Abstract
The aim of the study was to determine the efficacy of bioaugmentation in pyrene-contaminated soil based on microbial counts, colony development index (CD), ecophysiological diversity index (EP), soil enzyme activity, and an assay of residual pyrene levels in the soil. The soil samples were contaminated with pyrene doses of 100 and 1000 mg kg−1 DM soil. Two bacterial consortia were used in the study: P1 (Bacillus frigoritolerans Z2B-19, Bacillus simplex 2–134, and Bacillus thuringiensis ex4) and P2 (Bacillus pumilus Bp-11, Bacillus safensis L22, and Bacillus aerophilus KUDC1741). The following parameters were determined: counts of organotrophic bacteria, actinobacteria, and fungi; CD; EP; and the activity of soil enzymes. The pyrene degradation efficacy of the bioaugmentation was also established. Microbiological activity was influenced by the level of soil contamination with pyrene, the test time, and the type of consortium. Pyrene had a stimulatory effect on the microbial counts and was a diversifier of CD values, EP values, and enzyme activity levels in the soil. Bioaugmentation initially promoted the growth of microorganisms, but ultimately diminished the ecophysiological diversity and the activity of soil enzymes. The microorganisms used for bioaugmentation accelerated pyrene removal from the soil, by 24.6% and 16.4% in the case of P1 and P2 consortium, respectively. The use of bioaugmentation provides favorable conditions for the effective elimination of pyrene from soil. As the microbiological and biochemical properties of the soil were improved in the initial phase of the study, this method can be recommended for the bioremediation of pyrene-contaminated soils.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Environmental pollution with polycyclic aromatic hydrocarbons (PAHs) is one of the major issues faced by the modern world. Due to the persistence, toxicity, and carcinogenicity of PAHs, it is essential that their adverse impact is reduced by removing them from the environment. In view of the risks associated with PAHs, the US Environmental Protection Agency (US EPA) has designated 16 of them as priority pollutants targeted for elimination from the environment. Polycyclic aromatic hydrocarbons bioaccumulate in food chains and thus represent a threat to humans and animals (White 2002). The largest reservoirs for PAHs are: sewage sludge (Stefaniuk et al. 2018), bottom sediments (Dvořák et al. 2017), and urban/industrial soils (Hindersmann and Achten 2018; Liu et al. 2018).
Polycyclic aromatic hydrocarbons are comprised of two or more aromatic rings bonded in linear, angular, or cluster arrangements. They are grouped into two categories, based on the number of benzene rings: LMW (low molecular weight) and HMW (high molecular weight). The first group includes hydrocarbons having three or less rings, whereas the second group contains compounds with four and more benzene rings (Chauhan et al. 2008). Hydrocarbon molecules with a higher number of rings are less biodegradable, and thus can persist longer in the environment (Cao et al. 2009; Haritash and Kaushik 2009; Seo et al. 2009). Pyrene, the compound chosen for the present study, is a model example of an HMW molecule, with a 4-ring structure similar to those of other carcinogenic PAHs (Lu et al. 2014). It is toxic to aquatic organisms and animals, causing weight loss, neurological issues, and kidney damage (Coral and Karagoz 2005; Patri et al. 2009).
Industrial and urban soils are the largest reservoirs of PAHs, especially in the layers rich in organic matter, where hydrocarbons can persist for years. Therefore, PAHs have an appreciable effect on the physicochemical properties of soil (Griffiths and Philippot 2013), while also disrupting the diversity of microorganisms living therein (Lipińska et al. 2014; Borowik et al. 2017; Borowik et al. 2018; Wyszkowska et al. 2019). The soil-dwelling microbes are considered to be one of the best indicators of soil quality, as they are very sensitive to any changes in their environment (Andreoni et al. 2004). Soil contamination with polycyclic aromatic hydrocarbons significantly changes the structure of the microbial populations within. Such contamination often reduces the diversity of the microbial populations and upsets the ecological homeostasis. Biochemical activity is a good alternative means of assessing the impact of pollutants on soil quality (Baran et al. 2004; Labud et al. 2007; Futa et al. 2017), as it constitutes a reliable indicator of soil fertility and productivity. Furthermore, biochemical activity is the driver of all biochemical processes occurring within a given medium—processes like the decomposition of organic compounds or detoxification of xenobiotics (Margesin et al. 2000).
Bioremediation is one of the most effective biological methods of removing polycyclic aromatic hydrocarbons. Among bioremediation technologies, biostimulation and bioaugmentation are the most important ones. The former is considered to be an important strategy for supporting the bioremediation process by ensuring optimal conditions for microbial growth, thus potentially reducing the level of target pollution (Koshlaf et al. 2016; Haleyur et al. 2019). Bioaugmentation involves the introduction of individual strains or consortia of microorganisms with the desired catalytic capabilities. Microorganisms in consortia possess the diverse and extensive metabolic capabilities necessary for hydrocarbon degradation (Cerqueira et al. 2011; Shen et al. 2015; Varjani and Upasani 2017). Bioaugmentation is most often used when the microorganisms necessary for bioremediation to proceed are scarce or completely absent (Venkata Mohan et al. 2006). The introduction of additional microorganisms sometimes fails to produce tangible results due to the various conditions inhibitory to microbial growth. It should also be noted that a combination of biostimulation and bioaugmentation produces the best results for PAH degradation, as confirmed by multiple studies (Sun et al. 2012; Nikolopoulou et al. 2013; Varjani 2017). Bacteria that convert organic compounds and use pyrene as a carbon source are mostly Gram-negative, and include Mycobacterium (Seo et al. 2010), Pseudomonas (Deng et al. 2010), Bacillus, Achromobacter (Gielnik et al. 2019), Ralstonia, and Sphingomonas (Cao et al. 2009; Seo et al. 2009; Nzila et al. 2018).
The aim of the present study was to determine the efficacy of bioaugmentation in pyrene-contaminated soil based on microbial counts, colony development index, ecophysiological diversity index, and soil enzyme activity. The study also examined residual pyrene levels to establish the efficacy of the different consortia for hydrocarbon degradation.
2 Methodology
2.1 Research Subject
2.1.1 Pyrene
Pyrene was used in the experiment as a model HMW species. This hydrocarbon has a 4-ring structure similar to those of other carcinogenic PAHs. Owing to its low solubility in water, pyrene is classified as a persistent organic pollutant that accumulates in the soil (Table 1). As such, it has been listed by the US Environmental Protection Agency (US EPA) as a priority pollutant targeted for remediation.
In our study, we used pyrene doses of 0, 100, and 1000 mg kg−1 DM soil, being several times higher than the acceptable levels specified in the Regulation of the Minister of the Environment in September 1, 2016 on the conduct of the assessment of contamination of the surface of the earth (Polish Journal of Laws, item 1395). The soil was spiked with weighed portions of pyrene powder. The use of such high doses of pyrene was dictated by the level of PAHs released into the soil environment in petroleum product spills from traffic accidents (Park and Park 2011). Han et al. (2018) determined the PAH levels in diesel oil-contaminated soil in the immediate aftermath of a freight ship collision in India. The examination detected levels of 17,586 mg kg−1, with the soil samples collected in 6 and 62 days after the incident containing 4854 and 4016 mg kg−1 total PAHs, respectively.
2.1.2 Bacterial Consortium
In the first stage, the soil samples were spiked with pyrene doses of 4000 mg kg−1 DM soil and kept in an incubator for 32 weeks in the dark at a temperature 25 °C and stable humidity. Afterwards, the most characteristic indigenous bacteria were isolated from the soil samples. Before plating, 1 g of soil from the control sample was weighed out and the pyrene-contaminated soil sample was suspended in sterile saline at a ratio of 1:10. Following that, serial dilutions in saline were performed, and 1-cm3 extracts of the suspensions were plated in 3 replications onto the prepared PCA and Baird-Parker media. The Petri dishes with the PCA agar were plated through deep inoculation, after which the plates were placed in an incubator and kept at 37 °C for 96 h. The Petri dishes with the Baird-Parker agar were plated using the spread plate technique and then incubated at 37 °C for 48 h. In order to obtain pure microbial colonies, the most characteristic and fast-growing bacteria were purified through successive replating on PCA and Baird-Parker agars. The most characteristic and fast-growing colonies were subsequently recovered from the pyrene-contaminated soil samples and the control Petri dishes and labeled: P1 (PCA agar isolate) and P2 (Baird-Parker agar isolate). To obtain pure strain isolates, the P1 and P2 isolates were transferred to test tubes with the corresponding media. The cultures were incubated at 37 °C for 24 h and used to identify the bacteria.
An EXTRACTME DNA BACTERIA KIT (DNA Gdańsk) was used to identify the strains. The products of 16S–23S rDNA intergenic spacer region were amplified with PCR reagents from DNA Gdańsk (Poland). Forward (AGA GTT TGA TCC TGG CTC AG) and reversed (GTG TGA CGG GCG GTG TGT AC) primers were used for amplification, resulting in a PCR product of approximately 1400 bp. After PCR, the reaction mixtures were resolved on 1.5% agarose gel for quality control of the amplified products. The PCR product was excised from the gel and purified with the EXTRACTME DNA GEL-OUT KIT. A bidirectional sequence analysis was performed on the PCR products using a 3730 XL DNA Analyzer (Life Technologies). The results were BLAST analyzed against the NCBI database.
The bioaugmentation of the soil was performed using consortia that grow best in the pyrene-contaminated soil: P1 (Bacillus frigoritolerans Z2B-19, Bacillus simplex 2–134, Bacillus thuringiensis ex4) and P2 (Bacillus pumilus Bp-11, Bacillus safensis L22, Bacillus aerophilus KUDC1741). Individual species were deposited in the NCBI database (https://www.ncbi.nlm.nih.gov/nuccore/?term=MN845147%3AMN845159%5Baccn%5D). A phylogenetic tree was constructed to plot the evolutionary relationships between strains isolated from the PAH-contaminated soil (Fig. 1). The microbial preparation was obtained by transferring the isolates onto sterile, liquid culture media (PCA and Baird-Parker). Thus, prepared consortia were kept in a Memmert Perfect incubator at 28 °C for 72 h. One cubic centimeter of the suspension contained 6 ∙ 108 cells. The consortia consisted of: P1—B. frigoritolerans Z2B-19—34%, B. simplex 2–134—33%, B. thuringiensis ex4–33%, P2–B. pumilus Bp-11—34%, B. safensis L22–33%, B. aerophilus KUDC1741—33%.
2.1.3 Soil
The study was performed with loamy sand (LS) collected from the epipedon of the soil belonging to Eutric Cambisol (IUSS Working Group WRB 2015), with a soil texture of: sand fraction (grain diameter 0.05–2 mm) —72.42%; silt fraction (grain diameter 0.002–0.05 mm)—25.31%; and clay fraction (grain diameter < 0.002 mm) —2.27%. The soil was sampled from a depth of 0–20 cm from the surface layer of a farmland field at the Teaching and Research Centre in Tomaszkowo (north-eastern Poland, 53.7167° N, 20.4167° E). Prior to the experiment, the soil was air-dried and sieved through a screen with a mesh diameter of 2 mm. The soil had the following characteristics: pHKCl—7.15; hydrolytic acidity—0.48 cmol(+) kg−1; sum of exchangeable base cations − 27.9 cmol(+) kg−1; Corg content—13.1 g kg1; Norg content—1.1 g kg−1; available P content—200 mg kg−1; available K content—147 mg kg−1; and available Mg content—27 mg kg−1.
2.2 Procedure of Setting up the Experiment
The experiment was conducted in laboratory conditions and performed in 3 replications. After drying and sieving through a sieve with mesh size of 2 mm, the loamy sand was put into glass 150-cm3 beakers in portions of 100 g. To enrich the soil with nitrogen, each of the soil samples was amended with 1 cm3 of an NH4NO3 aqueous solution at a concentration of 100 mg N kg−1 DM soil. After the soil was thoroughly mixed with ammonium nitrate, the samples were spiked with pyrene doses of 100 and 1000 mg kg−1 DM soil and thoroughly mixed again. Pyrene-free soil served as the control. The soil was then inoculated with 1 cm3 of the P1 or P2 consortium preparation (with a density of 6 ∙ 108 cells). Samples of the soil were moisturized to 50% of the capillary water capacity, and subsequently kept in an incubator in the dark at 25 °C for 2 and 8 weeks. The temperature and humidity were controlled throughout the experiment, and water losses were completed with demineralized water. The study was thus conducted in two series – with and without bioaugmentation.
2.3 Microbiological Analysis
Microbiological analysis was conducted after 2 and 8 weeks to determine the association between the level of soil contamination with PAHs and microbial counts, colony development index (CD), and ecophysiological diversity index (EP). The counts of organotrophic bacteria, actinobacteria, and fungi were determined three times for each replicate. Ten grams of soil was weighed out from each soil sample and suspended in 90 cm3 of sterile saline. The samples were excited for half an hour using laboratory shakers, and then repeatedly diluted. One cubic centimeter of the preparation was plated onto prepared and sterilized Petri dishes. The following media were used for the different groups of microorganisms: organotrophic bacteria—Bunt and Rovira medium (Bunt and Rovira 1955); actinobacteria—Küster and Williams medium (Parkinson et al. 1971); and fungi—Martin’s medium (Martin 1950). After plating, the dishes were kept in an incubator at 28 °C. The microbial counts were performed in 3 replications. Organotrophic bacteria, actinobacteria, and fungi were counted daily over a period of 10 days. The counts of microorganisms were then determined and used to compute the colony development index (CD) and the ecophysiological diversity index (EP).
The CD index was expressed using a formula by Sarathchandra et al. (1997):
where CD—colony development index; N1, N2, N3, ..., N10—number of microbial colonies developing on day 1, 2, 3, ..., 10, as a proportion of total colonies.
The ecophysiological diversity index was computed using a formula by De Leij et al. (1994):
where EP—microbial diversity index; pi—number of microbial colonies developing on the given day, divided by total colonies.
2.4 Biochemical Analysis
After 2 and 8 weeks of incubation, the soil samples were analyzed to determine the activity levels of the following enzymes: dehydrogenases, catalase, urease, acidic phosphatase, alkaline phosphatase, arylsulfatase, and β-glucosidase. The activity of each enzyme was measured in 3 replications in soil samples from each replicate. The activity of dehydrogenases was established using Öhlinger’s (1996) method, whereas a method by Alef and Nannipieri (1998) was used for the activity levels of catalase, urease, acidic and alkaline phosphatase, arylsulfatase, and β-glucosidase. A detailed methodology for determination of activities of these enzymes is provided in a paper by Borowik et al. (2017). For all enzymes—with the exception of catalase—the measurements were performed using an Aquarius CE7500 spectrophotometer by CECIL Instruments. Catalase activity was determined using the titration method.
2.5 Determination of Pyrene Content
After 8 weeks of soil incubation, the extent of microbial pyrene degradation was established by chromatographic analysis of the residual pyrene levels. The first step of the analysis was to weigh out a 10-g soil sample. An internal standard of anhydrous sodium sulphate (to dry the soil) and 9 cm3 of acetone were then added to the sample. It was then homogenized in a horizontal shaker. A two-step extraction with hexane was performed in an ultrasonic bath. The resultant extract was dried with anhydrous sodium sulphate and transferred to a borosilicate glass test tube, where it was concentrated with a concentrator to 5 cm3 at 40 °C. One cubic centimeter of the extract was sampled and filtered through a column filled with 2 g of silica gel. The aliphatic hydrocarbon fractions were initially eluted with hexane. Polycyclic aromatic hydrocarbons were extracted with dichloromethane. The collected eluates were concentrated to 1 cm3 and analyzed with a 7890A GC gas chromatograph by Agilent Technologies. The pyrene content was computed using the added internal standard (consisting of a mixture of several perdeuterated PAHs) and via calibration. The results of the chromatographic analysis were entered into the calculation sheet and recalculated to accommodate for the weighted portion, soil dry matter content, and for the blank sample.
2.6 Statistical Analysis
The results were statistically treated using the Statistica 13.1 software package (Dell Inc. 2016). The proportion of variance was calculated by applying multi-way variance analysis (ANOVA), using the η2 method. The principal component analysis (PCA) was employed to plot the activity of soil enzymes in the bioaugmented pyrene-contaminated soil. A Ward’s dendrogram was constructed to illustrate the response of microorganisms to bioaugmentation using cluster analysis. Homogenous groups of the CD and EP indices for organotrophic bacteria, actinobacteria, and fungi were calculated with ANOVA, using the Tukey test, at p = 0.01. The phylogenetic tree was constructed using the neighbor-joining (NJ) method in Clustal W software (MEGA X tool).
3 Results
3.1 The Number and Variety of Microorganisms
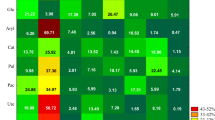
The counts of organotrophic bacteria and actinobacteria in the soil spiked with pyrene and amended with consortia were primarily determined by the pyrene dose, as reflected in the η2 percentages of the observed variability, which were 37.01% and 28.13% respectively (Fig. 2). In the control series (not amended with microorganisms), at all test times, the mean organotrophic bacteria courts increased with the pyrene levels in the soil, unlike the counts of actinobacteria and fungi, which varied (Table 2). The mean organotrophic bacteria, actinobacteria, and fungi counts in the soil samples spiked with pyrene doses of 1000 mg kg−1DM were, respectively, 138.00%, 26.62%, and 30.08% higher than in the pyrene-free samples.
The test time also had a significant effect on the counts organotrophic bacteria, actinobacteria, and fungi, accounting for 15.89%, 11.43%, and 7.39% of the variances, respectively (Fig. 2). At all pyrene doses and for both consortium types, the mean counts of organotrophic bacteria, actinobacteria, and fungi were 73%, 109%, and 57% higher in the second test time than in the first one (Table 2).
The consortium type made the least significant contribution to the variance in the organotrophic bacteria and actinobacteria counts. It had the biggest effect on the fungal counts, with a 10.20% η2 percentage of the observed variability (Fig. 2). At all test times and at all pyrene doses, soil bioaugmentation with the P2 consortium led to an 17.55% increase in the mean organotrophic bacteria count against the control (Table 2). The mean counts of actinobacteria and fungi increased by 17.10% after addition of the P2 consortium and by 29.70% after soil inoculation with P1.
The microbial response to the bioaugmentation is illustrated in a dendrogram constructed using the Ward’s method (Fig. 3). Two groups of microbes were distinguished whose response to the bioaugmentation was similar. The first group included fungi, which showed a more pronounced response to soil inoculation with supplemental consortia. The second group was represented by organotrophic bacteria and actinobacteria, which confirms their similar responses to bioaugmentation (Fig. 3).
Reaction of microorganisms on the use of bioaugmentation in soil contaminated with pyrene regardless of research term. Org, organotrophic bacteria; Act, actinomycetes; Fun, fungi; C, control (with pyrene, without consortia); P1, bacterial consortium (pyrene + Bacillus frigoritolerans Z2B-19, Bacillus simplex 2–134, Bacillus thuringiensis ex4); P2, bacterial consortium (pyrene + Bacillus pumilus Bp-11, Bacillus safensis L22, Bacillus aerophilus KUDC1741)
The colony development index values computed for organotrophic bacteria, actinobacteria, and fungi showed a large variation, and were linked with the pyrene dose, the test time, and the type of consortium (Table 3). At all test times and for all consortia types, the mean CD values were shown to decrease with the pyrene level increase in the soil. After 8 weeks of the experiment, the growth of organotrophic bacteria and actinobacteria in the control series was found to be lower than the corresponding value from the second week. At all test times and pyrene doses, soil bioaugmentation with the P2 consortium led to a slight increase in the development index values for organotrophic bacteria colonies, while soil samples inoculated with consortium P1 showed a minor increase in the CD values for fungi against the control (Fig. 4).
Average colony development indices (CD) of organotrophic bacteria (Org), actinomycetes (Act) and fungi (Fun) in soil contaminated with pyrene depending on the type of consortium. C, control (with pyrene, without consortia); P1, bacterial consortium (pyrene + Bacillus frigoritolerans Z2B-19, Bacillus simplex 2–134, Bacillus thuringiensis ex4); P2, bacterial consortium (pyrene + Bacillus pumilus Bp-11, Bacillus safensis L22, Bacillus aerophilus KUDC1741)
The ecophysiological diversity index values computed for organotrophic bacteria, actinobacteria, and fungi in the pyrene-contaminated soil were determined by the pyrene dose, the test time, and the type of consortium (Table 4). The mean EP values for all of the microorganism groups in the control series were lower after 8 weeks relative to the second week. Regardless of the pyrene dose and the test time, the addition of P1 consortium also caused a significant decrease in the mean EP values for organotrophic bacteria and actinobacteria. The EP values for these groups were 11.10% and 3.70% lower than in the non-inoculated series. After consortium P2 was added to the soil, the fungi EP values were found to increase significantly, i.e., by 7.55% against the control (Fig. 5).
Average ecophysiological diversity indices of organotrophic bacteria (Org), actinomycetes (Act), and fungi (Fun) in soil contaminated with pyren6e depending on the type of consortium. C, control (with pyrene, without consortia); P1, bacterial consortium (pyrene + Bacillus frigoritolerans Z2B-19, Bacillus simplex 2–134, Bacillus thuringiensis ex4); P2, bacterial consortium (pyrene + Bacillus pumilus Bp-11, Bacillus safensis L22, Bacillus aerophilus KUDC1741)
3.2 Biochemical Activity
The results of the present study reveal that the activity of soil enzymes exposed to pyrene and microbial consortia was varied and strongly linked with the dose of pyrene, the test time, and the type of consortium. The activity levels of dehydrogenases, catalase, acid phosphatase, and β-glucosidase were primarily determined by the test time, as reflected in the η2 percentages of the observed variability, which were 80.27%, 22.90%, 71.41%, and 49.06%, respectively. The activities of urease and arylsulfatase were primarily related to the type of consortium, whereas the activity of alkaline phosphatase correlated with the dose of pyrene (Fig. 6).
The mean activity levels of soil enzymes, presented as vectors of variables, were influenced by the dose of pyrene and the sampling time, and were illustrated in the PCA graph the form of cases (Fig. 7a). The PCA figure shows strong positive correlations between the catalase and β-glucosidase, as well as between the urease and acid phosphatase. The activity of soil enzymes was determined both by the dose of pyrene and the sampling period. At the first test time and for both consortium types, pyrene doses of 1000 mg kg−1 DM soil corresponded with the highest mean activity values for catalase and β-glucosidase, whereas the highest arylsulfatase activity was recorded in the soil samples spiked with a pyrene dose of 100 mg in kg−1 DM soil. At the same sampling period, the highest dehydrogenase activity was recorded in pyrene-free soil. At the second test time, soil contamination with a pyrene dose of 100 mg kg−1 DM soil led to the highest mean activities of acid phosphatase and alkaline phosphatase, whereas urease showed the highest response in the pyrene-free soil.
Enzymatic activity in soil contaminated with pyrene presented by PCA method regardless of the type of consortium (a) and the dose of pyrene (b); Deh, dehydrogenases; Cat, catalase; Ure, urease; Pac, acid phosphatase; Pal, alkaline phosphatase; Aryl, arylsulphatase; Glu, β-glucosidase; P, pyrene; 0, 100, 1000—doses of pyrene (mg kg−1 DM soil); 2, 8—research terms (in weeks); C, P1, P2— C—control (with pyrene, without consortia); P1, bacterial consortium (pyrene + Bacillus frigoritolerans Z2B-19, Bacillus simplex 2–134, Bacillus thuringiensis ex4); P2, bacterial consortium (pyrene + Bacillus pumilus Bp-11, Bacillus safensis L22, Bacillus aerophilus KUDC1741); Fuchsia Greek capital letter delta; blue Latin small letter “O” end of vectors; cases
The enzymatic activity was also determine by the type of consortium and the test time. Figure 7b shows the mean activities of soil enzymes when exposed to the consortia at the different test times as vectors of variance, which were presented in Fig. 7b as cases. The activity of soil enzymes was affected by soil bioaugmentation using bacterial consortia (Fig. 7b). The dehydrogenases and catalase formed the first group of enzymes based on their similar response to the soil inoculation. The highest activity of these enzymes was observed after soil inoculation with consortium P1 (for catalase) and P2 (for the dehydrogenases) during the first test time. At the second test time, the enzymes exposed to consortium P2 showed diminished activity, as reflected by the long distance between this case and the vectors of enzyme data variables observed in the figure. Urease and alkaline phosphatase formed the second group of enzymes, whose highest activity was recorded in the non-augmented soil after 8 weeks of the experiment. The addition of consortium P2 stimulated their activities in the first test time; however, both consortia were shown to inhibit their activities at the second test time. Acid phosphatase, arylsulfatase, and β-glucosidase responded differently to soil bioaugmentation. Both the P1 and P2 consortia bolstered the activity of acid phosphatase during the first test time. In contrast, during the second test time, its enhanced activity was shown only in the samples inoculated with P2. Both consortia had a stimulating effect on arylsulfatase activity after 2 and 8 weeks of soil incubation. β-Glucosidase showed suppressed activity after soil amendment with microorganisms at the first test time, but increased activity in the second period of measurement.
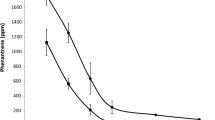
The pyrene degradation efficacy of the different consortia was determined on the basis of the detected levels of residual pyrene. The non-inoculated series demonstrates that pyrene is degraded by indigenous microorganisms in the soil. After 8 weeks of the experiment, 55.90% of the pyrene was degraded. Based on the rates of pyrene degradation after inoculation, consortium P1 proved to be the most effective pyrene degrader. Introduction of consortium P1 led to an increase in pyrene degradation from 55.90% to 80.50%. The P2 bacteria improved the degradation rate from 55.90 to 72.3%.
4 Discussion
4.1 The Number and Variety of Microorganisms
Bioaugmentation is one of the methods of bioremediation (that also includes biostimulation) wherein specific microorganisms are introduced into contaminated environments (soil, water) to increase the rate of degradation of organic pollutants, such as polycyclic aromatic hydrocarbons (Sarkar et al. 2016; Okparanma et al. 2017; Ramadass et al. 2018). Multiple experiments have shown that this method is effective in improving PAH degradation rates (Nikolopoluou et al. 2013; Varjani 2017; Borowik and Wyszkowska 2018). There are scientific reports describing the use of single strains of microorganisms in bioaugmentation of PAH-contaminated soils (Franzetti et al. 2009; Wu et al. 2016), and the use of microbial consortia for the same purpose (Abdulsalam et al. 2011; Wu et al. 2017; Rabodonirina et al. 2019). In many cases, consortia proved to be more effective than single strains, since the catabolic pathway intermediates of one strain can be further degraded by other strains possessing a suitable catabolic pathway (Heinaru et al. 2005).
The present study has shown that the application of microbial consortia isolated from PAH-contaminated soil affected the counts of organotrophic bacteria, actinobacteria, and fungi. At all test times, bioaugmentation of the soil with consortium P1 (B. frigoritolerans Z2B-19, B. simplex 2–134, B. thuringiensis ex4) promoted organotrophic bacteria growth and fungi growth, with increases of 5.08% and 29.70%, respectively (relative to the control). Inoculation of soil samples with consortium P2 (B. pumilus Bp-11, B. safensis L22, B. aerophilus KUDC1741), led to similar increases, with mean counts of organotrophic bacteria and actinobacteria being higher by, respectively, 17.55% and 17.10% than in the non-inoculated series.
A study by Wu et al. (2017) investigated the effect of bioaugmentation and biostimulation on hydrocarbon degradation and microbial activity in soil contaminated with diesel oil. The consortium used in that study consisted of: Pseudomonas stutzeri GQ-4 strain KF453954, Pseudomonas SZ-2 strain KF453956, Bacillus SQe2 strain KF453961, and Acinetobacter SZ-1 strain KF453955. After a period of 112 days, the use of bioaugmentation led to a small (5%) decrease in the total level of hydrocarbons (from the initial 61.000 mg kg−1DM soil). However, increased microbial activity was also observed. In turn, Rabodonirina et al. (2019) used a consortium of P. stutzeri, B. simplex, and B. pumilus in an experiment examining bioremediation of soils contaminated with 3-ring and 4-ring hydrocarbons. The study reported consortium efficacies of 65–86% and 86–95% for fluorene and phenanthrene degradation, respectively. HMW PAHs, such as pyrene and fluoranthene, were resistant to these microbial strains. A study by Abdulsalam et al. (2011) compared bioaugmentation and biostimulation in soil contaminated with diesel oil over a period of 40 years. A consortium of Bacillus subtilis and Pseudomonas aeruginosa was shown to increase the total heterotrophic bacteria count relative to the control. Moreover, use of microbes for soil remediation was shown to have an efficacy of 66% in reducing oil levels. According to these authors, the microbes needed approximately 10 days to adapt to the contaminated environment, and this adaptation period was followed with an increase in microbial counts. In line with the present study, just 2 weeks into the experiment, the microorganisms were able to harness the contaminants as metabolic substrates and rapidly grow, as confirmed by their increased counts. The increased counts of organotrophic bacteria, actinobacteria, and fungi in the 8th week of the study probably resulted from the degradation of organic substances by those microorganisms, which gradually released carbon from pyrene. The microorganisms can use xenobiotics as an additional source of carbon and energy by adapting to the new environmental conditions and boosting own metabolic processes (Zafra et al. 2014; Lipińska et al. 2015; Borowik and Wyszkowska 2018).
Polycyclic aromatic hydrocarbons not only affect microbial counts, but microbial diversity as well (Sun et al. 2012; Wyszkowska et al. 2015; Borowik et al. 2018). Our study showed variance in the colony development index and ecophysiological diversity index values for organotrophic bacteria, actinobacteria, and fungi. The growth of organotrophic bacteria colonies and actinobacteria colonies in the control series proved to be higher in the initial phase of the study. R strategists (fast-growing microorganisms with efficient metabolism, but susceptible to external agents) dominated in the first test time. They were later superseded by K strategists—microorganisms that adapt to contaminated environments and are specialized PAH degraders—which resulted in decreased CD values (De Leij et al. 1994). The reverse was observed for fungal colonies, which exhibited higher CD values during the second test time.
The mean EP values computed for all of the microorganism groups were lower after 8 weeks relative to the first test time. The variability of EP values in our study shows the impact of pyrene on the microbial structure. The ability of microorganisms to degrade hydrocarbons and adapt to function in an unfavorable environment is determined by, i.e., the properties and functions of microorganisms in the soil or the severity of the environmental stress (Wang et al. 2011). Ecophysiological diversity index (EP) describes how steady the microbial growth is over a given period of time. EP values can range from 0 to 1—the higher the value, the steadier the growth. Over the course of the study, the mean EP values computed for organotrophic bacteria, actinobacteria, and fungi decreased. The fast-growing microorganisms were replaced over time by the microorganisms that adapted to the pyrene contamination.
4.2 Biochemical Activity
Soil contamination with polycyclic aromatic hydrocarbons upsets soil homeostasis (Nannipieri et al. 2018; Borowik et al. 2019). One of the better ways to assess soil quality is to monitor the activity of soil enzymes, which are responsible for soil metabolism (which, in turn, is closely linked to nutrient activation and degradation of contaminants such as PAHs) (Lipińska et al. 2015; Borowik et al. 2017; Lipińska et al. 2019). Our study proved that various soil enzymes reacted differently to individual pyrene doses across the test times. The highest activity levels for dehydrogenases and urease were observed in pyrene-free soil at the first and second test times, respectively. Among the enzymes, dehydrogenases and urease are the least resistant to external agents (Lipińska et al. 2014; Lipińska et al. 2015; Wyszkowska et al. 2015), a susceptibility that usually manifests in lower activity levels after exposure to PAHs. The stimulating effect of pyrene doses of 100 mg kg−1 DM soil was the most pronounced for arylsulfatase activity in the second week of the study, and for acid/alkaline phosphatases in the eighth week of the study. By comparison, a pyrene dose of 1000 mg kg−1 DM soil showed the strongest stimulating effect with regard to catalase activity and β-glucosidase activity during the first test time. Pyrene is a HMW polycyclic aromatic hydrocarbon. Hydrocarbons with a higher number of rings are less biodegradable (Seo et al. 2009). Pyrene stimulates the activity of catalase, acid phosphatase, alkaline phosphatase, arylsulfatase, and β-glucosidase due to its degradation by microorganisms, which can utilize it as a source of carbon and energy (Wyszkowska and Wyszkowski 2010).
Our study showed that the activity of dehydrogenases, catalase, acid phosphatase, alkaline phosphatase, and arylsulfatase increased in the bioaugmented soil during the first test time. Conversely, the activity of urease and β-glucosidase decreased against the control. Dehydrogenases are the most sensitive among these enzymes, and thus indicate the presence of living and intact microbial cells (Andreoni et al. 2004). Our experiment showed that the activity of the enzymes increased in the first test time (the second week), and that the highest levels of produced TFF (25% more than the control) were recorded in the soil samples bioaugmented with consortium P2. A study by Patel et al. (2013) also noted increased activity of dehydrogenases due to soil amendment with a consortium of six strains: Bacillus sp. ASP1, Pseudomonas sp. ASP2, Stenotrophomonas maltophilia ASP3, Staphylococcus sp. ASP4, Geobacillus sp. ASP5, and Alcaligenes sp. ASP6. After 8 weeks of our experiment, decreased dehydrogenase activity was observed in the pyrene-spiked samples subjected to bioaugmentation. Dehydrogenases were not the only enzymes whose activity diminished due to bioaugmentation at the second test time—the same held true for catalase, urease, acid, and alkaline phosphatase. Given that the activity of those enzymes was higher in the control (non-inoculated) samples than in the inoculated batch, it is fair to assume that the main cause of this situation was unfavorable antagonistic interactions between indigenous microbes and the strains introduced into the soil (Yu et al. 2005).
The reduction in pyrene content by 55.90% in the control samples (not amended with consortia), observed after 8 weeks of soil incubation in our study, demonstrates that the microorganisms gradually utilized the hydrocarbons as part of the biodegradation processes, and adapted to the contaminated environment. A study by Macleod and Semple (2006) examined the capacity of microorganisms to mineralize pyrene after 4, 8, and 12 weeks. They found that the rate of pyrene degradation increased after 8 weeks of the experiment. The increased pyrene concentration may have contributed to the adaptation of microbial populations to degrade a given hydrocarbon at a given time. Another study, by Towell et al. (2011), demonstrated the microbial capacity to actively and quickly degrade aromatic hydrocarbons. Enzyme induction, genetic changes, and selective enrichment were the basic mechanisms used by microorganisms to adapt to organic pollutants. Among the genetic factors involved in microbial adaptation, gene amplification is the most important. This mechanism modifies the metabolism of microorganisms exposed to contaminants through gene enrichment and transfer (Leahy and Colwell 1990).
Based on the levels of PAHs in the soil, the consortium P1 (B. frigoritolerans Z2B-19, B. simplex 2–134, B. thuringiensis ex4) proved to be more effective in pyrene degradation, as its degradation rate increased from 55.90 to 80.5% in soil samples inoculated with these bacteria. Bacillus species are used for bioaugmentation of PAH-contaminated soils (Toledo et al. 2006; Seo et al. 2009; Maiti et al. 2012; Poi et al. 2017). As reported by Roy et al. (2018), bioaugmentation of soil with native Bacillus strains in tandem with biostimulation led to a decrease in the content of petroleum-based hydrocarbons by 57–75%. Oualha et al. (2019) showed that Bacillus sonorensis D1 played a crucial role in the process of PAH biodegradation. With optimum conditions established (a C:N:P ratio of 100:10:1, temperature of 37 °C, humidity of 10%, Tween 80 surfactant present), degradation of 32.4% of polycyclic aromatic hydrocarbons was recorded after 160 days of the experiment.
Although the use of bioaugmentation usually leads to a decrease in soil hydrocarbon levels, it has been reported by some authors (Yu et al. 2005) that the addition of non-indigenous microorganisms may inhibit PAH degradation. The reason for that are the competitive interactions that may emerge between indigenous and non-indigenous microbial communities.
5 Conclusions
Soil bioaugmentation with consortia isolated from pyrene-contaminated soil samples had an effect on counts of organotrophic bacteria, actinobacteria, and fungi; the colony development index (CD); the ecophysiological diversity index (EP); and the activity of soil enzymes. Initially, it had a positive effect on the growth of organotrophic bacteria, actinobacteria, and fungi, but ultimately diminished microbial growth, diversity, and the activity of soil enzymes. The microorganisms used for bioaugmentation accelerated pyrene removal from the soil. Consortia P1 and P2 led to accelerated pyrene degradation by 24.6% and 16.4% respectively. In general, the use of bioaugmentation provides favorable conditions for the effective elimination of pyrene from soil. This indicates that bacteria isolated from pyrene-contaminated soil are effective pyrene degraders. Bioaugmentation also has a beneficial effect on microbiological and biochemical properties of the soil, especially if recently contaminated.
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Abdulsalam, S., Bugaje, I. M., Adefila, S. S., & Ibrahim, S. (2011). Comparison of biostimulation and bioaugmentation for remediation of soil contaminated with spent motor oil. International journal of Environmental Science and Technology, 8(1), 187–194. https://doi.org/10.1007/BF03326208.
Alef, K., & Nannipieri, P. (1998). Methods in applied soil microbiology and biochemistry (pp. 316–320). London: Harcourt Brace & Company, Publishers.
Andreoni, V., Cavalca, L., Rao, M. A., Nocerino, G., Bernasconi, S., Dell'Amico, E., Colombo, M., & Gianfreda, L. (2004). Bacterial communities and enzyme activities of PAHs polluted soils. Chemosphere, 57, 401–412. https://doi.org/10.1016/j.chemosphere.2004.06.013.
Baran, S., Bielińska, J. E., & Oleszczuk, P. (2004). Enzymatic activity in an airfield soil polluted with polycyclic aromatic hydrocarbons. Geoderma, 118, 221–232. https://doi.org/10.1016/S0016-7061(03)00205-2.
Borowik, A., & Wyszkowska, J. (2018). Bioaugmentation of soil contaminated with diesel oil. Journal of Elementology, 23(4), 1161–1178. https://doi.org/10.5601/jelem.2018.23.1.1627.
Borowik, A., Wyszkowska, J., & Wyszkowski, M. (2017). Resistance of aerobic microorganisms and soil enzyme response to soil contamination with Ekodiesel Ultra fuel. Environmental Science and Pollution Research, 24(31), 24346–24363. https://doi.org/10.1007/s11356-017-0076-1.
Borowik, A., Wyszkowska, J., & Oszust, K. (2018). Changes in the functional diversity of bacterial communities in soil contaminated with diesel oil. Journal of Elementology, 23(3), 1099–1117. https://doi.org/10.5601/jelem.2018.23.1.1603.
Borowik, A., Wyszkowska, J., Gałązka, A., & Kucharski, J. (2019). Role of Festuca rubra and of the enzymatic activity in the soil polluted with diesel oil. Environmental Science and Pollution Research, 26, 27739–27751. https://doi.org/10.1007/s11356-019-05883-3.
Bunt, Y. S., & Rovira, A. D. (1955). Microbiological studies of some subarctic soils. Journal of Soil Science, 6, 119–128. https://doi.org/10.1111/j.1365-2389.1955.tb00836.x.
Cao, B., Nagarajan, K., & Loh, K. C. (2009). Biodegradation of aromatic compounds: current status and opportunities for biomolecular approaches. Applied Microbiology and Biotechnology, 85, 207–228. https://doi.org/10.1007/s00253-009-2192-4.
Cerqueira, V. S., Hollenbach, E. B., Maboni, F., Vainstein, M., Camargo, F., Do-Carmo, R. P. M., & Bento, F. M. (2011). Biodegradation potential of oily sludge by pure and mixed bacterial cultures. Bioresource Technology, 102(23), 11003–11010. https://doi.org/10.1016/j.biortech.2011.09.074.
Chauhan, A., Fazlurrahman Oakeshott, J. G., & Jain, R. K. (2008). Bacterial metabolism of polycyclic aromatic hydrocarbons: strategies for bioremediation. Indian Journal of Medical Microbiology, 48, 95–113. https://doi.org/10.1007/s12088-008-0010-9.
Coral, G., & Karagoz, S. (2005). Isolation and characterization of phenanthrene-degrading bacteria from a petroleum refinery soil. Annales de Microbiologie, 55, 255–259.
De Leij, F. A. A. M., Whipps, J. M., & Lynch, J. M. (1994). The use of colony development for the characterization of bacterial communities in soil and roots. Microbial Ecology, 27, 81–97. https://doi.org/10.1007/BF00170116.
Dell Inc. 2016. Dell Statistica (data analysis software system), version 13. software.dell.com.
Deng, Y., Zhang, Y., HeshamAel, L., Liu, R., & Yang, M. (2010). Cell surface properties of five polycyclic aromatic compound-degrading yeast strains. Applied Microbiology and Biotechnology, 86(6), 1933–1939. https://doi.org/10.1007/s00253-010-2477-7.
Dvořák, T., Száková, J., Vondráčková, S., Košnář, Z., Holečková, Z., & Najmanová, J. (2017). Content of inorganic and organic pollutants and their mobility in bottom sediment from the Orlík water reservoir (Vltava river, Czech Republic). Soil and Sediment Contamination, 26(6), 00–00. https://doi.org/10.1080/15320383.2017.1364222.
Franzetti, A., Caredda, P., Ruggeri, C., La Colla, L., Tamburini, E., Papacchini, M., & Bestetti, G. (2009). Potential application of surface active compounds by Gordonia sp. strain BS29 in soil remediation technologies. Chemosphere, 75, 801–807. https://doi.org/10.1016/j.chemosphere.2008.12.052.
Futa B, Bielińska EJ, Ligęza S, Chmielewski S, Wesołowska S, Patkowski K, Mocek-Płóciniak A (2017) Enzymatic activity and content of polycyclic aromatic hydrocarbons (PAHs) in soils under low-stack emission in Lublin. Pol J Soil Sci 50(1):63–74. doi:https://doi.org/10.17951/pjss/2017.50.1.63
Gielnik A, Pechaud Y, Huguenot D, Cébron A, Esposito G, D van Hullebusch E (2019) Bacterial seeding potential of digestate in bioremediation of diesel contaminated soil. Int Biodeter Biodegrad 143:104715. https://doi.org/10.1016/j.ibiod.2019.06.003.
Griffiths, B. S., & Philippot, L. (2013). Insights into the resistance and resilience of the soil microbial community. FEMS Microbiology Reviews, 37(2), 112–129. https://doi.org/10.1111/j.1574-6976.2012.00343.
Haleyur, N., Shahsavari, E., Jain, S. S., Koshlaf, E., Ravindran, V. B., Morrison, P. D., Osborn, A. M., & Ball, A. S. (2019). Influence of bioaugmentation and biostimulation on PAH degradation in aged contaminated soils: response and dynamics of the bacterial community. Journal of Environmental Management, 238, 49–58. https://doi.org/10.1016/j.jenvman.2019.02.115.
Han, Y., Nambi, I. M., & Clement, T. P. (2018). Environmental impacts of the Chennai oil spill accident – a case study. Sci Total Environ, 626, 795–806. https://doi.org/10.1016/j.scitotenv.2018.01.128.
Haritash, A. K., & Kaushik, C. P. (2009). Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. Journal of Hazardous Materials, 169, 1–15. https://doi.org/10.1016/j.jhazmat.2009.03.137.
Heinaru, E., Merimaa, M., Viggor, S., Lehiste, M., Leito, I., Truu, J., & Heinaru, A. (2005). Biodegradation efficiency of functionally important populations selected for bioaugmentation in phenol and oil-polluted area. FEMS Microbiology Ecology, 51, 363–373. https://doi.org/10.1016/j.femsec.2004.09.009.
Hindersmann B, Achten Ch (2018) Urban soils impacted by tailings from coal mining: PAH source identification by 59 PAHs, BPCA and alkylated PAHs. Environ Pollut 242 (Pt B):1217-1225. https://doi.org/10.1016/j.envpol.2018.08.014.
IUSS Working Group WRB, 2015. World reference base for soil resources 2014, update 2015. International soil classification system for naming soils and creating legends for soil maps. FAO, Rome, p. 182.World soil resources reports no. 106.
Koshlaf, E., Shahsavari, E., Aburto-Medina, A., Taha, M., Haleyur, N., Makadia, T. H., Morrison, P. D., & Ball, A. S. (2016). Bioremediation potential of diesel-contaminated Libyan soil. Exotoxicol Environ Safe, 133, 297–305. https://doi.org/10.1016/j.ecoenv.2016.07.027.
Labud, V., García, C., & Hernández, T. (2007). Effect of hydrocarbon pollution on the microbial properties of a sandy and a clay soil. Chemosphere, 66, 1863–1871. https://doi.org/10.1016/j.chemosphere.2006.08.021.
Leahy, J. G., & Colwell, R. R. (1990). Microbial degradation of hydrocarbons in the environment. Microbiological Reviews, 54(3), 305–315.
Lipińska, A., Kucharski, J., & Wyszkowska, J. (2014). The effect of polycyclic aromatic hydrocarbons on the structure of organotrophic bacteria and dehydrogenase activity in soil. Polycycl Aromat Comp, 34, 35–53. https://doi.org/10.1080/10406638.2013.844175.
Lipińska, A., Wyszkowska, J., & Kucharski, J. (2015). Diversity of organotrophic bacteria, activity of dehydrogenases and urease as well as seed germination and root growth Lepidium sativum, Sorghum saccharatum and Sinapis alba under the influence of polycyclic aromatic hydrocarbons. Environmental Science and Pollution Research, 22(23), 18519–18530. https://doi.org/10.1007/s11356-015-5329-2.
Lipińska, A., Kucharski, J., & Wyszkowska, J. (2019). Activity of phosphatases in soil contaminated with PAHs. Water, Air, and Soil Pollution, 230, 298. https://doi.org/10.1007/s11270-019-4344-1.
Liu, Y., Ga, P., Su, J., da Silva, E. B., de Oliveirac, L. M., Townsend, T., Xiang, P., & Ma, L. Q. (2018). PAHs in urban soils of two Florida cities: Background concentrations, distribution, and sources. Chemosphere, 214, 220–227. https://doi.org/10.1016/j.chemosphere.2018.09.119.
Lu, J., Guo, C., Zhang, M., Lu, G., & Dang, Z. (2014). Biodegradation of single pyrene and mixtures of pyrene by a fusant bacterial strain F14. Int Biodeter Biodegrad, 87, 75–80. https://doi.org/10.1016/j.ibiod.2013.11.004.
Macleod, C. J. A., & Semple, K. T. (2006). The influence of single and multiple applications of pyrene on the evolution of pyrene catabolism in soil. Environmental Pollution, 139, 455–460. https://doi.org/10.1016/j.envpol.2005.06.014.
Maiti, A., Das, S., & Bhattacharrya, N. (2012). Bioremediation of high molecular weight polycyclic aromatic hydrocarbons by Bacillus thuringiensis strain NA2. J Sci, 1(4), 72–75.
Margesin, R., Zimmerbauer, A., & Schinner, F. (2000). Monitoring of bioremediation by soil biological activities. Chemosphere, 40, 339–346. https://doi.org/10.1016/s0045-6535(99)00218-0.
Martin, J. (1950). Use of acid rose bengal and streptomycin in the plate method for estimating soil fungi. Soil Science, 69, 215–233.
Nannipieri, P., Trasar-Cepeda, C., & Dick, R. P. (2018). Soil enzyme activity: a brief story and biochemistry as a basis for appropriate interpretations and meta-analysis. Biol Fert Soils, 54, 11–19. https://doi.org/10.1007/s00374-017-1245-6.
Nikolopoulou, M., Pasadakis, N., & Kalogerakis, N. (2013). Evaluation of autochthonous bioaugmentation and biostimulation during microcosm-simulated oil spills. Marine Pollution Bulletin, 72(1), 165–173. https://doi.org/10.1016/j.marpolbul.2013.04.007.
Nzila, A., Ortega Ramirez, C., Musa, M. M., Sankara, S., & BasheerCh, L. Q. X. (2018). Pyrene biodegradation and proteomic analysis in Achromobacter xylosoxidans, PY4 strain. Int Biodeter Biodegrad, 130, 30–47. https://doi.org/10.1016/j.ibiod.2018.03.014.
Öhlinger R (1996) Dehydrogenase activity with the substrate TTC in: Methods in Soil Biology. Schinner F., Öhlinger R., Kandeler E., Margesin R. (eds), Spronger Verlag Berlin Heidelberg: 241-243.
Okparanma, R. N., Azuazu, I., & Ayotamuno, J. M. (2017). Assessment of the effectiveness of onsite exsitu remediation by enhanced natural attenuation in the Niger Delta region. Journal of Environmental Management, 204, 291–299. https://doi.org/10.1016/j.jenvman.2017.09.005.
Oualha, M., Al-Kaabi, N., Al-Ghouti, M., & Zouari, N. (2019). Identification and overcome of limitations of weathered oil hydrocarbons bioremediation by an adapted Bacillus sorensis strain. Journal of Environmental Management, 250, 109455. https://doi.org/10.1016/j.jenvman.2019.109455.
Park, I. S., & Park, J. W. (2011). Determination of a risk management primer at petroleum-contaminant sites: developing new human health risk assessment strategy. Journal of Hazardous Materials, 185(2–3), 1374–1380. https://doi.org/10.1016/j.jhazmat.2010.10.058.
Parkinson D, Gray FRG, Williams ST (1971) Methods for studying the ecology of soil microorganism. Blackwell Scientific Publication, Oxford, IBP Handbook 19.
Patel, V., Patel, J., & Madamwar, D. (2013). Biodegradation of phenanthrene in bioaugmented microcosm by consortium ASP developed from coastal sediment of Alang-Sosiya ship breaking yard. Marine Pollution Bulletin, 74(1), 199–2007. https://doi.org/10.1016/j.marpolbul.2013.07.001.
Patri, M., Padmini, A., & Babu, P. P. (2009). Polycyclic aromatic hydrocarbons in air and their neurotoxic potency in association with oxidative stress: a brief perspective. Annals of Neurosciences, 16(1), 22–30. https://doi.org/10.5214/ans.0972.7531.2009.160109.
Poi, G., Aburto-Medina, A., ChumMok, P., Ball, A. S., & Shahsavari, E. (2017). Large scale bioaugmentation of soil contaminated with petroleum hydrocarbons using a mixed microbial consortium. Ecological Engineering, 102, 64–71. https://doi.org/10.1016/j.ecoleng.2017.01.048.
Regulation of the Minister of Environment of 1 September 2016 for assessment of soil surface contamination (Polish Journal of Laws of 2016, item 1395).
Rabodonirina, S., Rasolomampianina, R., Krier, F., Drider, D., Merhaby, D., Net, S., & Ouddane, B. (2019). Degradation of floorene and phenanthrene in PAHs-contaminated soil using Pseudomonas and Bacillus strains isolated from oil spill sites. Journal of Environmental Management, 232, 1–7. https://doi.org/10.1016/j.jenvman.2018.11.005.
Ramadass, K., Megharaj, M., Venkateswarlu, K., & Naidu, R. (2018). Bioavailability of weathered hydrocarbons in engine oil-contaminated soil: impact of bioaugmentation mediated by Pseudomonas spp. on bioremediation. Sci Total Environ, 636, 968–974. https://doi.org/10.1016/j.scitotenv.2018.04.379.
Roy, A., Dutta, A., Pal, S., Gupta, A., Sarkar, J., Chatterjee, A., Saha, A., Sarkar, P., Sar, P., & Kazy, S. K. (2018). Biostimulation and bioaugmentation of native microbial community accelerated bioremediation of oil refinery sludge. Bioresource Technology, 253, 22–32. https://doi.org/10.1016/j.biortech.2018.01.004.
Sarathchandra, S. U., Burch, G., & Cox, N. R. (1997). Growth patterns of bacterial communities in the rhizoplane and rhizosphere of white clover (Trifolium repens L.) and perennial ryegrass (Lolium perenne L.) in long-term pasture. Applied Soil Ecology, 6, 293–299. https://doi.org/10.1016/S0929-1393(97)00015-2.
Sarkar, J., Kazy, S. K., Gupta, A., Dutta, A., Mohapatra, B., Roy, A., Bera, P., Mitra, A., & Sar, P. (2016). Biostimulation of indigenous microbial community for bioremediation of petroleum refinery sludge. Frontiers in Microbiology, 7(1407), 1–20. https://doi.org/10.3389/fmicb.2016.01407.
Seo, J. S., Keum, Y. S., & Li, Q. X. (2009). Bacterial degradation of aromatic compounds. International Journal of Environmental Research and Public Health, 6, 278–309. https://doi.org/10.3390/ijerph6010278.
Seo, J. S., Keum, Y. S., Kim, K., & Li, Q. X. (2010). Degradation of pyrene by Mycobacterium aromativorans strain JS19b1. Journal of Korean Society for Applied Biological Chemistry, 53, 323–329.
Shen, T., Pi, Y., Bao, M., Xu, N., Li, Y., & Lu, J. (2015). Biodegradation of different petroleum hydrocarbons by free and immobilized microbial consortia. Environ Sci: Process Impacts, 17, 2022–2033. https://doi.org/10.1039/c5em00318k.
Stefaniuk, M., Tsang, D. C. W., Yong, S. O., & Oleszczuk, P. (2018). A field study of bioavailable polycyclic aromatic hydrocarbons (PAHs) in sewage sludge and biochar amended soils. Journal of Hazardous Materials, 349, 29–34. https://doi.org/10.1016/j.jhazmat.2018.01.045.
Sun, G. D., Xu, Y., Jin, J. H., Zhong, Z. P., Liu, Y., Luo, M., & Liu, Z. P. (2012). Pilot scale ex-situ bioremediation of heavily PAHs-contaminated soil by indigenous microorganisms and bioaugmentation by a PAHs-degrading and bioemulsifier-producing strain. Journal of Hazardous Materials, 233-234, 72–78. https://doi.org/10.1016/j.jhazmat.2012.06.060.
Toledo, F. L., Calvo, C., Rodelas, B., & González-López, J. (2006). Selection and identification of bacteria isolated from waste crude oil with polycyclic aromatic hydrocarbons removal capacities. Systematic and Applied Microbiology, 29, 244–252. https://doi.org/10.1016/j.syapm.2005.09.003.
Towell, M. G., Bellarby, J., Paton, G. I., Coulon, F., Pollard, S. J. T., & Semple, K. T. (2011). Mineralisation of target hydrocarbons in three contaminated soils from former refinery facilities. Environmental Pollution, 159, 515–523. https://doi.org/10.1016/j.envpol.2010.10.015.
Varjani, S. J. (2017). Microbial degradation of petroleum hydrocarbons. Bioresource Technology, 23, 277–286. https://doi.org/10.1016/j.biortech.2016.10.037.
Varjani, S. J., & Upasani, V. N. (2017). A new look on factors affecting microbial degradation status and opportunities for biomolecular approaches. Applied Microbiology and Biotechnology, 85, 207–228. https://doi.org/10.1007/s00253-009-2192-4.
Venkata Mohan, S., Kisa, T., Ohkuma, T., Kanaly, R. A., & Shimizu, Y. (2006). Bioremediation technologies for treatment of PAH-contaminated soil and strategies to enhance process efficiency. Reviews in Environmental Science and Biotechnology, 5, 347–374. https://doi.org/10.1007/s11157-006-0004-1.
Wang, Q. F., Zhang, S. Y., Zou, L., & Xie, S. G. (2011). Impact of athracene addition on microbial community structure in soil microcosms from contaminated and uncontaminated sites. Biomedical and Environmental Sciences, 24(5), 543–549. https://doi.org/10.3967/0895-3988.2011.05.014.
White, P. A. (2002). The genotoxicity of priority polycyclic aromatic hydrocarbons in complex mixtures. Mut Res, 515(1–2), 85–98. https://doi.org/10.1016/S1383-5718(02)00017-7.
Wu, M., Dick, W. A., Li, W., Wang, X., Yang, Q., Wang, T., Xu, L., Zhang, M., & Chen, L. (2016). Bioaugmentation and biostimulation of hydrocarbon degradation and the microbial community in a petroleum-contaminated soil. International Biodeterioration & Biodegradation, 107, 158–164. https://doi.org/10.1016/j.ibiod.2015.11.019.
Wu, M., Ye, X., Chen, K., Li, W., Yuan, J., & Jiang, X. (2017). Bacterial community shift and hydrocarbon transformation during bioremediation of short-term petroleum-contaminated soil. Environmental Pollution, 223, 657–664. https://doi.org/10.1016/j.envpol.2017.01.079.
Wyszkowska, J., & Wyszkowski, M. (2010). Activity of soil dehydrogenases, urease and acid and alkaline phosphatases in soil polluted with petroleum. Journal of Toxicology and Environmental Health, 73, 1202–1210. https://doi.org/10.1080/15287394.2010.492004.
Wyszkowska J, Borowik A, Kucharski J (2015) Response of Avena sativa, microorganisms and enzymes to contamination of soil with diesel oil. Plant Soil Environ 61:483–488. https://doi.org/10.17221/463/2015-PSE.
Wyszkowska J, Borowik A, Olszewski J, Kucharski J (2019) Soil bacterial community and soil enzyme activity depending on the cultivation of Triticum aestivum, Brassica napus, and Pisum sativum spp. arvense. Diversity, 11(12):246. doi:https://doi.org/10.3390/d11120246.
Yu, K. S. H., Wong, A. H. Y., Yau, K. W. Y., Wong, Y. S., & Tam, N. F. Y. (2005). Natural attenuation, biostimulation and bioaugmentation on biodegradation of polycyclic aromatic hydrocarbons (PAHs) in mangrove sediments. Marine Pollution Bulletin, 51(8–12), 1071–1077. https://doi.org/10.1016/j.marpolbul.2005.06.006.
Zafra, G., Absalón, Á. E., Del Carmen Cuevas, M., & Cortés-Espinosa, D. V. (2014). Isolation and selection of a highly tolerant microbial consortium with potential for PAH biodegradation from heavy crude oil-contaminated soils. Water, Air Soil Pollut 225(2):1826. https://doi.org/10.1007/s11270-013-1826-4. https://www.ncbi.nlm.nih.gov/nuccore/?term=MN845147%3AMN845159%5Baccn%5D.
Funding
This paper was financed by the University of Warmia and Mazury in Olsztyn (Poland) as part of the topic No. 20.610.014-110. Project financially supported by Minister of Science and Higher Education in the range of the program entitled “Regional Initiative of Excellence” for the years 2019–2022, Project No. 010/RID/2018/19, amount of funding 12.000.000 PLN.”
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by AL, JW, and JK. The first draft of the manuscript was written by AL with the help of JW and JK; and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lipińska, A., Wyszkowska, J. & Kucharski, J. Microbiological and Biochemical Activity in Soil Contaminated with Pyrene Subjected to Bioaugmentation. Water Air Soil Pollut 232, 45 (2021). https://doi.org/10.1007/s11270-020-04950-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11270-020-04950-y