Abstract
Ethidium monoazide bromide (EMA) and propidium monoazide (PMA), in combination with amplicon-based sequencing (ABS) and quantitative polymerase chain reaction (qPCR) assays, were compared for the detection of viable bacterial species in rainwater. The ABS α- and β-diversity indices indicated that, in comparison to the untreated samples, both EMA and PMA reduced the detection of non-viable bacteria in the rainwater samples. However, while comparable results were obtained for the detection of the most abundant bacterial families and genera in the rainwater samples for both the EMA and PMA pre-treatments; the EMA pre-treatment produced highly significant differences in the relative abundance of the dominant bacterial families and genera, and significantly decreased the detection of “unclassified” bacteria in comparison to the untreated samples (i.e. unclassified genera: untreated 46.7% versus EMA-treated 39.7%). Additionally, the inclusion of EMA and PMA allowed for the increased detection of less abundant pathogenic bacteria in the rainwater samples, such as Clostridium, Listeria and Streptococcus spp. The qPCR assays also indicated that the EMA and PMA pre-treatments were comparable for the detection of intact and potentially viable Acinetobacter, Legionella and Pseudomonas spp. The combination of viability pre-treatments with ABS thus offers a comprehensive monitoring approach to detect less abundant bacterial genera and/or pathogens, which may pose a health risk to the end-users (prioritise target organisms) of unpiped environmental water sources. Moreover, the subsequent combination of the viability pre-treatments with qPCR allows for the specific detection and quantification of these pathogenic genera, which increases analysis sensitivity and allows for the application of risk assessment and water safety strategies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Recent advances in the use of high-throughput sequencing and next-generation sequencing (NGS) of the bacterial 16S ribosomal ribonucleic acid (rRNA) gene has greatly facilitated the elucidation of bacterial communities within diverse aquatic environments and has overcome the limitations associated with culture-dependent methods (Ahmed et al., 2017; Bibby et al., 2010; Ibekwe et al., 2013; Ye & Zhang, 2011). Moreover, pathogens that are generally present at low concentrations (< 1% of the sample composition) may be detected with high taxonomic resolution (Ahmed et al., 2017). However, one of the major limitations associated with amplicon-based sequencing (ABS) is its inability to distinguish the viable from the non-viable bacterial community. This information is essential, as various environmental conditions (e.g. nutrient availability) and microbial dynamics (e.g. predation by protozoa, predatory bacteria or bacteriophages) may influence the presence and viability of bacteria in a certain niche (e.g. environmental water sources). Consequently, it has been estimated that up to 44% of the bacterial deoxyribonucleic acid (DNA) within environmental water samples may be attributed to relic DNA (extracellular DNA or DNA from membrane-compromised/dead cells) (Lennon et al., 2018) that may negatively influence molecular-based analyses, as the detection or quantification of this DNA does not provide an indication of the viable microbial community. To circumvent this problem, nucleic acid binding dyes (also referred to as viability dyes) such as ethidium monoazide bromide (EMA) and propidium monoazide (PMA) have been utilised in conjunction with molecular techniques to quantify the intact and potentially viable bacterial species within water sources (Fittipaldi et al., 2012; Ni et al., 2020; Qin et al., 2012; Reyneke et al., 2016, 2017; Seinige et al., 2014).
Nucleic acid binding dyes selectively bind to extracellular DNA or the DNA in membrane-compromised cells upon photoactivation. This effectively removes the bound DNA from the sample, as it cannot be amplified using molecular-based methods [i.e. polymerase chain reaction (PCR) or sequencing based methods] and thus provides an indication of cell viability based on membrane integrity. Strauss et al. (2019) and Reyneke et al. (2020a) then reported on the use of a 6 µmol l−1 EMA viability pre-treatment in combination with Illumina ABS to monitor the total viable bacterial community in roof-harvested rainwater (RHRW) before and after treatment using a solar disinfection compound parabolic collector (SODIS-CPC) and a large-volume batch solar reactor system, respectively. Results indicated that the inclusion of the EMA treatment allowed for the detection of a shift in the viable bacterial community following treatment and the identification and prioritisation of organisms of interest for the health risk assessment of the untreated and treated RHRW (Reyneke et al., 2020a; Strauss et al., 2019). In comparison, Ni et al. (2020) utilised Illumina ABS in combination with PMA viability dye (final concentration of 0.1 mmol l−1) pre-treatment to monitor the viable microbiome in anaerobic sludge digesters. The PMA treatment significantly decreased the number of detected operational taxonomic units (OTUs) and the Chao1 and Shannon diversity indices in the sludge samples, as compared to the non-PMA-treated samples, thereby allowing for the identification of the active and core microbial members in the anaerobic sludge digesters.
However, discrepancies regarding the efficiency of the EMA and PMA nucleic acid binding dyes to detect Gram-positive versus Gram-negative bacteria (due to the structure, composition and complexity of the cell membrane); differences in the viability treatment procedure viz. dye concentration, incubation time and temperature; and exposure time and original sample matrix have been reported in literature (Fittipaldi et al., 2012; Kumar & Ghosh, 2019; Reyneke et al., 2017). For example, while it has been reported that PMA does not always sufficiently penetrate membrane compromised cells, possibly resulting in the overestimation of viability, EMA may penetrate viable cells and may exhibit a cytotoxic effect at high treatment concentrations (Fittipaldi et al., 2012).
Taking these factors into consideration, we previously assessed the efficiency of various EMA (6 to 50 µmol l−1) and PMA (25 to 100 µmol l−1) treatment concentrations for the selective detection of viable Gram-negative (i.e. Legionella pneumophila, Pseudomonas aeruginosa and Salmonella typhimurium) and Gram-positive (i.e. Enterococcus faecium and Staphylococcus aureus) bacterial species (Reyneke et al., 2017). Results from the study indicated that 6 µmol l−1 EMA and 50 µmol l−1 PMA treatment concentrations, in combination with quantitative PCR (qPCR) assays, were comparable and efficient for the selective detection of viable Gram-negative and Gram-positive bacterial species in untreated spiked water samples and spiked water samples heat-treated at 70 °C for 15 min (Reyneke et al., 2017). The current study therefore aimed to determine which optimised viability dye concentration (6 µmol l−1 EMA or 50 µmol l−1 PMA) may be better suited for monitoring mixed bacterial communities in RHRW, and effectively identify the primary bacterial contaminants. To achieve this aim, the total viable bacterial community in RHRW collected from a peri-urban housing complex was determined using Illumina (without viability dye pre-treatment), EMA-Illumina and PMA-Illumina ABS. Additionally, the most frequently detected bacterial genera known to contain pathogenic or opportunistic pathogenic species identified using the ABS analysis were enumerated in the RHRW samples using qPCR assays.
2 Materials and Methods
2.1 Description of Sampling Site and RHRW Sample Collection
In order to compare the efficiency of 6 µmol l−1 EMA and 50 µmol l−1 PMA pre-treatment for monitoring the total viable bacterial community in RHRW using ABS, 10 rainwater tanks were randomly sampled (n = 10) from a peri-urban housing complex (GPS co-ordinates: 34°20′ 11.81″S, 19°00′ 59.74″E) in the Overberg Municipality (Western Cape, South Africa). Each house had a 2000-L above-ground rainwater harvesting (RWH) tank connected to a tiled roof from which 10 L of rainwater was collected. Briefly, the tap connected directly to the RWH tank was sprayed with 70% ethanol, and the water was allowed to flow for approximately 15 s prior to filling two sterile 5 L polypropylene bottles [rinsed with distilled water, sterilised with 70% ethanol and dried overnight at 60 °C (Waso et al., 2018)] (Ahmed et al., 2017). The samples were transported to the laboratory, stored at 4 °C, and processed within 24 h of collection (Ahmed et al., 2017).
2.2 RHRW concentration, nucleic acid binding dye pre-treatments and DNA extractions
Two litres of the collected RHRW samples were concentrated through flocculation using 1 mol l−1 CaCl2 (Biolab, Merck, Wadeville, South Africa) and 1 mol l−1 Na2HPO4 (Saarchem, Durban, South Africa) and filtration as outlined in Dobrowsky et al. (2015). Briefly, 2 mL of 1 mol l−1 CaCl2 (Biolab, Merck) and 2 mL of 1 mol l−1 Na2HPO4 (Saarchem) was added to a 2 L water sample and the mixture stirred for 10 min using a magnetic stirrer to allow for flocculation, whereafter the samples were filtered through a 47-mm, 0.45-μm pore size non-charged mixed-ester membrane filter (Whatman GmbH, Dassel, Germany) at a flow rate of approximately ≥ 65 mL/min/cm2 at 0.7 bar (70 kPa). Following filtration, each membrane filter (Whatman GmbH, Germany) was washed with 3 mL of 0.3 mol l−1 citrate buffer (pH 3.5) to allow for the dissolution of the captured flocs from the membrane filter and each 3 mL concentrate was divided into three aliquots of 1 mL, respectively. Each aliquot was then centrifuged at 16,000 × g for 5 min and the resulting pellet resuspended in sterile saline. Subsequently, the protocol outlined in Reyneke et al. (2017) was followed to subject the first 1 mL aliquot to 6 µmol l−1 EMA (Biotium, Hayward, CA, USA) treatment and the second 1 mL aliquot to 50 µmol l−1 PMA (Biotium) treatment, with the third 1 mL aliquot left untreated (control sample).
Deoxyribonucleic acid was extracted from each of the untreated samples and the samples subjected to the respective viability treatments (EMA and PMA) using the Quick-DNA™ Fecal/Soil Microbe Miniprep Kit (Zymo Research, Irvine, USA) according to the manufacturer’s instructions. All DNA samples were cleaned and concentrated using a sodium acetate and ethanol precipitation protocol (Sambrook & Russell, 2001), whereafter the NanoDrop® ND-1000 (Nanodrop Technologies Inc., Wilmington, Delaware, USA) and Qubit™ fluorometer in conjunction with the Qubit™ dsDNA HS Assay Kit (Invitrogen Corporation, Carlsbad, California, USA) were used to determine the quality (A260/A280 and A260/A230 absorbance ratios) and the concentration of double-stranded DNA, respectively.
2.3 Illumina MiSeq ABS and Sequence Data Analysis
Following the quality control assessment, the DNA samples [n = 30; untreated/control (n = 10), EMA (n = 10) and PMA (n = 10)] were sent for Illumina MiSeq sequencing library construction and sequencing at the Centre for Proteomic and Genomic Research (CPGR), Cape Town, South Africa, as previously described by Strauss et al. (2019) and Reyneke et al. (2020a). Briefly, the sequencing library was prepared according to the Illumina 16S Metagenomic Sequencing Library Preparation guide by amplifying a ∼460 bp region located in the hypervariable V3–V4 region of the bacterial 16S rRNA gene using KAPA HiFi HotStart ReadyMix (Roche Diagnostics) and region of interest-specific primers with overhang Illumina adapter sequences (Klindworth et al., 2013). The 16S amplicon forward primer 5′- TCGTCGGCAGCGTCAG ATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG- 3′ and the 16S amplicon reverse primer 5′- GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC- 3′ were used where the underlined sequences refer to the 16S locus interest-specific sequence of the primer. The amplicons were purified from primer dimers and free primers using AMPure XP beads (Beckman Coulter Genomics, Danvers, MA, USA) followed by an Index PCR using the Nextera XT Index kit (Illumina, San Diego, CA, USA) to attach dual indices and Illumina sequencing adapters to the original amplicon. A final amplicon purification step was performed using AMPure XP beads (Beckman Coulter Genomics). Libraries were then normalised and pooled and sequenced on the Illumina MiSeq instrument using the MiSeq Reagent Kit v3 (Illumina).
The open-source bioinformatics program Mothur (version 1.42.0) was used to process the sequence reads (Schloss et al., 2009), while the standard operating procedure described by Kozich et al. (2013) was followed (https://mothur.org/wiki/MiSeq_SOP) with modifications as outlined by Strauss et al. (2019) and Reyneke et al. (2020a). Contigs were created with the sequence paired-end reads and trimmed to 470 nucleotides. Sequences with homopolymers of eight nucleotides or ambiguous bases were removed and high-quality sequences were aligned using the SILVA (v.128) reference database (Pruesse et al., 2007). A 1% pre-cluster error and chimaera removal was conducted using VSEARCH in order to quality trim the sequences. Assignment of OTUs was completed at a 97% identity using the OptiClust algorithm. Taxonomic assignments were made against the Ribosomal Database Project (RDP; version 16) (Cole et al., 2008). The method described by Wang et al. (2013) was used for the classification (bootstrap cut-off of 80%). Following the taxonomic assignments, sequences from individual samples were rarefied to equal sample size based on the sample with fewest sequences which equalled 9680.
Mothur (version 1.42.0) was used to calculate α-diversity indices, including sample coverage, the number of observed OTUs, species richness, Shannon diversity index and the abundance-based coverage estimate (ACE) (Ahmed et al., 2017). In order to assess the differences in bacterial community composition [beta (β)-diversity] between the untreated (control), EMA-treated and PMA-treated RHRW samples, permutational multivariate analysis of variance (PERMANOVA) was performed and the results visualised using non-metric multidimensional scaling (NMDS) using the computer software RStudio (version 1.0.153) (Library = Vegan) (Strauss et al., 2019). Additionally, relative abundances at the family and genus level were calculated using RStudio and was visualised using the computer software Microsoft® Excel (version 15.39), while the number of shared bacterial families/genera between the sample types were visualised using Euler diagrams (Micallef & Rodgers, 2014). The normality of all the generated data was assessed using the D’Agostino-Pearson normality test, while significant differences between the untreated/control, EMA-treated and PMA-treated samples were assessed using one or two-way ANOVA and Tukey’s multiple comparisons test on GraphPad Prism (version 7.04).
2.4 Quantification of the Most Readily Detected Bacterial Genera Known to Contain Pathogenic or Opportunistic Pathogenic Species
Based on the ABS results, qPCR was conducted to quantify the most readily detected bacterial genera known to contain pathogenic or opportunistic pathogenic species (viz. Acinetobacter spp., Legionella spp. and Pseudomonas spp.) in the untreated DNA samples as well as the samples that were subjected to treatment with 6 µmol l−1EMA or 50 µmol l−1 PMA pre-treatment. All qPCR assays were performed using a LightCycler® 96 (Roche Diagnostics) instrument with the primers and cycling parameters as outlined in Table 1. For all qPCR assays, the reaction mixture (final volume of 20 μL) consisted of 10 µL FastStart Essential DNA Green Master Mix (1 ×), 5 μL template DNA and 0.4 µL of each primer (0.2 μM). All DNA samples were diluted (tenfold) prior to analysis with the respective qPCR assays (minimise PCR inhibitors) and all analysis was conducted in duplicate. Melt curve analysis was included for all of the SYBR green real-time PCR assays in order to verify the specificity of the primer set by ramping the temperature from 65 to 97 °C at a rate of 0.2 °C/s with continuous fluorescent signal acquisition at 5 readings/°C.
To generate a standard curve for the quantification of Acinetobacter spp., Legionella spp. and Pseudomonas spp., the target genes (i.e. Acinetobacter spp. = rpoB; Legionella spp. = 23S rRNA; Pseudomonas spp. = oprI) were first amplified by using the respective primers (Table 1) in conventional PCR assays on DNA extracted from Acinetobacter baumannii ATCC 19606, Legionella pneumophila ATCC 33152 and Pseudomonas aeruginosa ATCC 27853, respectively. All PCR mixtures consisted of a final volume of 25 µL and contained Green GoTaq® Flexi buffer (Promega, Madison, WI, USA), MgCl2 (Promega), dNTP mix (Thermo Scientific, Hudson, NH, USA), the respective forward and reverse primers, GoTaq® Flexi DNA polymerase (Promega) and 2 µL template DNA, as outlined in Table 1. Amplification was performed on a T100™ Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA) using the PCR cycling parameters outlined in Table 1. For each PCR assay, sterile milliQ was used as a negative control.
The obtained PCR products were cleaned and concentrated using the Wizard® SV Gel and PCR Clean-up System (Promega, Madison, WI, USA) as per manufacturer’s instructions. Following DNA concentration determination using a NanoDrop® ND-1000 (Nanodrop Technologies Inc.), the DNA concentration and gene product size were used to calculate the dilution required to obtain a final DNA concentration of 109 gene copies/µL (Dobrowsky et al., 2016). Serial tenfold dilutions (108 to 100) of the PCR products (Acinetobacter spp., Legionella spp. and Pseudomonas spp.) were then prepared in order to generate a standard curve (1.00 × 108 gene copies/µL to 1.00 × 100 gene copies/µL). The lower limit of detection (LLOD) for all qPCR assays was determined as the lowest number of gene copies consistently detected within the respective standard curves.
The gene copies detected in the samples utilising the qPCR assays were converted to gene copies per 100 mL of the original RHRW sample as outlined by Waso et al. (2018). The gene copy numbers (gene copies per 100 mL) were then converted to cell equivalents (cells per 100 mL) by utilising the number of copies of the target gene present within the target host (Table S1, Supporting Information) (Reyneke et al., 2020a).
3 Results
3.1 Bacterial α-Diversity Analysis of the RHRW Samples
The ABS analysis of the untreated RHRW samples yielded a total of 290,400 sequences, which were further clustered into a total of 8361 OTUs, at a similarity of 97%. Overall, the sequencing analysis resulted in a 95.4 ± 0.9% mean sample coverage for the untreated RHRW samples, suggesting a nearly complete characterisation of the bacterial communities (Table 2). Analysis of the corresponding RHRW samples which were EMA- and PMA-treated indicated that sample coverage was not significantly influenced (p > 0.05) by the respective viability treatments (Table 2). Similarly, the recorded species richness [observed richness (Sobs)], Shannon diversity and ACE did not differ significantly (p > 0.05) between the untreated, EMA-treated and PMA-treated RHRW samples (Table 2). Comparison of the NMDS ordination plots of the untreated, EMA-treated and PMA-treated RHRW samples indicated distinct groupings for these samples (Fig. 1).
Non-metric multidimensional scaling (NMDS) ordination plot depicting the relative differences in OTU community composition in the untreated (Greek capital letter delta), EMA-treated (Greek capital letter omicron) and PMA-treated ( +) RHRW samples. The Bray–Curtis distance metric was used to quantify the similarity between the observed patterns. Ellipses indicate a 95% confidence interval
3.2 Characterisation and Comparison of the Bacterial Families Detected in the RHRW Samples
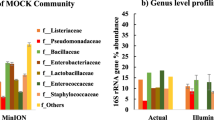
Taxonomic diversity analysis revealed that 61,365 sequences were classified into 119 unique families for the untreated RHRW samples. Based on the relative abundance, the overall bacterial population in the untreated RHRW samples were predominantly comprised of the families Comamonadaceae (10.9%), Oxalobacteraceae (7.3%), Chitinophagaceae (5.6%) and Pseudomonadaceae (5.3%), with 36.5% of the OTUs not classified into any known bacterial family (Fig. 2a; Table S2, Supporting Information).
Mean relative abundance of the primary bacterial a families and b genera detected in the untreated, EMA-treated and PMA-treated RHRW samples. Sequences with a homology ≥ 80% were used and samples with an abundance > 1% are presented. The presented data is also outlined in Table S2 and S3 (Supporting Information)
For the EMA-treated samples, 66,005 sequences were classified into 120 unique families, with the overall relative abundance of the viable bacterial population in the RHRW samples predominantly comprised of the families Oxalobacteraceae (12.0%), Pseudomonadaceae (8.9%) and Sphingomonadaceae (7.2%) (Fig. 2a; Table S2, Supporting Information). Additionally, 31.7% of the OTUs could not be classified into any known bacterial family, which was less than was observed for the corresponding untreated RHRW samples (Fig. 2a; Table S2, Supporting Information). Comparison of the EMA-treated and untreated RHRW samples indicated that the relative abundance of the Chitinophagaceae, Comamonadaceae, Oxalobacteraceae, Planctomycetaceae, Pseudomonadaceae, Rubrobacteraceae and Sphingomonadaceae families differed significantly (p < 0.05) following EMA treatment (Fig. 3a).
Based on relative abundance, the overall viable bacterial population in the RHRW samples which were PMA-treated (65,947 sequences classified into 117 unique families) was predominantly comprised of the families Sphingomonadaceae (10.6%), Oxalobacteraceae (7.7%), Chitinophagaceae (7.4%), Pseudomonadaceae (6.7%) and Moraxellaceae (5.3%) (Fig. 2a; Table S2, Supporting Information). Additionally, 31.8% of the OTUs could not be classified into any known bacterial family, which was also less than was observed for the corresponding untreated RHRW samples (Fig. 2a; Table S2, Supporting Information). Comparison of the PMA-treated and untreated RHRW samples indicated that the relative abundance of the Comamonadaceae and Sphingomonadaceae differed significantly (p < 0.05) following PMA treatment (Fig. 3a), while the relative abundance of the Chitinophagaceae, Comamonadaceae, Oxalobacteraceae, Pseudomonadaceae, and Sphingomonadaceae in the PMA-treated samples differed significantly (p < 0.05) from the corresponding EMA-treated samples (Fig. 3a).
Overall, the untreated, EMA-treated and PMA-treated RHRW samples had 93 bacterial families in common, while 13, 10 and 11 bacterial families were only observed in the untreated, EMA-treated and PMA-treated RHRW samples, respectively (Fig. S1, Supporting Information). Additionally, seven bacterial families were only detected in the untreated and EMA-treated RHRW samples, five bacterial families were only detected in the untreated and PMA-treated RHRW samples, while seven bacterial families were only observed in the EMA-treated and PMA-treated RHRW samples (Fig. S1, Supporting Information).
3.3 Characterisation and Comparison of the Bacterial Genera Detected in the RHRW Samples
At the genus level, for the untreated RHRW samples, the taxonomic diversity analysis revealed that 49,625 sequences were classified into 218 unique genera. Based on the relative abundance, the overall bacterial population detected in the untreated RHRW samples were predominantly comprised of the genera Limnohabitans (8.6%), Pseudomonas (5.2%) and Sediminibacterium (4.1%), with 46.7% of the OTUs not classified into any known bacterial genera (Fig. 2b; Table S3, Supporting Information).
At the genus level, for the EMA-treated RHRW samples, 54,466 sequences were classified into 239 unique genera, with the overall relative abundance of the viable bacterial population predominantly comprised of the genera Janthinobacterium (10.7%), Pseudomonas (8.9%), Acinetobacter (4.8%) and Rubrobacter (4.7%) (Fig. 2b; Table S3, Supporting Information). Additionally, 39.7% of the OTUs could not be classified into any known bacterial genera, which was less than was observed for the corresponding untreated RHRW samples (Fig. 2b; Table S3, Supporting Information). Comparison of the EMA-treated and untreated RHRW samples indicated that the relative abundance of the Janthinobacterium, Limnohabitans, Opitutus, Pseudomonas, Reyranella, Rubrobacter, Sediminibacterium and Sphingobium genera differed significantly (p < 0.05) following EMA treatment (Fig. 3b).
At the genus level, the overall viable bacterial population detected in RHRW samples which were PMA-treated (55,156 sequences classified into 217 unique genera), were predominantly comprised of Pseudomonas (6.6%), Janthinobacterium (5.6%), Sediminibacterium (5.4%) and Acinetobacter (5.2%) (Fig. 2b; Table S3, Supporting Information). Additionally, 41.0% of the OTUs could not be classified into any known bacterial genera, which was less than was observed for the corresponding untreated RHRW samples (Fig. 2b; Table S3, Supporting Information). Comparison of the PMA-treated and untreated RHRW samples indicated that the relative abundance of the Janthinobacterium, Limnohabitans, Novosphingobium, Opitutus and Sphingorhabdus differed significantly (p < 0.05) following PMA treatment (Fig. 3b), while the relative abundance of the Janthinobacterium, Limnohabitans, Novosphingobium, Pseudomonas, Reyranella, Rubrobacter, Sediminibacterium and Sphingorhabdus in the PMA-treated samples differed significantly (p < 0.05) from the corresponding EMA-treated samples (Fig. 3b).
Overall, the untreated, EMA-treated and PMA-treated RHRW samples had 165 bacterial genera in common, while 23, 33 and 24 bacterial genera were only observed in the untreated, EMA-treated and PMA-treated RHRW samples, respectively [Fig. S2(a), Supporting Information]. Additionally, 21 bacterial genera were only detected in the untreated and EMA-treated RHRW samples, while eight bacterial genera were only detected in the untreated and PMA-treated RHRW samples. Furthermore, 21 bacterial genera were only observed in the EMA-treated and PMA-treated RHRW samples [Fig. S2(a), Supporting Information]. Overall, an increase in the detection of Gram-negative bacterial genera was observed following EMA treatment (172 genera) in comparison to the untreated RHRW (167 genera), while a decrease in the detection of Gram-negative bacterial genera was observed following PMA treatment (162 genera) in comparison to the untreated RHRW [Fig. S2(b), Supporting Information]. Additionally, an increase in the detection of Gram-positive bacterial genera was observed following EMA treatment (64 genera) and PMA treatment (50 genera), in comparison to the untreated RHRW (47 genera) [Fig. S2(b), Supporting Information].
3.4 Detection of Bacterial Genera Containing Pathogenic and/or Opportunistic Pathogenic Species
The presence of viable opportunistic pathogens and pathogenic genera in the RHRW samples were determined through taxonomic assignment using the SILVA and RDP databases. Data analysis identified a total of 20 viable bacterial genera containing pathogenic or opportunistic pathogenic species in the RHRW samples (Fig. 4). Acinetobacter spp., Legionella spp. and Pseudomonas spp. were present in all (100%) the untreated, EMA- and PMA-treated RHRW samples. High detection frequency percentages were also obtained for Mycobacterium spp., which was present in 90% of the untreated, 100% of the EMA-treated and 90% of the PMA-treated RHRW samples (Fig. 4). In contrast, while Clostridium sensu stricto and Serratia spp. were present in 90% and 100% of the EMA-treated RHRW samples, respectively, these genera were only detected in 60% and 40% of the untreated samples and 50% (for both genera) of the PMA-treated RHRW samples (Fig. 4).
It should also be noted that certain genera were only detected in the RHRW samples when EMA or PMA pre-treatment was used in combination with Illumina ABS (Fig. 4). For example, Chlamydia spp. and Streptococcus spp. were only detected in 10% (for each genus) and Listeria spp. in 20% of the EMA-treated RHRW samples, while Corynebacterium spp. were only detected in 10% of the PMA-treated RHRW samples. Similarly, Clostridium III was only detected in 10% of both the EMA-treated and PMA-treated RHRW samples, while Clostridium XI was detected in 30% of the EMA-treated RHRW samples and 10% of the PMA treated RHRW samples. Lastly, Streptomyces spp. were detected in 40% of the EMA-treated RHRW samples and 20% of the PMA-treated RHRW samples.
3.5 Assessment of the EMA and PMA Pre-treatments for the Quantification of Opportunistic Pathogens
Quantitative PCR was conducted to enumerate Acinetobacter, Legionella and Pseudomonas spp. in the untreated, EMA-treated and PMA-treated samples, as they were the most frequently detected bacterial genera known to contain pathogenic or opportunistic pathogenic species as indicated by the ABS analysis (Fig. 4). The performance characteristics of the respective qPCR assays are provided in Table S1 (Supporting Information). For the quantification of Acinetobacter cells in the untreated (no viability treatment control) RHRW samples, the concentration ranged from 2.60 × 103 cells/100 mL to 2.13 × 106 cells/100 mL, with a mean concentration of 5.20 × 105 cells/100 mL recorded (Fig. 5a). A mean Acinetobacter concentration of 3.69 × 105 cells/100 mL (ranging from 2.63 × 103 cells/100 mL to 2.33 × 106 cells/100 mL) was then recorded in the corresponding EMA-treated samples, while a mean Acinetobacter concentration of 4.46 × 105 cells/100 mL (ranging from 2.37 × 103 cells/100 mL to 1.49 × 106 cells/100 mL) (Fig. 5a) was obtained in the corresponding PMA-treated samples. For the quantification of Legionella cells in the untreated (no viability treatment control) RHRW samples, the concentration ranged from 8 cells/100 mL to 1.04 × 103 cells/100 mL, with a mean concentration of 1.84 × 102 cells/100 mL recorded (Fig. 5b). In comparison, a mean Legionella concentration of 1.48 × 102 cells/100 mL (ranging from 4 cells/100 mL to 5.95 × 102 cells/100 mL) was obtained in the corresponding EMA-treated samples, while in the corresponding PMA-treated samples a mean Legionella concentration of 1.33 × 102 cells/100 mL (ranging from 6 cells/100 mL to 5.57 × 102 cells/100 mL) (Fig. 5b) was recorded. For the quantification of Pseudomonas cells in the untreated (no viability treatment control) RHRW samples, the concentration ranged from 2.90 × 103 cells/100 mL to 8.17 × 104 cells/100 mL, with a mean concentration of 4.10 × 104 cells/100 mL recorded (Fig. 5c). A mean Pseudomonas concentration of 2.43 × 104 cells/100 mL (ranging from 4.01 × 102 cells/100 mL to 6.50 × 104 cells/100 mL) was then obtained in the corresponding EMA-treated samples, while a mean Pseudomonas concentration of 1.91 × 104 cells/100 mL (ranging from 1.37 × 103 cells/100 mL to 4.12 × 104 cells/100 mL) was recorded in the corresponding PMA-treated samples (Fig. 5c).
Quantification and distribution of a Acinetobacter spp., b Legionella spp. and c Pseudomonas spp. in the RHRW samples [
 = untreated (n = 10);
= untreated (n = 10);
 = EMA-treated (n = 10);
= EMA-treated (n = 10);
 = PMA-treated (n = 10)]. The whiskers at the end of each box indicate the minimum and maximum values, while the box is defined by the lower and upper quartiles and the mean value. The percentage indicates the mean difference between the untreated and corresponding EMA- or PMA-treated samples, respectively
= PMA-treated (n = 10)]. The whiskers at the end of each box indicate the minimum and maximum values, while the box is defined by the lower and upper quartiles and the mean value. The percentage indicates the mean difference between the untreated and corresponding EMA- or PMA-treated samples, respectively
4 Discussion
The use of whole community ABS displays great potential in providing information on the presence and abundance of microorganisms and community characteristics in environmental water sources (Chiao et al., 2014). Additionally, sample pre-treatment with nucleic acid binding dyes (e.g. EMA and PMA) allows for the selective detection of the viable portion (based on membrane integrity) of the microbial community. The current study therefore aimed to assess and compare the use of ABS in combination with EMA or PMA pre-treatment, to monitor the viable bacterial community in RHRW. Comparison of the α-diversity indices obtained for the EMA and PMA viability pre-treatments, in combination with the ABS, indicated that, while not statistically significant, both dyes were able to reduce the detection of non-viable bacteria in the RHRW samples (reduced number of species and diversity detected in the viability treated samples in comparison to the untreated samples), with lower overall diversity scores obtained for the EMA-treated samples, in comparison to the PMA-treated samples. Similarly, Reyneke et al. (2020a) reported lower (albeit not significant) diversity scores [i.e. Sobs, Chao1 (emphasises the presence of low abundance/rare species to estimate community diversity) and Shannon diversity indices] between untreated and EMA-treated RHRW samples, while Guo and Zhang (2014) reported lower (albeit not significant) diversity scores (i.e. Shannon and Simpson diversity indices) between untreated and PMA-treated activated sludge samples. In comparison, Ni et al. (2020) reported that PMA treatment of anaerobic sludge samples, prior to ABS, resulted in a significant decrease in the number of OTU’s, Shannon diversity index and Chao1 diversity index, in comparison to the non-PMA treated samples. In the current study, the α-diversity results were however, corroborated by the NMDS ordination plots as close yet distinct groupings were observed for the untreated (no viability treatment control), EMA-treated and PMA-treated RHRW samples. This indicates that the viability treatments may have non-selectively reduced the overall detection of the non-viable bacterial cells [i.e. the ability of EMA and PMA to distinguish between intact (viable) and membrane-compromised cells (dead) was not limited to a specific group of organisms].
Classification of the detected OTUs into bacterial families and genera revealed that overall, the dominant bacterial genera detected corresponded to the dominant bacterial families in the untreated, EMA-treated and PMA-treated samples. For example, Janthinobacterium and Pseudomonas belong to the Oxalobacteraceae and Pseudomonadaceae families, respectively, and were detected at a high mean relative abundance in the RHRW samples analysed (Fig. 2). Additionally, both the EMA and PMA pre-treatment, prior to ABS, significantly influenced the relative abundance of certain dominant bacterial families (e.g. Comamonadaceae, Oxalobacteraceae, Pseudomonadaceae and Sphingomonadaceae) and genera (e.g. Limnohabitans, Novosphingobium, Reyranella and Sediminibacterium) in the collected RHRW samples (Fig. 3). For example, at the genus level, the relative abundance of Limnohabitans significantly decreased following EMA and PMA treatment, while the relative abundance of Janthinobacterium significantly increased following EMA and PMA treatment. It should however, be noted that in comparison to PMA pre-treatment, highly significant p-values were obtained in the relative abundance of the dominant bacterial genera following EMA treatment (Fig. 3). Additionally, the EMA and PMA pre-treatments decreased the detection of “unclassified” bacterial families and genera within the RHRW samples, which corresponded with the findings from Htwe et al. (2015) and Ni et al. (2020), who similarly reported that the inclusion of the nucleic acid binding dye pre-treatment resulted in significant changes (both increases and decreases) in the relative abundance of the dominant bacteria.
In comparison to the untreated (no viability treatment control) RHRW samples, 10 and 11 less abundant bacterial families were then detected in the EMA-treated or PMA-treated samples, respectively, while 33 and 24 less abundant bacterial genera were detected in the EMA-treated or PMA-treated samples, respectively. Approximately 44% of the bacterial DNA within environmental water samples have been reported to be relic DNA (Lennon et al., 2018), while extracellular bacterial DNA has been shown to persist in environmental water samples for 25 days (Li et al., 2017). The exclusion of relic DNA during culture-independent water analysis may significantly improve the elucidation of the total viable bacterial community, including the identification of rare/less abundant pathogenic bacterial genera, which may not have been identified and screened for in the water source during routine analysis (Reyneke et al., 2020a). The ability of the nucleic acid binding dyes to effectively exclude the detection of relic DNA (extracellular DNA or DNA from membrane-compromised cells) within the RHRW samples, may thus have increased the sensitivity of the analysis, leading to the detection of rare/less abundant bacterial families and genera in the collected RHRW samples.
The collected RHRW samples were subsequently screened for the presence of bacterial genera known to contain pathogenic or opportunistic pathogenic species in order to identify the primary genera that may potentially pose a health risk to end-users. In total, 20 bacterial genera that are known to contain pathogenic and/or opportunistic pathogenic species were identified in the RHRW samples. Acinetobacter, Legionella, Mycobacterium and Pseudomonas were identified as the most frequently detected genera, irrespective of whether the RHRW was untreated (no viability treatment control), EMA-treated or PMA-treated. Quantitative PCR analyses was subsequently conducted to analyse the mean cell counts of Acinetobacter spp., Legionella spp. and Pseudomonas spp., in the RHRW samples, with comparable results obtained for the EMA and PMA pre-treatments. However, in comparison to the untreated RHRW samples, decreased cell concentrations (albeit not significantly) of 20 to 41% (for EMA) and 14 to 53% (for PMA), respectively, were recorded for the quantification of Acinetobacter spp., Legionella spp. and Pseudomonas spp. Previous studies have indicated that EMA or PMA pre-treatments improved the detection of the viable/active microbial community within monitored samples, with the most pronounced influence of the viability pre-treatment observed in samples that were subjected to a disinfection strategy (e.g. ozonation, chlorination, heat treatment, ultraviolet inactivation), and containing a high proportion of membrane-compromised cells and extracellular DNA (Chiao et al., 2014; Nocker et al., 2010). However, the reduced concentrations recorded in the EMA and PMA treated samples supports the theory that viability dye treatment in combination with qPCR (and ABS) may reduce the detection of relic DNA (extracellular DNA or DNA from membrane-compromised/dead cells), particularly in environmental water samples that have not been disinfected or purified. Moreover, the viability dye pre-treatment increased the detection frequency of certain less abundant genera known to contain pathogenic and/or opportunistic pathogenic species, which may also pose a health risk to end-users (Fig. 4). For example, the detection frequency of Chlamydia, Clostridium XI, Enterococcus, Escherichia/Shigella, Listeria, Nocardia, Streptococcus, Clostridium sensu stricto, Serratia and Streptomyces were higher in the EMA-treated samples in comparison to the untreated RHRW samples; while the detection frequency of Bordetella, Clostridium XI, Corynebacterium, Escherichia/Shigella, Serratia, Staphylococcus and Streptomyces were higher in the PMA-treated samples in comparison to the untreated RHRW samples.
Conflicting conclusions have however, been reported in literature regarding the efficiency of the EMA and PMA nucleic acid binding dyes to detect Gram-positive versus Gram-negative bacteria (due to the structure, composition and complexity of the cell membrane) with EMA reported to potentially result in an under estimation of viability (ability to penetrate viable cells) (Fittipaldi et al., 2012; Kumar & Ghosh, 2019; Nocker et al., 2006; Reyneke et al., 2017), while the use of PMA may result in an overestimation of viability (inability to adequately penetrate membrane-compromised cells) (Fittipaldi et al., 2012; Kumar & Ghosh, 2019; Lee & Levin, 2009; Li et al., 2017; LØvdal et al., 2011; Reyneke et al., 2017). Moreover, research has indicated that differences in the viability treatment procedure i.e. dye concentration, incubation time and temperature, exposure time and original sample matrix may contribute to these reported discrepancies (Fittipaldi et al., 2012; Kumar & Ghosh, 2019; Reyneke et al., 2017). In order to overcome the drawbacks associated with the use of EMA, it has been recommended that lower EMA concentrations should be used with shorter incubation and exposure times, while the incubation step should also be performed on ice to decrease membrane permeability and reduce the entry of EMA into viable cells (Fittipaldi et al., 2012). In contrast, to overcome the drawbacks associated with the use of PMA, it has been recommended that higher PMA concentrations should be used with longer incubation times. Additionally, the incubation step should be performed at room temperature (higher temperature as compared to EMA) to facilitate the entry of PMA into membrane compromised bacterial cells (Fittipaldi et al., 2012). Taking these recommendations into consideration, our previous work indicated that 6 µmol l−1 EMA and 50 µmol l−1 PMA treatment concentrations, in combination with qPCR assays, were comparable and efficient for the selective detection of viable Gram-negative and Gram-positive bacterial species (Reyneke et al., 2017). The optimised concentrations as outlined in Reyneke et al. (2017) were thus employed in the current study for the EMA and PMA pre-treatment protocol and may elucidate why significant discrepancies between the detection of the bacterial families and genera in the EMA and PMA-treated RHRW samples were not observed, as has been reported in previous studies (Fittipaldi et al., 2012). Results from the current study also indicated that the EMA treatment did not negatively influence the detection of Gram-negative versus Gram-positive bacterial genera, as the detection of both groups increased following EMA treatment (Gram-negative = 172 genera; Gram-positive = 64 genera), as compared to the untreated (no viability treatment control) RHRW samples (Gram-negative = 167 genera; Gram-positive = 47 genera). However, in comparison to the untreated RHRW samples, the detection of Gram-negative genera then decreased (n = 162 genera) and Gram-positive genera increased (n = 50 genera) following PMA treatment.
In conclusion, the results from the current study indicated that at the concentrations analysed, both EMA and PMA were efficient in reducing the detection of the non-viable bacterial community within the RHRW in comparison to the untreated control. However, while the EMA and PMA concentrations used in the current study were previously optimised for the detection of various bacterial species in RHRW (Reyneke et al., 2017), it is recommended that the EMA or PMA dye concentrations be optimised for the sample matrix it will be applied to (e.g. higher dye treatment concentrations and treatment times may be required for high turbidity environmental water samples, such as river water and wastewater) to ensure the accuracy of results obtained. Less abundant pathogenic and opportunistic pathogenic bacterial genera were also more readily detected in the EMA- or PMA-treated samples, in comparison to the untreated samples. Thus, by excluding relic DNA, the detection of less abundant genera e.g. pathogenic bacteria in environmental water samples, may be increased, whereafter qPCR analysis may be used to monitor the cell concentration of a specific target organism. However, in comparison to the bacterial families and genera detected in the untreated and PMA-treated RHRW samples, the EMA pre-treatment produced highly significant differences in the relative abundance of the dominant bacterial families and genera. Additionally, EMA-treatment significantly decreased the detection of “unclassified” bacteria and allowed for the increased detection of less abundant bacteria in the RHRW samples, as compared to the PMA-treated RHRW samples. Thus, based on the required treatment concentrations and associated dye costs, the use of EMA may be more efficient for the routine analysis of environmental water samples as compared to PMA. Furthermore, recent advances in the development of alternative viability dyes that target membrane integrity and/or metabolic activity, such as DyeTox13 Green C-2 Azide or PEMAX, necessitates the investigation of these dyes in combination with NGS platforms to elucidate the viable microbial community in diverse water environments.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Ahmed, W., Staley, C., Hamilton, K. A., Beale, D. J., Sadowsky, M. J., Toze, S., & Haas, C. N. (2017). Amplicon-based taxonomic characterization of bacteria in urban and peri-urban roof-harvested rainwater stored in tanks. Science of the Total Environment, 576, 326–334. https://doi.org/10.1016/j.scitotenv.2016.10.090.
Bibby, K., Viau, E., & Peccia, J. (2010). Pyrosequencing of the 16S rRNA gene to reveal bacterial pathogen diversity in biosolids. Water Research, 44, 4252–4260. https://doi.org/10.1016/j.watres.2010.05.039.
Bouvresse, S., Socolovshi, C., Berdjane, Z., Durand, R., Izri, A., Raoult, D., Chosidow, O., & Brouqui, P. (2011). No evidence of Bartonella quintana but detection of Acinetobacter baumannii in head lice from elementary schoolchildren in Paris. Comparative Immunology, Microbiology and Infectious Diseases, 34, 475–477. https://doi.org/10.1016/j.cimid.2011.08.007.
Chiao, T.-H., Clancy, T. M., Pinto, A., Xi, C., & Raskin, L. (2014). Differential resistance of drinking water bacterial populations to monochloramine disinfection. Environmental Science and Technology, 48, 4038–4047. https://doi.org/10.1021/es4055725.
Cole, J. R., Wang, Q., Cardenas, E., Fish, J., Chai, B., Farris, R. J., Kulam-Syed-Mohideen, A. S., McGarrell, D. M., Marsh, T., Garrity, G. M., & Tiedje, J. M. (2008). The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Research, 37, D141–D145. https://doi.org/10.1093/nar/gkn879.
Dobrowsky, P. H., Lombard, M., Cloete, W. J., Saayman, M., Cloete, T. E., Carstens, M., Khan, S., & Khan, W. (2015). Efficiency of microfiltration systems for the removal of bacterial and viral contaminants from surface and rainwater. Water, Air, and Soil Pollution, 226, 33. https://doi.org/10.1007/s11270-015-2317-6.
Dobrowsky, P. H., Khan, S., Cloete, T. E., & Khan, W. (2016). Molecular detection of Acanthamoeba spp., Naegleria fowleri and Vermamoeba (Hartmannella) vermiformis as vectors for Legionella spp. in untreated and solar pasteurized harvested rainwater. Parasites and Vectors, 9, 539–552. https://doi.org/10.1186/s13071-016-1829-2.
Fittipaldi, M., Nocker, A., & Codony, F. (2012). Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification. Journal of Microbiological Methods, 91, 276–289. https://doi.org/10.1016/j.mimet.2012.08.007.
Guo, F., & Zhang, T. (2014). Detecting the nonviable and heat-tolerant bacteria in activated sludge by minimizing DNA from dead cells. Microbial Ecology, 67, 829–836. https://doi.org/10.1007/s00248-014-0389-2.
Herpers, B. L., De Jongh, B. M., Van Der Zwaluw, K., & Van Hannen, E. J. (2003). Real-time PCR assay targets the 23S–5S spacer for direct detection and differentiation of Legionella spp. and Legionella pneumophila. Journal of Clinical Microbiology, 41, 4815–4816. https://doi.org/10.1128/jcm.41.10.4815-4816.2003.
Htwe, N. H. N., Miyanaga, K., Rollon, A. P., & Tanji, Y. (2015). Metagenomic analysis of bacteria community in activated sludge of domestic wastewater treatment plant. International Proceedings of Chemical, Biological and Environmental Engineering, 90, 127–131. https://doi.org/10.7763/IPCBEE.2015.V90.20.
Ibekwe, A. M., Leddy, M., & Murinda, S. E. (2013). Potential human pathogenic bacteria in a mixed urban watershed as revealed by pyrosequencing. PLoS ONE, 8, e79490. https://doi.org/10.1371/journal.pone.0079490.
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., & Glöckner, F. O. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Research, 41, e1. https://doi.org/10.1093/nar/gks808.
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., & Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Applied and Environmental Microbiology, 79, 5112–5120. https://doi.org/10.1128/AEM.01043-13.
Kumar, S. S., & Ghosh, A. R. (2019). Assessment of bacterial viability: A comprehensive review on recent advances and challenges. Microbiology, 165, 593–610. https://doi.org/10.1099/mic.0.000786.
Lee, J. L., & Levin, R. E. (2009). A comparative study of the ability of EMA and PMA to distinguish viable from heat killed bacterial flora from fish fillets. Journal of Microbiological Methods, 76, 93–96. https://doi.org/10.1016/j.mimet.2008.08.008.
Lennon, J. T., Muscarella, M. E., Placella, S. A., & Lehmkuhl, B. K. (2018). How, when, and where relic DNA affects microbial diversity. mBio, 9, e00637-18. https://doi.org/10.1128/mBio.00637-18.
Li, R., Min Tun, H., Jahan, M., Zhang, Z., Kumar, A., Dilantha Fernando, W. G., Farenhorst, A., & Khafipour, E. (2017). Comparison of DNA-, PMA-, and RNA-based 16S rRNA Illumina sequencing for detection of live bacteria in water. Scientific Reports, 7, 5752. https://doi.org/10.1038/s41598-017-02516-3.
LØvdal, T., Hovda, M. B., Björkblom, B., & MØller, S.G. (2011). Propidium monoazide combined with real-time quantitative PCR underestimates heat-killed Listeria innocua. Journal of Microbiological Methods, 85, 164–169. https://doi.org/10.1016/j.mimet.2011.01.027.
Micallef, L., & Rodgers, P. (2014). eulerAPE: Drawing area-proportional 3-venn diagrams using ellipses. PLoS ONE, 9, e101717. https://doi.org/10.1371/journal.pone.010717.
Ni, J., Hatori, S., Wang, Y., Li, Y.-Y., & Kubota, K. (2020). Uncovering viable microbiome in anaerobic sludge digesters by propidium monoazide (PMA)-PCR. Microbiol Ecology, 79, 925–932. https://doi.org/10.1007/s00248-019-01449-w.
Nocker, A., Cheung, C. Y., & Camper, A. K. (2006). Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. Journal of Microbiological Methods, 67, 310–320. https://doi.org/10.1016/j.mimet.2006.04.015.
Nocker, A., Richter-Heitmann, T., Montjin, R., Schuren, F., & Kort, R. (2010). Discrimination between live and dead cells in bacterial communities from environmental water samples analyzed by 454 pyrosequencing. International Microbiology, 13, 59–65. https://doi.org/10.2436/20.1501.01.111.
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W., Peplies, J., & Glöckner, F. O. (2007). SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Research, 35, 7188–7196. https://doi.org/10.1093/nar/gkm864.
Qin, T., Tian, Z., Ren, H., Hu, G., Zhou, H., Lu, J., Luo, C., Liu, Z., & Shao, Z. (2012). Application of EMA-qPCR as a complementary tool for the detection and monitoring of Legionella in different water systems. World Journal of Microbiology and Biotechnology, 28, 1881–1890. https://doi.org/10.1007/s11274-011-0986-x.
Reyneke, B., Dobrowsky, P. H., Ndlovu, T., Khan, S., & Khan, W. (2016). EMA-qPCR to monitor the efficiency of a closed-coupled solar pasteurization system in reducing Legionella contamination of roof-harvested rainwater. Science of the Total Environment, 553, 662–670. https://doi.org/10.1016/j.scitotenv.2016.02.108.
Reyneke, B., Ndlovu, T., Khan, S., & Khan, W. (2017). Comparison of EMA-, PMA- and DNase qPCR for the determination of microbial cell viability. Applied Microbiology and Biotechnology, 101, 7371–7383. https://doi.org/10.1007/s00253-017-8471-6.
Reyneke, B., Hamilton, K. A., Fernández-Ibáñez, P., Polo-López, M. I., McGuigan, K. G., Khan, S., & Khan, W. (2020a). EMA-amplicon-based sequencing informs risk assessment analysis of water treatment systems. Science of the Total Environment, 743, 140717. https://doi.org/10.1016/j.scitotenv.2020.140717.
Reyneke, B., Ndlovu, T., Vincent, M. B., Martínez-García, A., Polo-López, M. I., Fernández-Ibáñez, P., Ferrero, G., Khan, S., McGuigan, K. G., & Khan, W. (2020b). Validation of large-volume batch solar reactors for the treatment of rainwater in field trials in sub-Saharan Africa. Science of the Total Environment, 717, 137223. https://doi.org/10.1016/j.scitotenv.2020.137223.
Roosa, S., Wauven, C. V., Billon, G., Matthijs, S., Wattiez, R., & Gillan, D. C. (2014). The Pseudomonas community in metal contaminated sediments as revealed by quantitative PCR: A link with metal bioavailability. Research in Microbiology, 165, 647–656. https://doi.org/10.1016/j.resmic.2014.07.011.
Sambrook, J., & Russell, D. W. (2001). Molecular cloning: A laboratory manual (3rd ed., Vol. 1). Cold Spring Harbor Laboratory Press.
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., Lesniewski, R. A., Oakley, B. B., Parks, D. H., Robinson, C. J., Sahl, J. W., Stres, B., Thallinger, G. G., Van Horn, D. J., & Weber, C. F. (2009). Introducing Mothur: Open-source, platform-independent, community supported software for describing and comparing microbial communities. Applied and Environmental Microbiology, 75, 7537–7541. https://doi.org/10.1128/AEM.01541-09.
Seinige, D., Von Köckritz-Blickwede, M., Krischek, C., Klein, G., & Kehrenberg, C. (2014). Influencing factors and applicability of the viability EMA-qPCR for a detection and quantification of Campylobacter cells from water samples. PLoS ONE, 9, e113812. https://doi.org/10.1371/journal.pone.0113812.
Strauss, A., Reyneke, B., Waso, M., Ndlovu, T., Brink, C., Khan, S., & Khan, W. (2019). EMA-amplicon-based taxonomic characterisation of the viable bacterial community present in untreated and SODIS treated roof-harvested rainwater. Environmental Science: Water Research and Technology, 5, 91–101. https://doi.org/10.1039/C8EW00613J.
Wang, J., McLenachan, P. A., Biggs, P. J., Winder, L. H., Schoenfeld, B. I., Narayan, V. V., Phiri, B. J., & Lockhart, P. J. (2013). Environmental bio-monitoring with high-throughput sequencing. Briefings in Bioinformatics, 14, 575–588. https://doi.org/10.1093/bib/bbt032.
Waso, M., Khan, S., & Khan, W. (2018). Microbial source tracking markers associated with domestic rainwater harvesting systems: Correlation to indicator organisms. Environmental Research, 161, 446–455. https://doi.org/10.1016/j.envres.2017.11.043.
Ye, L., & Zhang, T. (2011). Pathogenic bacteria in sewage treatment plants as revealed by 454 pyrosequencing. Environmental Science and Technology, 45, 7173–7179. https://doi.org/10.1021/es201045e.
Acknowledgements
The authors wish to thank the Centre for Proteomic and Genomic Research (CPGR), Illumina and Whitehead Scientific for funding the Illumina sequencing through their Core Lab Competition. The authors would particularly like to thank Drs. Shane Murray and Jeanne Korsman from CPGR for their assistance during the Illumina NGS and Dr. Ferdinand Postma for assistance with the sequence data and statistical analysis.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: BR and WK. Performed the experiments: BR, MW, TN, TC and BH. Analysed the data: BR. Contributed reagents/materials/analysis tools: WK and SK. Compiled the manuscript: BR, MW, TN, TC, BH and WK. Edited the manuscript: SK.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Reyneke, B., Waso, M., Ndlovu, T. et al. EMA- Versus PMA-Amplicon-Based Sequencing to Elucidate the Viable Bacterial Community in Rainwater. Water Air Soil Pollut 233, 103 (2022). https://doi.org/10.1007/s11270-022-05578-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11270-022-05578-w







 ; n = 10), EMA-treated (E or
; n = 10), EMA-treated (E or
 ; n = 10) and PMA-treated (P or
; n = 10) and PMA-treated (P or
 ; n = 10) RHRW samples. Significance: * = p < 0.05; ** = p < 0.005; *** = p < 0.0005; **** p < 0.0001
; n = 10) RHRW samples. Significance: * = p < 0.05; ** = p < 0.005; *** = p < 0.0005; **** p < 0.0001
 = untreated (n = 10);
= untreated (n = 10);
 = EMA-treated (n = 10);
= EMA-treated (n = 10);
 = PMA-treated (n = 10)]
= PMA-treated (n = 10)]