Abstract
This study aims to expand the knowledge about fecal contamination by humans and animals using Adenovirus (AdV) as bioindicators in different water sources from rural areas, to evaluate the viral infectivity, and to compare the different techniques used to detect the Human mastadenovirus (HAdV). For that, 124 samples were collected (86 from groundwater and 38 from surface water) along the Rio dos Sinos Basin. Escherichia coli count was carried out, and the samples were submitted for the detection and characterization tests of AdV by different methods (qPCR, multiplex qPCR, and nested PCR). In addition, the viral infectivity was realized by integrated cell culture quantitative PCR (ICC-qPCR). E. coli was detected in 63% of groundwater samples (geometric mean of 16.7 MPN/100 mL) and 68% (geometric mean: 5.08×102 MPN/100 mL) in surface waters. Among the viral indicator in the groundwater, the HAdV was detected in 49% of the samples, followed by Canine mastadenovirus (CAV, 20%), Bovine mastadenovirus (BAdV, 17%), Aviadenovirus (AvAdV, 15%), and Porcine mastadenovirus (PAdV, 03%). In surface water, HAdV was detected in 45%, followed by CAV (42%), BAdV (29%), and PAdV and AvAdV (13%). The quantification of genomic copies per liter ranged from 9.40×104 to 5.54×1010 gc/L. In groundwater samples, it was possible to observe infectious adenovirus in 12% of the samples, as well as in surface water for 18%. The results showed an increase in the sensitivity of positive samples when a combined set of techniques were used for HAdV detection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The provision of drinking water was one of the most successful measures achieved by humanity in terms of disease control and prevention (Ashbolt 2015). The access to basic sanitation, as well as drinking water, contribute significantly to disease and death reduction, mainly among children (UNICEF and WHO 2015; WHO 2017). Globally, in rural areas, the drinking water cover is 84%, while in urban areas, the cover of such services can reach 96% (WHO/UNICEF 2015). However, in Brazil, according to the demographic census of 2010, 93% of households in urban areas receive drinking water and only 29% in rural areas (IBGE 2010a). This high contingent of households on the margins of the distribution of treated water makes the population living at these locations dependent on other water sources, as well as water springs, artesian wells, or shallow wells. Usually, these water sources are of doubtful quality because they do not receive adequate treatment and are not even routinely monitored, representing a danger to animal and human health that enjoy these waters (Perdomo et al. 2006).
Enteric viruses are microorganisms associated with environmental contamination and infections caused by consumption of contaminated water and food (Bosch et al. 2008). These pathogens are eliminated in large quantities in the feces of humans and animals, and they have easy dispersion into the environment; so, these viruses are frequently found in rivers, groundwater, recreation water, drinking water, wastewater, and sewerage (Bosch et al. 2008; Staggemeier et al. 2015). Among enteric viruses, it stands out the Adenoviridae family that is composed of five genera: Atadenovirus, Aviadenovirus, Ichtadenovirus, Mastadenovirus, and Siadenovirus. Adenovirus (AdV) is a non-enveloped virus of icosahedral capsid, it has the approximate size of 70 to 90 nm diameter and has trait projections, and there is a single linear molecule of dsDNA (ICTV 2011). Besides human beings, species-specific AdVs can also infect a wide range of animal species, including mammals, birds, reptiles, amphibians, and fish (Benkő et al. 2002). As these pathogens are ubiquitous in the environment, studies have shown the great importance of research and insertion of fecal indicators of viral origin in evaluation of water pollution (Hewitt et al. 2013; Rames et al. 2016). Different species of enteric viruses can be found in aquatic environments even when these waters are in accordance to the standards of bacterial indicators (Dalla Vecchia et al. 2015; Fong et al. 2005). The viruses are host-specific allowing a better characterization of contamination sources than E. coli which is an invariable bacterium found in humans and animals excreta (Fong et al. 2005; Gotkowska-Płachta et al. 2016). AdVs are highlighted due to the structural characteristics that give them greater stability in adverse conditions, including temperature and pH changes and physical and chemical treatments, such as filtration and chlorination processes, and these methods are often used in water treatment nowadays (Barardi et al. 2012; Fong et al. 2005; Thurston-Enriquez et al. 2002). In addition, they tend to be more resistant to ultraviolet radiation (UV) when compared to RNA viruses, considering that HAdV can use host cell enzymes to repair their DNA damage, thus allowing them a greater persistence in remaining infectious in the environment for a longer period (Ko et al. 2005; Thurston-Enriquez et al. 2002).
This research aimed to use different molecular methods to analyze the microbiological quality of groundwater and surface water in rural areas. In addition to the use of traditional parameters, such as E. coli, this study proposes the investigation of AdV of different species in order to characterize the main source of pollution in these places.
2 Material and Methods
2.1 Sampling
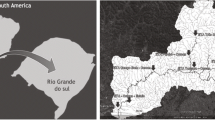
The Rio dos Sinos Basin (Bacia Hidrográfica do Rio dos Sinos - BHRS) is located at the east of the State of Rio Grande do Sul (RS), Brazil (Fig. 1). It covers 32 municipalities and is used for human and animal supply (cattle, horses, pigs, sheep, and birds), rice irrigation, and industrial uses.
On the left of a, map of Brazil and in green the Rio Grande do Sul state. On the right of a, map of RS state and in gray the BHRS. In b highlighted in red, the urban areas spot in the region of Vale do Rio dos Sinos. In c the fluvial map of BHRS. It is possible to observe in panels c and b the distribution of rural properties in the region of Vale of Rio dos Sinos. High stretch: properties 01 and 02 belong to the city of Caraá; properties 03 to 06 belong to the city of Santo Antônio da Patrulha; properties 07 to 09 refer to the city of Riozinho; properties 10 and 11 to the city of Rolante; and properties 21 to 23 the city of Taquara. Middle stretch: properties 17 to 20 to the city of Araricá and properties 24 to 26 to the city of Igrejinha. Lower stretch: properties 12 to 16 to the city of Sapiranga; properties 27 to 29 to the city of Nova Santa Rita; property 30 to the city of São Leopoldo; and properties 31 to 34 to the city of Campo Bom
The Basin is currently divided into three distinct, being the following: (a) high stretch, represented by areas of small rural properties with low population density, where agriculture and small dairy, pigs and poultry farms predominate; (b) medium stretch, higher population density, still with characteristics of rural areas, but not as predominant as in the upper part; and (c) lower stretch, represented by large industrial and population volume (PRÓ-SINOS n.d.), the lower reaches of the Rio dos Sinos Valley are located where there is a greater demand of water for agricultural purposes. That of a total of 3.81 m3/s water, 85% is aimed for irrigation of rice in the low stretch, 13% for the municipalities of the high stretch, and only 2% goes to the middle stretch (COMITESINOS 2015). Thus, this shows that the lower part of the Basin is not only composed of urban and industrial areas. Table 1 shows the density of human and animal population of research participant municipalities, and there are not dog population estimate because no official data were found. However, in Brazil, it is common that the people have dogs such as domestic animal, mainly in rural areas.
The samplings were carried out on November and December of 2015, in 34 rural properties of 11 municipalities located along the BHRS (Fig. 1). Water samples (500 mL) were collected in sterile bottles, being that 86 were from groundwater (spring and artesian well) and 38 were from surface water (streams, weir, and river), totalizing 124 samples. This research received the support of Technical Assistance and Rural Extension Company of Rio Grande do Sul State Government (Empresa de Assistência Técnica e Extensão Rural do Governo do RS - EMATER/RS), which is an institution that has the goal to promote sustainable rural development in the State of Rio Grande do Sul. Because of this, the selected rural proprieties are in sites where projects have been carried out by EMATER.
2.2 E. coli Detection and Quantification
Detection and quantification of E. coli were performed using the Colilert® (IDDEX) kit, following the manufacturer’s instructions. In the cases where the most probable number (MPN) of bacteria exceeded the detection limit of the method, the samples were diluted within 24 h after collection, and the technique was applied again.
2.3 Viral Concentration by Ultracentrifugation and Nucleic Acid Extraction
All water samples were concentrated by ultracentrifugation method (Girardi et al. 2018). Aliquots of 36 mL were centrifuged (Sigma® 3-30KS equipment, Germany, rotor 12150-H) at a speed of 41.000Xg, at 8°C for 3 h. After this step, the precipitates were resuspended and vigorously homogenized in the vortex for 1 min with 1 mL of Tris-EDTA buffer (pH 8.0). The DNA of concentrated samples were extracted using the BioPur® kit, following the instructions of the manufacturer. The final elution (60 μL) was performed in nuclease-free microtubes.
2.4 Detection and Quantification by Real-Time Polymerase Chain Reaction (qPCR)
Two different qPCR reactions were performed: first, using the VTB1 oligonucleotides pair targeting HAdV-F and, second, using the VTB2 oligonucleotides pair targeting HAdV-C (Table 2). For a final reaction volume of 25 μL, 12.5 μL of SYBR® Green SuperMix (Invitrogen™ Platinum® SYBR® Green qPCR SuperMix-UDG), 5.5 μL nuclease-free water, 1 μL of each of each pair of oligonucleotides and 5 μL of extracted DNA. The conditions of the qPCR were an initial incubation of 2 min at 50°C, 10 min at 95°C, 45 cycles of 20 s at 95°C for denaturation, and 1 min at 55°C for annealing. After this step, a denaturing curve was made to check the specificity of the amplification products (melting step between 55 and 95°C). The sensitivity of the reactions was 6.2×101 gc/5 μL (Dalla Vecchia et al. 2015). The assays were performed in 96-well plates, and all samples were tested in duplicates, including negative controls and a five-point standard curve formed by serial dilution of positive control with known quantification. When required, the samples were diluted 1:10 (DNA) in nuclease-free water in order to reduce the PCR inhibitory substances present in the samples.
2.5 Detection and Quantification by Multiplex qPCR
The multiplex qPCR was performed using the AdV oligonucleotides pair with HAdV, CAV, BAdV, PAdV, and AvAdV targets (Table 2). The volume and concentration of reagents were same used in qPCR, as well as the qPCR conditions; however, the annealing temperature was 58°C. The identity of final PCR products was confirmed by high resolution melting curves (HRM). The melting temperatures of each species of AdV are as follows: HAdV (88 °C), BAV (85.5 °C), PAdV (83.5 °C), CAV (82 °C), and AvAdV (80.5 °C) (Luz et al. 2015).
2.6 Detection by Nested Polymerase Chain Reaction
Nested polymerase chain reaction (nested PCR) was performed to detect the presence of the AdV genome of different hosts. To a final volume of 50 μL, it was used 25 μL of Master Mix (GoTaq® Colorless Master Mix - PROMEGA), 18 μL nuclease-free water, 1 μL of each oligonucleotide (Pol-F 5′-CAGCCKCKGTTRTGYAGGGT-3′ and Pol-R 5′-GCHACCATYAGCTCCAACTC-3′), and 5 μL of extracted DNA. Assay conditions were an initial incubation step at 94°C for 5 min, 40 cycles of amplification were carried out, consisting of denaturation for 30 s at 94°C, annealing at 50°C for 30 s (−0.5°C per cycle), and extension at 72°C for 1 min and a final extension stage at 72°C for 10 min. The second PCR reaction was performed using the same reagents and quantities of the first step, as well as the amplification cycles, but, just replacing the oligonucleotide pair using in the second the Pol-nF 5′-GGGCTCRTTRGTCCAGCA-3′ and Pol-nR 5′-TAYGACATCTGYGGCATGTA-3′, and the 5 μL of extracted DNA that was replaced by the first PCR products (Li et al. 2010). At the end of the second amplification, the products were submitted to 2% of agarose gel electrophoresis with 0.5 mg of ethidium bromide/mL. And the molecular sizes of the products were compared to a DNA standard of 100 bp (Ludwig). After those amplicons were visualized, they were purified using a QiaQuick DNA purification kit (Qiagen) and sent to sequencing.
2.7 Genetic Sequencing and Phylogenetic Analysis
The DNA sequencing of amplicons obtained from nested PCR was performed by the Sanger method. To identify the DNA fragments, nucleotide sequences were assembled using the CAP3 computer program implemented in the BioEdit 7.0.5 suite. This way, the obtained sequences were compared with other nucleotide fragments available in GenBank according to the neighbor-joining methodology (Saitou and Nei 1987). Afterwards, the phylogenetic tree was elaborated from the calculation of evolutionary distances using the Kimura-parameter 2 method (Kimura 1980) and using Molecular Evolutionary Genetics Analysis version 5 (MEGA5) software (Tamura et al. 2007).
2.8 Detection of HAdV Viability by Integrated Cell Culture Quantitative PCR
To detect the presence of infectious viruses in the samples, lineage cell A549 (human lung carcinoma) was used based on its permission to HAdV; such cells were maintained on Eagle’s minimum essential medium (E-MEM), supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (10,000 IU/mL–10 mg/mL). Cells were cultured in 24-well plates, and after approximately 24 h, the supernatant E-MEM was removed and replaced with 200 μL of the concentrated water samples previously diluted (1:2) with E-MEM and filtered with a membrane of 0.22 μL. The plates were kept in incubator for 2 h with uniform stirring every 15 min. After this incubation period, the inoculum was removed, and 1 mL of cell maintenance medium was added, but without FBS. Plates were maintained in incubator at 37°C with 5% CO2 atmosphere for 5 days, and after this period, the plates were frozen at −80°C. For the second passage in cells, the plate from the first passage was thawed three times, and the previous process procedure was repeated, except for the filtering step, as this time the inoculum used was 200 μL of the result of the first passage. This process was repeated until the third passage. To assess whether there were viable particles, the first and the third passage samples were treated with DNAse. Subsequently, the same samples were submitted to the extraction of the nucleic acids and to qPCR using the oligonucleotide pair VTB1 and VTB2 for the detection of HAdV genomes.
2.9 Comparison HAdV Detection Techniques
A Venn diagram was elaborated in order to obtain the best view of different results obtained with three different tests; the three tests are able to detect HAdV. For that, only the results that used AdV, VTB2, and DNApol oligonucleotides were considered.
3 Results
3.1 E. coli
E. coli was an indicator most abundant in this study; in groundwater samples, the detection rate was 63% (54/86), ranging from 01 to 1.99×103 MPN/100 mL. In surface water, E. coli was detected in 68% (26/38) of the samples with the counting ranging from 05 to 4.35×104 MPN/100 mL.
3.2 Viral Analyses in Groundwater and Surface Water Samples
In groundwater HAdV was the viral indicator with the higher detection rate, which was present in 49% of the samples. In those positive samples, the minimum quantification was 9.40×104 genomic copies per liter (gc/L), and the maximum quantification was 4.51×1010 gc/L, followed by CAV 20% (5.85×107–5.54×1010 gc/L), AvAdV 15% (9.07×106–6.52×108 gc/L), BAdV 17% (7.02×106–3.57×109 gc/L), and PAdV 03% (9.07×106–2.92×108 gc/L) respectively (Fig. 2). In the surface water samples, HAdV was presented with the highest occurrence as well, being detected in 45% with quantification between 5.81×105 and 1.55x1010 gc/L, followed by CAV 39% (4.13×107–1.55×1010 gc/L), BAdV 29% (1.65×107–1.55×1010 gc/L), and finally, with the lowest detection rate, PAdV and AvAdV, where both were found in 13% ( 2.85×108–2.74×109 gc/L and 7.38×107–9.77×109 gc/L, respectively) (Fig. 2). In Fig. 2 it is possible to observe the different detection rates and the quantification of the viral indicators comparing them with E. coli in groundwater and surface water from different stretches of BHRS, and in Table 3, there are the absolute and the detection rate of positive samples.
In a it is possible to observe different detection rates in groundwater and surface water of the viral indicators comparing them with E. coli. In b, c, and d, the quantification minimum (dark gray), maximum (light gray), and de geometric mean (diamond) in high, medium, and low stretch of Rio dos Sinos Basin, respectively
3.3 Phylogenetic Analysis
The phylogenetic analysis demonstrated that were found three different species of AdV (HAdV-C, HAdV-F, and BAdV), both belonging to the Mastadenovirus genus, being the C species the most predominant (Fig. 3).
Phylogenetic analysis of AdVs. Neighbor-joining tree was built using AdV sequences obtained through Sanger sequencing and the reference strains from the NCBI GenBank database. The samples analyzed in this study are highlighted with a diamond for groundwater and a square for surface water. Bootstrap values are indicated at each tree root
3.4 Viral Infectivity Analysis
In groundwater samples, it was possible to observe 10/86 (11.6%) infectious samples, being five in the first passage in cells and six in the third passage. However, only one sample of the first passage remained infectious in the third passage as well. In surface water, the total of infectious samples were 07/38 (18.4%), but only two samples were infectious in the first passage and five in the third. None of the samples of the first passage remained infectious in the third passage (Table 4). There were infectious samples only for HAdV-C, none of the samples was infectious for HAdV-F, and no infectivity tests for animal viruses were performed. Only one sample was detected after the third passage with 3.81×102 gc/5μL and in the tenth passage the quantification was 1.35×104. The isolate LMM 2836 did not provide any cytopathic effect despite being detected by molecular techniques.
3.5 Comparison for HAdV Detection Techniques
The reaction that obtained the highest number of positive samples for HAdV was the one used in the AdV oligonucleotide pairs, totaling 32 (26%) positive samples, followed by the reaction using the DNApol oligonucleotide pairs and detected 26 (22%) positive samples and lastly the reaction with the VTB2 oligonucleotide pairs which obtained 21 (17%) positive samples.
4 Discussion
E. coli was the indicator most abundant found in the samples; however, the concentration of E. coli was determined using the Colilert® (IDDEX); this way, only viable bacterium could be detected. In this study, of the 32 negative samples for E. coli, 21 (66%) were positive for one or more viral indicators in groundwater samples. In surface water, all the samples (12) that were negative for E. coli have detected some type of viral indicator. Here, it is not possible to compare the MPN with the unit of measure (genomics copies) used for viral quantification. However, the results of this study corroborate the findings in others that have been shown that the use of bacterial indicators alone is not enough to attest to the microbiological quality of water, because there is no correlation between the presence of E.coli and enteric viruses (Dalla Vecchia et al. 2015; Peteffi et al. 2018; Skraber et al. 2004). Although, using E. coli in combination with viral indicators can be a great strategy for the most complete microbiological water monitoring.
In rural regions, groundwaters are the main sources of water used to drink and other household purposes (such as cleaning, bathing, and cooking). Even if preventive measures as representative evaluation of water quality are rarely taken (Guo et al. 2011), there is still a belief that the groundwaters from rural environments are safer and free of pathogenic microorganisms; this way, it is often considered potable for human consumption (Amaral 2004). However, besides the high rate of E. coli (63%), the HAdV was detected in almost half the sample. This is a risky situation because the detection rates of genomic copies were sometimes higher than the rates found in surface waters and the number of genomic copies was very similar in both matrices, being the maximum difference of just one log.
The dissemination of enteric pathogens in groundwater can occur due to the displacement of the virus from septic tanks (Futch et al. 2010). However, it is known that the safe distance from wastewater disposal to aquifers is difficult to be reached, as demonstrated by Blaschke et al. (2016), who in assessing small biological effluent systems in decentralized locations without access to sewage systems, they observed that for a reduction of up to 12 logs of the total number of enteric viruses, the horizontal setback distance ranges from 39 to 144 m in sand aquifers, 66 to 289 m in gravel aquifers, and from 1 to 2 m, 5km in coarse gravel aquifers.
Due to the lack of basic sanitation services in rural areas, it is observed that the direct disposal of waste in streams by rudimentary channeling and the use of septic tanks in households are rare (Spilki et al. 2013). These facts explain the high detection rate found in the studied regions and underlie the high concentration of genomic copies that were detected, since, in this study, the geometric mean resembles the concentration of wastewater and sewage samples from the developed countries (Dong et al. 2010; Fong et al. 2010). However, in Brazil, it was not the first time that these concentrations were observed in surface water. In the city of Rio de Janeiro, the concentration of HAdV-C was up to 109 gc/L (Staggemeier et al. 2017), and in the surface water streams from BHRS region, it was already detected concentration up to 3.28×108 gc/L (Peteffi et al. 2018).
HAdV was the viral indicator most abundant showing a mean rate of detection in surface water and groundwater of 45%. Considering the 11 municipalities of this study, the human population overcomes the production animal population. Even though a rural area, this area is characterized by only small farms and modest animal production. The animal population is only greater in the upper stretch (Table 1). In the high stretch’s groundwater, the HAdV was detected in 50% of the samples; although it is expected to lower levels of human origin contamination in areas that have the lowest population density, as it is the case of the high stretch of the Basin, the great impact of poor sanitation in this region can change that (Spilki et al. 2013). This situation was already observed in a previous study; on assessing groundwater from the in the same stretch, it was found a detection rate of 83% for HAdV (Staggemeier et al. 2015).
Among the viral indicators from animal origin, CAV was with the highest detection rate that can be explained by the high presence of domestic animals, in which, especially, dogs have been considered a reservoir for many infectious agents and it is believed that they are more affected than the wild animals (Fiorello et al. 2004). The dogs are closely associated with the presence of humans and are often found in high numbers in rural properties; therefore, their proximity disturbs the natural habitat of wild species. Given that the great detection rate of CAV found in surface water shows that this virus circulates in this region, often occurring infections in subclinical forms in domestic species, but it can be fatal in wild populations. Because the viral agents may travel large distances, until reaching wildlife habitats and be a threatening for these species and conserving biodiversity, the lack of antibodies in wild canids can lead to increased mortality and lower fertility when they are exposed to this viral agent (Cleaveland et al. 2000; de Almeida Curi et al. 2010; Fiorello et al. 2004).
BAdV was found in an expressive form in surface water, mainly in the high stretch (50%), and it was expected because this region is represented by small dairy and by poor sanitation strategies. In the five municipalities from this region, there is a bovine population of almost 5 thousand cattle heads (Table 1). However, research on its presence in environmental samples are still limited, even knowing that this virus is ubiquitous in cattle and its occurrence can cause everything from subclinical infections to abortion and cardiopathies (Spilki et al. 2009; Wong and Xagoraraki 2010). A common practice in the rural environment is to use the cattle manure as fertilizer, but this can be a dangerous practice. In this regard, Wong and Xagoraraki (2010) found significant levels of BAdV in manure, feces, and drainage water, which may indicate a high potential for surface water pollution by manure when applied to agricultural fields. The indicators of avian and porcine origin were the ones with the lowest detection rate. This occurrence can be because of the production of these animals which is not the main activity in this region, as the example, the pig farming that is mainly concentrated in the Northeastern and Eastern central region of the State of Rio Grande do Sul (IBGE 2017)
Overall, the majority of AdV from animal origin had a higher detection rate in surface water. These water matrices have been suffering a sharp deterioration in their quality through diffuse sources that are difficult to identify in order to adequate management and remediation planning. Among them, urban, forest, and livestock runoff, the latter, occurs mainly through livestock and other animal access to rivers and streams as well as their solid waste (Amaral 2004; Knappett et al. 2012; Staggemeier et al. 2015). In rural areas, these situations are accentuated because the use of biodigesters has not been widely adopted yet, being common the use of landfills and direct disposal of wastewater in the soil and water resources (Amaral 2004; de Oliveira et al. 2012; Spilki et al. 2013).
Phylogenetic analysis was possible to observe the predominance of HAdV-C, corroborating with other studies that showed that this specie is often found in environmental samples (Gularte et al. 2019; Lin and Singh 2015; Staggemeier et al. 2017). The performance of Sanger sequencing reduces a possible diversity when there is a specie most abundant in a sample because this method consistently identifies the most abundant sequence (Paul et al. 2018). In the HAdV-C sample positives, the majority of their strains are almost identical, and it can be explained because of the oligonucleotides used in this study amplification in the region encoding the DNA polymerase. It is well known that those genes are highly conserved between all HAdV-C (Dhingra et al. 2019).
For the viruses of human origin, it was performed the detection of viral viability, and considering the first and third passages in cells from groundwater and surface water, the rate of samples that presented HAdV viable was 13.7% (17/124). In the surface waters of the city of Rio de Janeiro, in Brazil, the rate of samples with infectious particles of HAdV was 17% (Staggemeier et al. 2017). These values are lower than those seen in tap water intended for human consumption in Korea, where the rate sample with of the HAdV infectious particles was 39.1% (Lee and Kim 2002). However, even with a low viability rate found in this study, one should not discard the risk. Because the viruses evaluated are wild and sometimes they cannot adapt to cell culture, this can generate false-negative results. In addition, from 17 infectious samples, it was possible to isolate only one, namely, the isolate LMM 2863(HAdV-C). Probably, in other samples, the HAdV was below the detection limit of qPCR, but this evidence shows that it is difficult to adapt wild viruses in cell culture.
With many available methods to detect viruses in water, it is difficult to know which test is the most appropriate, given that the result values may double when used a set of technics instead of only one. In this study, analyzing each reaction individually, the differences among them do not seem so big, but considering the three reactions, the number of positive samples is 58 (47%). This is because within the three reactions that can detect HAdV genome, 41 samples were positive individually just in one reaction, 12 in two reactions, and only four samples were positive in three reactions, as demonstrated in Fig. 4. In addition, after submitted negative samples to cell culture, nine samples that were negative before passage in cell culture became positive. This can be explained by two hypotheses: (a) the virus was able to replicate in vitro and exceeded the minimum threshold of the qPCR technique, and (b) it was possible to reduce the number of qPCR reaction inhibitors after the sample passage in the cell. Therefore, out of a total of 124 samples, 67 (54%) were HAdV positive. HAdVs (46.7%) detection rate is of considerable value. However, this number could be obtained only because it was used for more than one technique and, in addition, different pairs of oligonucleotides with the same target (HAdV) were used, but with the different annealing regions. By the way, by doing the samples water passages at cells, followed by molecular detection, it was possible to observe that the number of positive samples became more expressive (54%). In this comparison, the VTB1 oligonucleotide pairs were not considered because in this essay, only one sample was positive.
5 Conclusion
To detect and identify different species of AdV can contribute to models for microbial source tracking. As in this study, it was possible to observe that the biggest contamination occurred by human, canine, and bovine origin; thus, those data may help to manage plans for river basins. In this research, the detection rate of infectious samples was not so expressive, but it is dangerous to say that those water present no risk health infection, as a high concentration of genomic copies has been found and it is already expected that some viruses from environment do not adapt to cell cultures. In addition, these results demonstrate that rural areas deserve more attention from government agencies because it is evident that basic sanitation is neglected in these locations.
Finally, AdVs are considered excellent candidates to be used in conjunction with current indicators; however, there is not a standard protocol yet. The differences in the results found using different protocols to detect HAdV show an important variation in the detection. Thus, it is necessary more research about sensibility tests among the different techniques used to detect HAdV in environmental samples, to verify if the divergence among the detection is in the protocols or in the sample quality, as well as to find a procedure that is fast and reliable and that outweighs the disadvantages of using only one diagnostic tool.
References
Amaral, L. A. (2004). Drinking water as a risk factor to poultry health. Brazilian Journal of Poultry Science, 6(4), 191–199. https://doi.org/10.1590/S1516-635X2004000400001.
Ashbolt, N. J. (2015). Microbial contamination of drinking water and human health from community water systems. Current Environmental Health Reports, 2(1), 95–106. https://doi.org/10.1007/s40572-014-0037-5.
Barardi, C. R. ., Viancelli, A., Rigotto, C., Corrêa, A. A., Moresco, V., Souza, D. S. ., et al. (2012). Monitoring viruses in environmental samples. International Journal of Environmental Science and Engineering Research, 3(3), 1–9.
Benkő, M., Élő, P., Ursu, K., Ahne, W., LaPatra, S. E., Thomson, D., & Harrach, B. (2002). First molecular evidence for the existence of distinct fish and snake adenoviruses. Journal of Virology, 76(19), 10056–10059. https://doi.org/10.1128/JVI.76.19.10056-10059.2002.
Blaschke, A. P., Derx, J., Zessner, M., Kirnbauer, R., Kavka, G., Strelec, H., et al. (2016). Setback distances between small biological wastewater treatment systems and drinking water wells against virus contamination in alluvial aquifers. Science of the Total Environment, 573, 278–289. https://doi.org/10.1016/j.scitotenv.2016.08.075.
Bosch, A., Guix, S., Sano, D., & Pintó, R. M. (2008). New tools for the study and direct surveillance of viral pathogens in water. Current Opinion in Biotechnology, 19(3), 295–301. https://doi.org/10.1016/j.copbio.2008.04.006.
Cleaveland, S., Appel, M. G., Chalmers, W. S., Chillingworth, C., Kaare, M., & Dye, C. (2000). Serological and demographic evidence for domestic dogs as a source of canine distemper virus infection for Serengeti wildlife. Veterinary Microbiology, 72(3–4), 217–227. https://doi.org/10.1016/S0378-1135(99)00207-2.
COMITESINOS. (2015). Balanço Hídrico da Bacia Hidrográfica do Rio dos Sinos. http://www.comitesinos.com.br/arquivos/balanco-hidrico-da-bacia-hidrografica-do-rio-dos-sinos%2D%2D-jun-2015%2D%2Drelatorio%2D%2D%2D%2Dautor%2D%2Dsidnei-agra-2016-03-02-1495030574.pdf. Accessed 10 June 2020
Dalla Vecchia, A., Rigotto, C., Staggemeier, R., Soliman, M. C., Gil de Souza, F., Henzel, A., et al. (2015). Surface water quality in the Sinos River basin, in Southern Brazil: Tracking microbiological contamination and correlation with physicochemical parameters. Environmental Science and Pollution Research, 22(13), 9899–9911. https://doi.org/10.1007/s11356-015-4175-6.
de Almeida Curi, N. H., Araújo, A. S., Campos, F. S., Lobato, Z. I. P., Gennari, S. M., Marvulo, M. F. V., et al. (2010). Wild canids, domestic dogs and their pathogens in Southeast Brazil: Disease threats for canid conservation. Biodiversity and Conservation, 19(12), 3513–3524. https://doi.org/10.1007/s10531-010-9911-0.
de Oliveira, L. K., Fleck, J. D., Comerlato, J., Kluge, M., Bergamaschi, B., Fabres, R. B., et al. (2012). Enteric viruses in water samples from Brazilian dairy farms. Agricultural Water Management, 111, 34–39. https://doi.org/10.1016/j.agwat.2012.05.001.
Dhingra, A., Hage, E., Ganzenmueller, T., Böttcher, S., Hofmann, J., Hamprecht, K., et al. (2019). Molecular evolution of human adenovirus (HAdV) species C. Scientific Reports, 9(1), 1039. https://doi.org/10.1038/s41598-018-37249-4.
Dong, Y., Kim, J., & Lewis, G. D. (2010). Evaluation of methodology for detection of human adenoviruses in wastewater, drinking water, stream water and recreational waters. Journal of Applied Microbiology, 108(3), 800–809. https://doi.org/10.1111/j.1365-2672.2009.04477.x.
Fiorello, C. V., Deem, S. L., Gompper, M. E., & Dubovi, E. J. (2004). Seroprevalence of pathogens in domestic carnivores on the border of Madidi National Park, Bolivia. Animal Conservation, 7(1), 45–54. https://doi.org/10.1017/S1367943003001197.
Fong, T.-T., Griffin, D. W., & Lipp, E. K. (2005). Molecular assays for targeting human and bovine enteric viruses in coastal waters and their application for library-independent source tracking. Applied and Environmental Microbiology, 71(4), 2070–2078. https://doi.org/10.1128/AEM.71.4.2070-2078.2005.
Fong, T.-T., Phanikumar, M. S., Xagoraraki, I., & Rose, J. B. (2010). Quantitative detection of human adenoviruses in wastewater and combined sewer overflows influencing a Michigan river. Applied and Environmental Microbiology, 76(3), 715–723. https://doi.org/10.1128/AEM.01316-09.
Futch, J. C., Griffin, D. W., & Lipp, E. K. (2010). Human enteric viruses in groundwater indicate offshore transport of human sewage to coral reefs of the Upper Florida Keys. Environmental Microbiology, 12(4), 964–974. https://doi.org/10.1111/j.1462-2920.2010.02141.x.
Girardi, V., Demoliner, M., Rigotto, C., Schneider, V. E., Paesi, S., & Spilki, F. R. (2018). Assessment of diversity of adenovirus DNA polymerase gene in recreational waters facilitated by ultracentrifugal concentration. Journal of Water and Health, 16(1), 102–111. https://doi.org/10.2166/wh.2017.144.
Gotkowska-Płachta, A., Gołaś, I., Korzeniewska, E., Koc, J., Rochwerger, A., & Solarski, K. (2016). Evaluation of the distribution of fecal indicator bacteria in a river system depending on different types of land use in the southern watershed of the Baltic Sea. Environmental Science and Pollution Research, 23(5), 4073–4085. https://doi.org/10.1007/s11356-015-4442-6.
Gularte, J. S., Girardi, V., Demoliner, M., de Souza, F. G., Filippi, M., Eisen, A. K. A., et al. (2019). Human mastadenovirus in water, sediment, sea surface microlayer, and bivalve mollusk from southern Brazilian beaches. Marine Pollution Bulletin, 142(December 2018), 335–349. https://doi.org/10.1016/j.marpolbul.2018.12.046.
Guo, Y., Schissler, J. T., Opryszko, M. C., Gibson, K. E., & Schwab, K. J. (2011). Evaluation of human enteric viruses in surface water and drinking water resources in Southern Ghana. The American Journal of Tropical Medicine and Hygiene, 84(1), 20–29. https://doi.org/10.4269/ajtmh.2011.10-0389.
Hewitt, J., Greening, G. E., Leonard, M., & Lewis, G. D. (2013). Evaluation of human adenovirus and human polyomavirus as indicators of human sewage contamination in the aquatic environment. Water Research, 47(17), 6750–6761. https://doi.org/10.1016/j.watres.2013.09.001.
IBGE. (2010a). Censo 2010: Rede geral de abastecimento de água avança mais na zona rural. https://censo2010.ibge.gov.br/noticias-censo.html?idnoticia=2017&view=noticia. Accessed 2 May 2019.
IBGE. (2010b). Conheça cidades e estados do Brasil. https://cidades.ibge.gov.br/. Accessed 2 Jan 2021.
IBGE. (2017). Tabela 3939 - Efetivo dos rebanhos, por tipo de rebanho. https://sidra.ibge.gov.br/. Accessed 7 July 2020.
ICTV. (2011). ICTV 9th Report (2011) - Adenoviridae. https://talk.ictvonline.org/ictv-reports/ictv_9th_report/dsdna-viruses-2011/w/dsdna_viruses/93/adenoviridae. Accessed 2 May 2019
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16(2), 111–120. https://doi.org/10.1007/BF01731581.
Knappett, P. S. K., Mckay, L. D., Layton, A., Williams, D. E., Alam, J., Huq, R., et al. (2012). Implications of fecal bacteria input from latrine-polluted ponds for wells in sandy aquifers. Environmental Science & Technology, 46(3), 1361–1370. https://doi.org/10.1021/es202773w.
Ko, G., Cromeans, T. L., & Sobsey, M. D. (2005). UV inactivation of adenovirus type 41 measured by cell culture mRNA RT-PCR. Water Research, 39(15), 3643–3649. https://doi.org/10.1016/j.watres.2005.06.013.
Lee, S.-H., & Kim, S.-J. (2002). Detection of infectious enteroviruses and adenoviruses in tap water in urban areas in Korea. Water Research, 36(1), 248–256. https://doi.org/10.1016/S0043-1354(01)00199-3.
Li, Y., Ge, X., Zhang, H., Zhou, P., Zhu, Y., Zhang, Y., et al. (2010). Host range, prevalence, and genetic diversity of adenoviruses in bats. Journal of Virology, 84(8), 3889–3897. https://doi.org/10.1128/JVI.02497-09.
Lin, J., & Singh, A. (2015). Detection of human enteric viruses in Umgeni River, Durban, South Africa. Journal of Water and Health, 13(4), 1098–1112. https://doi.org/10.2166/wh.2015.238.
Luz, R. B., Staggemeier, R., Fabres, R. B., Soliman, M. C., Souza F. G., Gonçalves R., et al. (2015). Bioaccumulation of animal adenoviruses in the pink shrimp. Brazilian Journal of Microbiology, 46(3), 715–723. https://doi.org/10.1590/S1517-838246320140323.
Paul, F., Otte, J., Schmitt, I., & Dal Grande, F. (2018). Comparing Sanger sequencing and high-throughput metabarcoding for inferring photobiont diversity in lichens. Scientific Reports, 8(1), 8624. https://doi.org/10.1038/s41598-018-26947-8.
Perdomo, D. M. X., de Castro, F. P., Durigon, V., Tavares, G. M. D., & de Medeiros, J. T. (2006). Avaliação da qualidade da água consumida na zona rural da região centro do estado do Rio Grande do Sul. Infarma - Ciências Farmaceuticas, 18(09/10), 3–6.
Peteffi, G. P., Fleck, J. D., Kael, I. M., Girardi, V., Bündchen, R., Krajeski, D. M., et al. (2018). Caffeine levels as a predictor of Human mastadenovirus presence in surface waters—A case study in the Sinos River basin—Brazil. Environmental Science and Pollution Research, 25(16), 15774–15784. https://doi.org/10.1007/s11356-018-1649-3.
PRÓ-SINOS. (n.d.). Plano Sinos - Plano de Gerenciamento da Bacia Hidrográfica do Rio dos Sinos. https://iwaponline.com/jwh/article/16/1/102/37990/Assessment-of-diversity-of-adenovirus-DNA. Accessed 16 July 2020.
Rames, E., Roiko, A., Stratton, H., & Macdonald, J. (2016). Technical aspects of using human adenovirus as a viral water quality indicator. Water Research, 96, 308–326. https://doi.org/10.1016/j.watres.2016.03.042.
Saitou, N., & Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4(4), 406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454.
Skraber, S., Gassilloud, B., Schwartzbrod, L., & Gantzer, C. (2004). Survival of infectious Poliovirus-1 in river water compared to the persistence of somatic coliphages, thermotolerant coliforms and Poliovirus-1 genome. Water Research, 38(12), 2927–2933. https://doi.org/10.1016/j.watres.2004.03.041.
Spilki, F. R., de Oliveira, L. K., Vecchia, A. D., Comerlato, J., Frezza, R., & Silva, J. V. d. S. (2009). Detecção e desinfecção de vírus em dejetos de ruminantes. Revista Conhecimento Online, 1, 1–11.
Spilki, F. R., da Luz, R. B., Fabres, R. B., Soliman, M. C., Kluge, M., Fleck, J. D., et al. (2013). Detection of human adenovirus, rotavirus and enterovirus in water samples collected on dairy farms from Tenente Portela, Northwest Of Rio Grande do Sul, Brazil. Brazilian Journal of Microbiology, 44(3), 953–957. https://doi.org/10.1590/S1517-83822013000300046.
Staggemeier, R., Bortoluzzi, M., da Silva Heck, T. M., da Luz, R. B., Fabres, R. B., Soliman, M. C., et al. (2015). Animal and human enteric viruses in water and sediment samples from dairy farms. Agricultural Water Management, 152, 135–141. https://doi.org/10.1016/j.agwat.2015.01.010.
Staggemeier, R., Heck, T. M. S., Demoliner, M., Ritzel, R. G. F., Röhnelt, N. M. S., Girardi, V., et al. (2017). Enteric viruses and adenovirus diversity in waters from 2016 Olympic venues. Science of the Total Environment, 586, 304–312. https://doi.org/10.1016/j.scitotenv.2017.01.223.
Tamura, K., Dudley, J., Nei, M., & Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Molecular Biology and Evolution, 24(8), 1596–1599. https://doi.org/10.1093/molbev/msm092.
Thurston-Enriquez, J. A., Haas, C. N., Jacangelo, J., Riley, K., & Gerba, C. P. (2002). Inactivation of Feline calicivirus and adenovirus type 40 by UV radiation. Proceedings of the Water Environment Federation, 2002(1), 502–516 https://doi.org/10.2175/193864702785033572.
UNICEF and WHO. (2015). Progress on sanitation and drinking water: 2015 update and MDG assessment. https://www.unicef.org/publications/index_82419.html. Accessed 20 May 2020.
WHO. (2017). Diarrhoeal disease. https://www.who.int/en/news-room/fact-sheets/detail/diarrhoeal-disease. Accessed 20 May 2020.
Wolf, S., Hewitt, J., & Greening, G.E. (2010). Viral multiplex quantitative PCR assays for tracking sources of fecal contamination. Applied and environmental microbiology, 76(5), 1388–1394. https://doi.org/10.1128/AEM.02249-09
Wong, K., & Xagoraraki, I. (2010). Quantitative PCR assays to survey the bovine adenovirus levels in environmental samples. Journal of Applied Microbiology, 109(2), 605–612. https://doi.org/10.1111/j.1365-2672.2010.04684.x.
Acknowledgements
F. R. Spilki is a fellow of the Brazilian National Council for Scientific and Technological Development (Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq). M. Demoliner is recipient of a master scholarship of the Brazilian Higher Education Personnel Improvement Coordination (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - CAPES). The authors are grateful for the financial and structural support from Feevale University, CAPES, CNPq, Research Support Foundation of the State of Rio Grande do Sul (FAPERGS), and Verde Sinos Project (Petrobras) and the collaboration of Empresa de Assistência Técnica e Extensão Rural do Governo do RS (EMATER/RS).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Demoliner, M., Gularte, J.S., Girardi, V. et al. Microbial Source Tracking in Small Farms: Use of Different Methods for Adenovirus Detection. Water Air Soil Pollut 232, 63 (2021). https://doi.org/10.1007/s11270-021-05011-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11270-021-05011-8