV absorbing bands and was separated by PTLC (silica gel 230–400 mesh) and further purified on Sephadex LH-20 (Lipophilic Sephadex; Amersham Biosciences, Ltd., Sigma-Aldrich Chemie, Steinheim, Germany) to afford bafilomycins A1 (1), B1 (2), and D (3). Fraction three (Fraction III) was subjected to Sephadex LH-20 (MeOH) and gave nonactic acid (4) and Bostrycoidin-9-methyl ether (5).
Structure determination of the compounds
NMR spectra were recorded on Varian Unity 300 (300.145 MHz) and Varian Inova 500 (499.876 MHz) spectrometers. ESI-MS was recorded on a Finnigan LCQ with quaternary pump Rheos 4000 (Flux Instrument). ESI-HR mass spectra were measured on a Micromass LCT spectrometer coupled with an HP 1100 HPLC with a diode array detector.
Results
Streptomyces sp. strain ZDB showed rectiflexible substrate mycelia and hyphae under a light microscope. The mycelium was branched; the aerial mycelium was monopodially branched with broom-shape of sporophores. Cultural characteristics of the strain are shown in Table 1. The strain showed moderate to abundant growth on most media except tryptone yeast extract agar and glycerol asparagine agar. Typically, the colonies were convex and some part of the aerial mycelium and spore chains could be observed around the colony edge. The aerial mycelium was initially white; greenish to brownish depending on the medium used and gradually darkened in old cultures. Likewise, the vegetative mycelium was colorless on all medium. No diffusible pigment was observed. Growth occurred between 15 and 37 °C and at pH values ranging from 5 to 11. Growth occurred in the presence of 0.1% phenol (v/v), 0.1% sodium propionate (w/v), 3% NaCl (w/v) but not in the presence of 5% NaCl (w/v). Nitrate was reduced. The strain was also able to degrade Tween 20 but unable to degrade Tween 40, 60, 80, casein, chitin, gelatin, testosterone, and xylan. This isolate was sensitive to most antibiotics including (µg/mL): neomycin (50), novobiocin (50), oleandomycin (50), rifampicin (50), and streptomycin (100). But was resistant to penicillin G (10 i.u./mL). The physiological properties tested showed positive results for utilization of many carbon sources including l-adonitol, l-arabinose, d-cellobiose, d-fructose, d-fucose, d-glucose, l-lactose, d-mannitol, raffinose, l-rhamnose, sucrose, and d-trehalose but was unable to utilize arabitol, d-galactose, d-melezitol, salicin, d-sorbitol, and xylitol. For nitrogen source utilization, d,l-alanine, l-arginine, l-asparagine, l-histidine, l-leucine, l-methionine, d,l-norleucine, l-threonine and l-tryptophan were shown to be positive, while d,l-α-amino-n-butyric acid, l-cysteine, and l-phenylalanine were not utilized.
Chemotaxonomic tests of strain ZDB showed that the whole-cell hydrolysate was rich in l,l-diaminopimelic acid (ll-DAP) with no characteristic sugar pattern. With the morphological characteristics of spore chains, cell wall chemotype I with no characteristic sugar, it was clear that the strain ZDB belongs to the genus Streptomyces.
The assignment of this strain to the genus Streptomyces was further supported by its 16S rRNA gene sequence analysis (1467 nucleotides), which was submitted to the DNA Data Bank of Japan (DDBJ) under the accession number LC270811. Comparison of the sequence of strain ZDB with those of representative strains of the genus Streptomyces showed that this organism formed a distinct phylogenetic line with a clade encompassed by the Streptomyces mobaraensis (Fig. 2). Moreover, the strain showed a 16S rRNA gene sequence similarity of 98.64% with S. mobaraensis, which is equivalent to a difference of 19 nucleotides out of 1399 positions, and a 98.64% difference with Streptomyces abikoensis (19 nucleotide differences out of 1393 positions).
Neighbor-joining tree based on almost complete 16S rRNA gene sequence showing phylogenetic relationships between Streptomyces sp. strain ZDB and related members of the genus Streptomyces. Numbers at nodes indicate the level of bootstrap support based on a neighbor-joining analysis of 1000 resampled data sets; only values above 50% are given. Scale bar indicates 0.005 substitution per nucleotide position
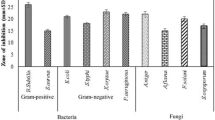
The terrestrial Streptomyces sp. strain ZDB was cultivated in four Erlenmeyer flasks each containing 250 mL of the ISP2 medium at 28 °C for 7 days. The culture was extracted with ethyl acetate and the extract was used for the primary screenings. The crude extract showed inhibitory activity against E. coli, B. subtilis, S. aureus, C. vulgaris, and C. sorokiniana (Table 2).
The strain also displayed strong cytotoxicity against larvae of A. salina. At a dose of 100 µg/mL, the crude extract caused a 100% mortality rate of the larvae. The chemical screening showed UV absorbing bands, which gave brown and red spots on spraying with anisaldehyde/sulphuric acid. Further workup on the strain led to the isolation of bafilomycins A1 (1), B1 (2) and D (3), nonactic acid (4), and bostrycoidin-9-methyl ether (5) (Fig. 3).
The major fraction obtained after separation of the crude extract on a silica gel column showed UV absorbing dark spots on TLC. Separation by PTLC and further purification by Sephadex LH-20 chromatography afforded bafilomycins A (1), B (2), and D (3) as the major metabolites. While Bafilomycin B1 (2) was isolated as a yellowish solid, bafilomycins A1 (1) and D (3) were isolated as colourless solid. All the three gave a brown color reaction on TLC with anisaldehyde/sulphuric acid. The 1H NMR spectrum of bafilomycin B1 showed H/D exchangeable protons, olefinic methines, methoxy groups, multiplets in the range of δ 4.15–3.10 for methine groups connected to oxygen, signals for methyl groups bound to a double bond and methyl doublets in the range of δ 1.05–0.6. ESI-MS gave an ion peak at m/z 838 [M+Na]+ indicating a molecular weight of 815 Dalton. These data together with the 13C NMR spectrum were used to search for a related structure in the microbial natural product database, the AntiBase (Laatsch 2014), which gave bafilomycin B1 (1) as the structure of the compound. Bafilomycins A1 and D also displayed similar proton NMR spectra as that of bafilomycin B1, however, some methylene signals in the aliphatic region, the doublets observed in the aromatic region and the two H/D exchangeable protons seen in the spectrum of bafilomycins B1 were absent in the proton spectrum of both bafilomycins A1(1) and D (3). Their structure was also confirmed based on NMR and mass spectral data.
Nonactic acid (4) was isolated as a colorless oil and identified with its proton and mass spectral data. It is frequently isolated from bacteria. Bostrycoidin-9-methyl ether (5) on the other hand, was isolated as a red solid. The ESI mass spectrum showed ion signals for m/z 322 [M+Na]+ and 298 [M−H]− respectively giving a molecular weight of 299 Dalton. Both compounds were identified based on spectral analysis and comparison with authentic spectra from the AntiBase database (Laatsch 2014).
Discussion
Bafilomycins are macrolide antibiotics possessing a 16-membered lactone ring and were isolated for the first time from a culture of Streptomyces griseus (Werner et al. 1984). These antibiotics are potent inhibitors of vacuolar V-type ATPase, an enzyme responsible for maintaining vesicular acidification and for their broad antibacterial and antifungal activities (Bowman et al. 1988). Studies on the anthelminthic properties of bafilomycins revealed that they are active against nematode strains resistant to the known benzimidazole, levamisole and avermectin anthelmintics (Lacey et al. 1995). In our bioassay, they showed strong toxicity against larvae of A. salina accounting for the activity exhibited by the crude extract of the strain. On the other hand, the antibiotic bostrycoidin-9-methyl ether (5) was discovered by Steyn et al. (1979) from the fungus Fusarium moniliforme. Its antibiotic activity against S. aureus has also been reported (Baker et al. 1990). While looking for new antibiotics, studies show that testing the already known bioactive metabolites against new targets is showing promising results (Dame et al. 2016). Therefore, testing these macrolides against various pathogens could reveal novel bioactivities.
References
Baker RA, Tatum JH, Nemec S (1990) Antimicrobial activity of naphthoquinones from Fusaria. Mycopathologia 1:9–15
Bowman EJ, Siebers A, Altendorf K (1988) Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc Natl Acad Sci USA 85:7972–7976
Cowen LE (2008) The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nat Rev Microbiol 6:187–198.
Dame ZT, Suwannarach N, Lumyong S, Laatsch H (2015) A new citrinin dimer isolated from Aspergillus terreus strain ZDF21. Nat Prod Commun 10:623–624
Dame ZT, Islam MT, Helmke E, von Tiedemann A, Laatsch H (2016) Oligomycins and pamamycin homologs impair motility and induce lysis of zoospores of the grapevine downy mildew pathogen, Plasmopara viticola. FEMS Microbiol Lett 363(16):fnw167.
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 3:1792–1797
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Hancock REW (2007) The complexities of antibiotic action. Mol Syst Biol 3:142–142. doi:10.1038/msb4100184
Hasegawa T, Takizawa M, Tanida S (1983) A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol 29:319–322
Hensler ME et al (2014) Anthracimycin activity against contemporary methicillin-resistant Staphylococcus aureus. J Antibiot 67:549–553
Jang KH, Nam S-J, Locke JB, Kauffman CA, Beatty DS, Paul LA, Fenical W (2013) Anthracimycin, a potent anthrax antibiotic from a marine-derived actinomycete. Angew Chem Int Ed 52:7822–7824
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Monro HN (ed) Mammalian protein metabolism, vol 3. Academic Press, New York, pp 21–123
Katz ML, Mueller LV, Polyakov M, Weinstock SF (2006) Where have all the antibiotic patents gone? Nat Biotechnol 24:1529–1531
Kim O-S, Chi Y-J, Lee K, Yoon S-H, Kim M, Na H, Park S-C, Jeon YS Lee J-K, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16 S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
KL K (1958) Centroid notations for the revised ISCC-NBS color name blocks. J Res Natl Bur Stand 61:472
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Laatsch H (2014) AntiBase 2014: the natural compound identifier. Wiley, Weinheim
Lacey E, Gill JH, Power ML, Rickards RW, O’Shea MG, Rothschild JM (1995) Bafilolides, potent inhibitors of the motility and development of the free-living stages of parasitic nematodes. Int J Parasitol 25:349–357
Ling LL et al (2015) A new antibiotic kills pathogens without detectable resistance. Nature 517:455–459
Ruanpanun P, Tangchitsomkid N, Hyde K, Lumyong S (2010) Actinomycetes and fungi isolated from plant-parasitic nematode infested soils: screening of the effective biocontrol potential, indole-3-acetic acid and siderophore production. World J Microbiol Biotechnol 26:1569–1578
Ruanpanun P, Laatsch H, Tangchitsomkid N, Lumyong S (2011) Nematicidal activity of fervenulin isolated from a nematicidal actinomycete, Streptomyces sp. CMU-MH021, on Meloidogyne incognita. World J Microbiol Biotechnol 27:1373–1380
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Steyn PS, Wessels PL, Marasas WF (1979) Pigments from Fusarium moniliforme Sheldon: structure and 13C nuclear magnetic resonance assignments of an azaanthraquinone and three naphthoquinones. Tetrahedron 12:1551–1555
Strobel G, Daisy B (2003) Bioprospecting for microbial endophytes and their natural products. Microbiol Mol Biol Rev 67:491–502
Strobel G, Daisy B, Castillo U, Harper J (2004) Natural products from endophytic microorganisms. J Nat Prod 67:257–268
Werner G, Hagenmaier H, Drautz H, Baumgartner A, Zahner H (1984) Metabolic products of microorganisms. 224. Bafilomycins, a new group of macrolide antibiotics. Production, isolation, chemical structure and biological activity. J Antibiot 37:110–117
Williams ST, Goodfellow M, Alderson G, Wellington EM, Sneath PH, Sackin MJ (1983) Numerical classification of Streptomyces and related genera. J Gen Microbiol 129:1743–1813
Acknowledgements
Prof. Dr. Hartmut Laatsch (Göttingen, Germany) is thanked for the opportunity to do this work in his laboratory. ZTD wish to thank DAAD for Grant Number A/07/80161. We also thank Mr. Machinek for the NMR and Dr. H. Frauendorf for the mass spectra, respectively, F. Lissy for microbiological work and A. Kohl for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dame, Z.T., Ruanpanun, P. Production of macrolide antibiotics from a cytotoxic soil Streptomyces sp. strain ZDB. World J Microbiol Biotechnol 33, 139 (2017). https://doi.org/10.1007/s11274-017-2306-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-017-2306-6