Abstract
Tetrachlorobisphenol A (TCBPA) is a widely used flame retardant and a potential endocrine disruptor. We estimated the role of the microbial community in degradation of TCBPA in river sediment from the vicinity of an E-waste processing facility. The effects of different anaerobic conditions on degradation efficiency of TCBPA were investigated, and differences in bacterial communities among these conditions were analyzed by 16S ribosomal RNA (rRNA) gene sequencing. The most effective dechlorination of TCBPA occurred under methanogenic conditions followed by electron donor-enhanced conditions and sulfate-reducing conditions with initial sulfate concentrations of 1, 10, and 20 mM. The extent of TCBPA removal under these conditions mentioned above was 65, 44, 43, 23, and 23%, respectively. 16S rRNA gene sequence analysis indicated that five dominant genera in the phylum Chloroflexi and another five species of Bacteroidetes, Chlorobi, and Firmicutes in these five systems were largely involved in TCBPA dechlorination. The initial sample had a total relative abundance of autochtonous potential dechlorinating bacteria of 12%. After 160 days, these values increased to 29–43% under above conditions. Addition of TCBPA decreased bacterial diversity. Efficiency of TCBPA degradation depends on the abundance and metabolism of dechlorinating bacterial guilds. The effectiveness of dechlorinating microbes in degradation of TCBPA was reduced by high sulfate concentrations.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
TCBPA is widely used as a flame retardant additive in polymers, paints, epoxy and polycarbonate resins, high impact polystyrene, phenolic resins, and adhesives used in synthetic textiles, building materials, epoxy resin electronic circuit boards, and other electronic equipments (Horikoshi et al. 2008; Kalasekar et al. 2015). During the production, processing, and life cycles of these products, free and combined TCBPA can be released into the environment. Electronic wastes (e-wastes), especially printed circuit boards, contain several hazardous materials like halogenated flame retardants, metals and alloy compounds, and polyvinyl chloride (Tsydenova and Bengtsson 2011). Acid leaching is the main method of extraction of valuable metals from e-wastes, and this is a major source of flame retardants in e-waste disposal areas (Leung et al. 2007). TCBPA is a chemically stable and strongly hydrophobic compound. It easily absorbs to the particulate matter, sediment, and aquatic organisms and residues persist in the environment. TCBPA has been detected in many environmental matrices such as coastal lagoons (Casatta et al. 2015), influent and effluent of sewage treatment plants (Yang et al. 2014), effluent of wastepaper recycling plants (Fukazawa et al. 2001; Gallart-Ayala et al. 2010), river water (Blanco et al. 2005), drinking water (Fan et al. 2013), sewage sludge (Chu et al. 2005; Song et al. 2014), sediment (Chu et al. 2005), and agricultural and industrial soil (Sánchez-Brunete et al. 2009). TCBPA is chemically similar to the thyroid hormone, so it is a potential endocrine disruptor in humans and wildlife (Ruan et al. 2015; Terasaki et al. 2011).
Efficient degradation of TCBPA in the environment is the key to eliminating its biological hazards. Removal of TCBPA has focused on photochemical transformation (Eriksson et al. 2004; Horikoshi et al. 2008), chemical oxidation (Bastos et al. 2008), and microbial degradation (Liu et al. 2014; Voordeckers et al. 2002; Yuan et al. 2011). The anaerobic degradation rate of TCBPA in sediment of the Erren River was enhanced by adding yeast extract, cellulose, sodium chloride, non-ionic detergents (Brij 30 and Brij 35), rhamnolipid, or surfactin. Anaerobic degradation was inhibited by addition of phthalic esters, nonylphenol, and heavy metals (Yuan et al. 2011). Bacillus megaterium and Pseudomonas putida possessed the best biodegradation potential (Yuan et al. 2010). The dehalogenation rate of TCBPA under methanogenic conditions was higher than that of sulfate reducing conditions (Liu et al. 2014; Voordeckers et al. 2002) which also was considered to promoting dechlorination. In addition, co-amendment of sediment with 2, 6-dichlorophenol can shorten the lag time of TCBPA dechlorination (Voordeckers et al. 2002).
Sediments and soils near e-waste recycling sites are often polluted by TCBPA. The study of TCBPA biodegradation by indigenous microorganism in sediment/soil has long-term sustained practical significance for in situ remediation of field polluted by this contaminant. However, current research on TCBPA is mainly focused on the level of environmental detection and its toxicological properties. Few publications have reported bioremediation methods for TCBPA contaminated areas and the TCBPA-degrading bacteria involved in this process. Variation of the bacterial communities in different anaerobic systems of TCBPA degradation has also been overlooked. Longtang town is a typical e-waste polluted area with 40-year history of e-waste recycling where heavily polluted by halogenated flame retardants, heavy metals, and high concentration sulfate in the soil/sediment (Lin et al. 2015). And there is controversy existed in the effects of sulfate and its content on dehalogenation (Liu et al. 2014; Panagiotakis et al. 2014). Some studies found that dechlorination capacity of sulfate reducing bacteria (SRB) and sulfate reducing condition were provided with applicable oxidation reduction potential for dechlorination (Liu et al. 2014). Nonetheless, SRB were more compentitive for electron donor than dehalogenating bacteria which had a better dechlorination performance under low concentration sulfate condition (Panagiotakis et al. 2014). Therefore, we studied (a) the TCBPA degradation ability of Longtang River sediment microorganisms under electron donor enhanced conditions, methanogenic, and sulfate reducing conditions (b) the effect of different initial sulfate concentrations on TCBPA degradation, and (c) the bacterial community using 16S ribosomal RNA (rRNA) gene sequencing. Our results provide a basis for in situ remediation of TCBPA or other halogenated organic compounds contaminating soil/sediment.
2 Materials and Methods
Chemicals
TCBPA was purchased from J&K Scientific (Beijing, China) and was >98% pure. The reagents for medium preparation were purchased from Aladdin Industrial Inc. (Shanghai, China). The solvents for extractions and high-performance liquid chromatography (HPLC) analyses were purchased from ANPEL Laboratory Technologies Inc. (Shanghai, China). The materials for molecular biology experiments were purchased from Sangon Biotech (Shanghai, China). Individual stock solutions of TCBPA dissolved in dichloromethane were made at 10 mM and then diluted to desired concentrations before use.
Sediment
Surface (0–15 cm) and deep (>15 cm) sediment samples were collected, using a gravity corer, from the Longtang River near an e-waste recycling establishment located in Qingyuan City (Guangdong Province, China). All sediment samples were taken randomly, in triplicate, from an area of approximately 1 m2 at the center of each sediment site. All of the sediment samples had equal volumes and they were mixed evenly and transferred to sterile glass jars, which were then sealed and transported back to the laboratory and stored at 4 °C until use.
TCBPA Added to Sediment
One gram of air-dried sediment was homogenized and sieved through a 0.22-mm screen and then autoclaved at 121 °C for 30 min on three successive days before adding to the serum bottle. The autoclaved sediment was completely saturated by 1 ml of the TCBPA dichloromethane stock solution (10 mM) to yield a 200 μM nominal concentration. Solvent was allowed to evaporate from vented bottles under a fume hood overnight and the surface of dry sediment particles was coated by TCBPA.
Sediment Slurries
Sediment slurry was prepared as follows: 2000 ml of mineral salt medium prepared as previously described (Voordeckers et al. 2002) was autoclaved at 121 °C (30 min, three consecutive days) and cooled to room temperature under a purified N2 gas stream. Then, Longtang Town sediment was added to 2000-ml medium to achieve a final 2700 ml sediment slurry to which 1 ml of 1% resazurin was added as a redox indicator.
Sediment microcosms of 25% biologically active Longtang town sediment were prepared under deoxygenated conditions (purged with a stream of purified N2 gas for 20–30 min) by dispensing 50 ml of the sediment slurry into 100-ml serum bottles containing the sediment powder coated with TCBPA using a purged sterile pipet, then sealed by PTFE-covered butyl rubber gasket aluminum cap with a headspace of N2. The bottles were shaken vigorously to mix the sediment powder into the slurry.
Experimental Treatments
Except for the un-amended and sterile controls, 1 ml equimolar acetate, propionate, butyrate, and lactate stock solution (200 mM) was added to the other treatments via deoxygenated sterile syringes so that the final cultures contained 4-mM electron donor mixture. Additionally, the methanogenic and sulfate reducing conditions were obtained by adding 20 mM NaHCO3 and 1, 10, or 20 mM Na2SO4, respectively. The sterile control was autoclaved at 121 °C for 30 min on three consecutive days after the addition of TCBPA. All above sediment slurries were set up in triplicate and incubated at 30 °C in continuous darkness without shaking.
Sampling
Sampling was done once a week and after 42 days once about every 40 days. Serum bottles were agitated vigorously to ensure uniform sampling while 3.5 ml N2 was injected and then an equal volume of sediment slurry was withdrawn using a deoxygenated springe fitted with a 16-gauge needle. For the 3.5-ml slurry, 1.5 ml was stored at −20 °C for molecular analysis, while the remainder was placed in a 5.5-cm diameter culture dish and air-dried under a fume hood for determination of TCBPA concentration.
TCBPA Extraction and HPLC Analyses
Air-dried sediment samples (about 0.5 g) were milled and transferred into 8 ml glass vials. The samples were extracted overnight with 2 ml dichloromethane in a 4-D rotation mixer and then exposed to an ultrasonic water bath for 10 min. The supernatant was then transferred to an 8 ml glass bottle after centrifuging for 5 min at 10000 rpm. This extraction process was repeated twice and extraction efficiency was >80%. The combined supernatant containing TCBPA was mixed and filtered through a 0.22 μm Millipore nylon filter and analyzed by HPLC (Shimadzu LC20A; SPD-M20A UV-vis Detector; SIL-20A Auto sampler; Eclipse XDB column, 4.6 × 150 mm, 5 μm). The mobile phase was 80% methanol and 20% ultrapure water at a flow rate of 1 ml/min. The UV-vis detector and the analytical column temperature were set at 238 nm and 40 °C, respectively. The retention time of TCBPA was 3.5 min. Concentrations were calculated by comparing to five point external standard curves and the detection limit was 0.03 ng/g.
Bacterial Community Analysis
Genomic DNA was extracted with a Soil DNA Isolation Kit (Omega Biotec, USA) according to the manufacturer’s instructions. DNA integrity was assessed by 1% agarose gel electrophoresis.
Genomic DNA was analyzed by a Qubit® dsDNA Assay Kit (Life technologies, USA) for determining the amount of DNA used for PCR. The V3-V4 region of the bacterial 16S rRNA gene was amplified using a T100TM Thermal Cycler PCR System (BIO-RAD, USA) using primers 341 F (5′-barcode-CCTACGGGNGGCWGCAG-3′) and 805R (5′-GACTACHVGGGTATCTAATCC-3′) (Herlemann et al. 2011), where the barcode was a six-base sequence unique to each sample. Each 50 μL PCR reaction mixture contained 5 μL of 10× PCR buffer, 0.5 μL of 10 mM dNTPs, 10 ng of genomic DNA, 0.5 μL of 50 μM primer, and 0.5 μL of 5 U/μL Plantium® Taq DNA Polymerase. The PCR process was conducted as follows: 94 °C for 2 min, followed by 5 cycles at 94 °C for 30 s, 45 °C for 20 s, 65 °C for 30 s and then 20 cycles at 94 °C for 20 s, 55 °C for 20 s, 72 °C for 30 s and final extension at 72 °C for 5 min. After PCR, amplicons extracted from 2% agarose gels were purified using the SanPrep Column DNA Gel Extraction Kit (Sangon Biotech, China) and then quantified accurately using a Qubit® dsDNA Assay Kit (Life technologies, USA).
Bacterial Community Composition
A total of eight DNA samples were sequenced, including DNA extracted from sediment prior to incubation (called initial sediment) and sediment after 160 days incubation under methanogenic condition, sulfate reducing conditions, electron donor-enhanced condition, and un-amended control. All of the amplicons were mixed in equimolar amounts and pooled in the Illumina MiSeq platform for paried-end sequencing. Raw sequencing data obtained from this study were deposited to the NCBI Sequence Read Archive database with accession nos. SRP5109774, SRR5109777, SRP5109779-SRP5109781, SRP5109783, SRP5109784, and SRP5109787. The preprocessing of sequencing data was as follows: On the basis of the overlap relationship between the paired-end read data in the fastq file format, the paired reads were joined together by FLASH version 1.2.3 (Magoc and Salzberg 2011) and then according to the barcode. Contiguous sequences were classified followed by quality control.
The overlap field of contiguous sequences with a mismatch ratio below 0.1, short fragment (<50 bp), low-quality fragment (tail quality of joined reads below 20 bases), and low complexity sequences were filtered by quality control using Prinseq version 0.20.4 (Schmieder and Edwards 2011). After the non-target amplification area sequences were eliminated, sequences were checked for sequencing error using pseudo-single linkage in Pre.cluster of Mothur and chimeras were removed by Chimeras. UCHIME (Edgar et al. 2011).
Analyses Based on OTUs and Taxonomy
Sequences were classified into operational taxonomic units (OTUs) with similarity of 0.97 using Uclust version 1.1.579. Then, based on OTUs, taxonomy, alpha diversity, and beta diversity were analyzed. Rarefaction curves, Chao1, Ace, Coverage, Shannon, and Simpson values were determined for all samples using the Mothur program version 1.30.1, and sample distance and cluster were calculated according to the Unifrac metric (Yatsunenko et al. 2012). Taxonomic classifications of trimmed fasta sequences of the V3-V4 region were assessed by BLAST analysis against the SILVA database (Quast et al. 2013) for 16S reads with a confidence threshold of 0.9. Relative abundances of bacteria in taxonomies of all samples were analyzed using MEGAN version 5.7.1.
3 Results and Discussion
Transformation of TCBPA in Anaerobic Sediment Microcosms
The pH values under various conditions were circumneutral and all above 7.0 during cultivation. Sulfate loss (H2S production) was detected and no methane was observed under sulfate-reducing conditions, implying that sulfate-reducing conditions were promoted. Similarly, CH4 and CO2 were detected under methanogenic condition, verifying that methanogenic condition established. TCBPA degradation during 160 days under these conditions is shown in Fig. 1. There were no significant changes of TCBPA levels in the autoclaved control and the un-amended control, while loss of TCBPA in the amended treatments was detected. This indicates that the microbes can affect TCBPA degradation, but this ability is weak without amendments. Our previous study showed that anaerobic degradation of TCBPA in river sediments was dechlorinated in order of 3,3’,5-trichlorobisphenol A, 3,3’-dichlorobisphenol A, 3-monochlorobisphenol A and bisphenol A (BPA), with no further degradation of BPA (Li 2014). This was consistent with dechlorination pathway of TCBPA in river sediment proposed by Yuan et al. (2011). Voordeckers et al. (2002) found that metabolites of TCBPA anaerobic degradation in estuarine sediments were mainly dichlorobisphenol A and a small amount of monochlorobisphenol A while no BPA was detected. Hence, TCBPA transformation in anaerobic sediment was mainly from dechlorination, and the extent of dechlorination was affected by variation in the environmental conditions or the bacteria involved (Voordeckers et al. 2002; Yuan et al. 2011). Under the amended treatments, the chlorine atoms on TCBPA can be dechlorinated completely.
In the electron donor-amended cultures, 44% loss of 200 μM was observed after 160 days including an initial lag period of 21 days. Addition of acetate, propionate, butyrate, and lactate enhanced biotransformation of TCBPA and shortened the degradation period compared to un-amended control. As carbon sources, these low-molecular-weight organic acids could be utilized directly by microbes. H2, the electron donor for transformation, was produced and therefore improved activity of microbes, promoted reductive dechlorination, and accelerated degradation (Chang et al. 2012; Nies and Vogel 1990; Yuan et al. 2011).
In this study, the best degradation effect of TCBPA was seen in the methanogenic sediment. Significant degradation of 200 μM TCBPA commenced at 14 days and finished in 160 days with 65% loss, a disappearance rate constant of 0.0064 day−1, and a half-life period of 112 days. Panagiotakis et al. (2014) found that different species of Dehalococcoides and other dechlorinating bacteria performed better under methanogenic conditions or at low sulfate concentrations (0.31 mM) than high sulfate concentration (4.2 mM).
Given 9.5–321 mg/kg (0.03–1 mM) sulfate in Longtang river sediment, 1, 10, and 20 mM sulfate were set up, respectively, in three levels of sulfate reducing conditions for investigating impact of sulfate on dechlorination. And under these three conditions, the lag period and removal rate of TCBPA were 21 days, 43%; 42 days, 23%, and 42 days, 23% at 160 days, respectively. Compared with high concentration sulfate, TCBPA in sediment with a low sulfate concentration has higher degradation efficiency and a shorter lag time. Panagiotakis et al. (2014) found that dechlorinating Dehalococcoides spp. predominate in mixed anaerobic microbial communities regardless of the magnitude of sulfate concentration while performed better in the presence of low sulfate concentrations (<0.31 mM) than in the presence of higher sulfate concentrations (>4.2 mM). Through investigating the effect of sulfate reduction on anaerobic dechlorination of pentachloroaniline (PCA), competition between sulfate reduction and dechlorination for electron donor postponed degradation of PCA (Ismail and Pavlostathis 2010). The rate of sulfate reduction rather than sulfate concentration alone dictated the outcome of the competition between sulfate reducers and either dechlorinators or methanogens. Aulenta et al. (2008) showed that the rapid and competitive utilization of electron donor by sulfate reducing bacteria (SRB) had an adverse impact on the rate of reductive dechlorination. Liu et al. (2014) found that the number of SRB in the process of TCBPA degradation decreased after the initial increase and SRB played a positive role in the fate of TCBPA. Bao et al. (2012) suggested that reduced sulfur compounds produced by classical sulfate reducing bacterium Clostridium sp. BXM have the ability to induce dechlorination of p,p'-DDTs. In this study, we found that SRB promote dechlorination, but the competition between SRB and dechlorinators for electron donors leads to transient inhibition of dechlorination. The activity of dechlorinators can therefore be suppressed by high sulfate concentrations. Under sulfate reducing conditions (with electron donors), dechlorination efficiency of TCBPA was moderate among the amended treatments, where the removal rate of TCBPA was less than that of the electron donor alone enhanced system.
Microbial Diversity
Reductive dechlorination of TCBPA in sediment slurries was attributed to microorganisms. Dominant microbial species, especially dechlorinators, were the key factor leading to the differences of TCBPA dechlorination efficiency under different treatments. Therefore, it is necessary to analyze bacterial communities at 0 day (initial sediment) and after 160-day incubation under different conditions. The bacterial community diversity and structures in the sediment slurries were determined by Illunmia 16S rRNA gene MiSeq paired-end sequencing. From eight samples, 282641 high-quality 16S rRNA gene sequences with an average length of 414 bp were generated from 300119 raw sequences following quality control and removal of chimeras and non-target sequences (Table S1). These sequences were assigned into 5107–5853 OTUs with a 0.97 similarity level (Table 1).
The richness of the samples was analyzed by OTUs through the rarefaction curve (Fig. 2). The rarefaction curves tended to plateau with an increase of sequences, suggesting the majority of OTUs were represented (Jayamani and Cupples 2015). The coverage index indicated that the number of sequences was sufficient to cover 87–94% of the bacterial communities (Table 1). Therefore, this sequencing result is representative of the actual sample composition. The OTU numbers of the microcosm samples with each 160-day inoculation were lower than that of the initial sediment (Fig. 2). This indicated that bacterial diversity of sediment microcosms declined after the addition of TCBPA.
Bacterial community diversity can be represented by the alpha diversity index (Table 1). The abundance-based coverage estimator (ACE) and Chao1 index expressed community richness, Shannon and Simpson index signified community diversity, and Coverage index indicated sequencing depth. The values of ACE and Chao1 are all in direct proportion to the richness as is the Shannon index for diversity. The value of Simpson index is negatively correlated with diversity. The highest Shannon index and lowest Simpson index obtained in initial sediment samples illustrated that community diversity decreased during the time of culture regardless of the anaerobic sediment treatments used for TCBPA degradation. At 160 days, sediment samples spiked with TCBPA have higher ACE and Chao1 indices than these of initial sediment. This suggests that addition of TCBPA in sediment microcosms led to increases of community richness.
Bacterial Community Analysis at Higher Taxonomic Levels
In the present study, nearly all sequences (99.5%) were classified to bacteria and few sequences belonged to archaea. Phyla with a relative abundance of >1% in at least one sample were classified as dominant. A total of 30 identified phyla were observed among the test samples, and the major phyla for each sample are shown in Fig. 3a. The most abundant phyla were Proteobacteria, Chloroflexi, Firmicutes, Bacteroidetes, Planctomycetes, Nitrospirae, Verrucomicrobia, Acidobacteria, Gemmatimonadetes, and Chlorobi, and their relative abundance occupied over 97% of all the sequences. The compositions of phyla under different conditions were almost the same while abundance of phyla in different systems was significantly different. Since no degradation of TCBPA was observed in the un-amended control, bacteria responsible for dechlorination could preliminary proposed based on comparison of relative abundance between amended treatments and initial sediment.
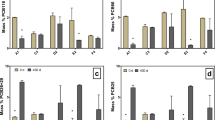
At both 0 and 160 days, the most dominant phylum in all treatments was Proteobacteria. Its relative abundance decreased from 48% at 0 day to about 40% at 160 days except for un-amended control and 20 mM sulfate treatment whose increased to >50%. However, it is noteworthy that experimental group with 20 mM sulfate performed lower TCBPA dechlorination efficiency than 1 mM sulfate, and the un-amended control had no effect on the TCBPA dechlorination as mentioned before. Proteobacteria may not include in the groups directly capable of TCBPA elimination in anaerobic degradation systems. However, it should be noted that some non-dechlorinating bacteria are known to have important roles to support other dechlorinating bacteria, such as Desulfovibrio vulgaris cable of ferment lactate to provide H2 and acetate for Dehalococcoides ethenogenes strain 195 growth and dechlorination (Men et al. 2012). Similarly, Planctomycetes, Acidobacteria, Gemmatimonadetes, and Chlamydiae made no contribution to TCBPA dechlorination because of lower relative abundance and low abundance (<1%) in amended conditions at 160 days compared to the initial sediment and the un-amended control at 160 days. Due to the relative abundances of Nitrospirae and Verrucomicrobia in the unamended control and other amended treatments increased significantly from 0 to 160 days, the effect of them on TCBPA dechlorination needs advance estimation.
Chloroflexi plays a key role in dechlorination of TCBPA. Relative abundance of Chloroflexi increased by 19% (electron donor), 21% (TCBPA + electron donor), 24% (methanogenic condition), 17% (1 mM SO4 2−), 24% (10 mM SO4 2−), and 11% (20 mM SO4 2−) while only 5% for the un-amended control. Chloroflexi are ubiquitous in many environments (Yamada and Sekiguchi 2009), and previous works have documented the dechlorination ability of Chloroflexi, especially the genus Dehalococcoides (Bunge et al. 2003; Fennell et al. 2004; Jayachandran et al. 2003). In this study, no Dehalococcoides were identified, but other bacteria in the Chloroflexi may have the ability to dechlorinate TCBPA. These groups include the Anaerolinea, Bellilinea, Leptolinea, and Longilinea (Fang et al. 2014; Praveckova et al. 2015).
Besides, Bacteroidetes and Firmicutes which have been reported as important constituents of other dechlorinating systems and include some dechlorination strains (Pérez-de-Mora et al. 2014; Zhang et al. 2015). Firmicutes is capable of dechlorination and is involved in anaerobic dechlorinating processing of chlorinated organic compounds, such as polychlorinated biphenyls (Gomes et al. 2014; Suzuki et al. 2013), pentachlorophenol (Liu et al. 2013), and chlorinated ethylene (Miura et al. 2015; Ziv-El et al. 2011). Relative abundance of Firmicutes remained about 8% during incubation. Chlorobi belongs to obligate anaerobic bacteria using the reduction of sulfur and hydrogen molecules as electron donors (Podosokorskaya et al. 2013). Besides, Chlorobi grow with CO2 as the sole carbon source and also assimilate simple organic acides such as acetate, are commonly found in stratified, anoxic sediments and aquatic environments. In this study, Chlorobi (green sulfur bacteria) in methanogenic condition was the most abundance among all the amended treatment.
Figure 3b illustrates that 21 identified classes with relative abundance of >1% in at least in one samples accounted for >96% of all sequences. The dominant class at 160 days was Anaerolineae (Chloroflexi), followed by β-proteobacteria, α-proteobacteria, δ-proteobacteria and γ-proteobacteria (Proteobacteria), Clostridia (Firmicutes), and Bacteroidia (Bacteroidetes). And an obvious shift of dominant bacterial was taken from α-proteobacteria at 0 day to Anaerolineae at 160 days (Fig. 3b). The relative abundance of Anaerolineae increased from 12% at 0 day to 31% (electron donor), 17% (un-amended control), 33% (TCBPA + electron donor), 34% (methanogenic condition), 28% (1 mM SO4 2−), 35% (10 mM SO4 2−), and 23% (20 mM SO4 2−) at 160 days. The relative abundance of Anaerolineae may have been promoted by electron donors but partially suppressed by high sulfate concentrations. Relative abundance of δ-proteobacter was decreased under methanogenic conditions at 160 days compared with the initial sediment, while there was an increase under the other conditions and a positive correlation with sulfate concentration (11, 11, and 14% at 1, 10, and 20 mM SO4 2−, respectively) under sulfate reducing conditions. Most haloalkaliphilic SRBs reported belong to the phylum Proteobacteria (Zhou et al. 2015) and some groups have the ability to induce dechlorination of chlorinated organic compounds (Bao et al. 2012). Members of the class δ-proteobacteria of Proteobacteia in this study may be capable of dechlorinating TCBPA. While because of considerable levels of β-proteobacteria, α-proteobacteria, and γ-proteobacteria in the un-amended control in which no TCBPA degradation was observed, these three classes had no apparent effects directly on the dechlorination of TCBPA.
At the same time, Bacteroidia increased from 0.6% at 0 day to 4.4% (electron donor), 3.1% (un-amended control), 4.2% (TCBPA + electron donor), 4.7% (methanogenic condition), 4.3% (20 mM SO4 2−), 3.8% (10 mM SO4 2−), and 5.8% (1 mM SO4 2−) at 160 days. The relative abundance of Ignavibacteria in phylum Chlorobi under the methanogenic condition at 160 days was at least four times that of the other conditions. It is worth noting that there were few Dehalococcoidia in sediment microcosms at 0 and 160 days. Known dechlorinating bacteria belong to the Dehalococcoidia class of the Chloroflexi (Loffler et al. 2013). Some of these, such as Dehalococcoides mccartyi, can dechlorinate chloroethylene and many other chlorinated aromatic compounds.
Bacterial Community Analysis at the Genus Level
Among 895 genera found in the eight samples, the heat map of the top 50 with relative abundance >1% in at least one sample is shown in Fig. 4. The maximum relative abundance occupied by unclassified Anaerolineaceae, rose from 5.7% in initial sediment to 14% (electron donor), 7.3% (un-amended control), 16% (TCBPA + electron donor), 15% (methanogenic condition), 13% (1 mM SO4 2−), 17% (10 mM SO4 2−), and 9.5% (20 mM SO4 2−) (Table 2). Besides, other genera including Longilinea (1.6–8.0%), Leptolinea (2.5–6.3%), Azospira (2.6–4.1%), Anaerolinea (0.7–3.7%), unclassified Gallionellaceae (0.2–5.9%), vadinBC27 (0.1–3.7%), Thiohalophilus (0.04–8.5%), unclassified Xanthomonadales (0.7–4.1%), Clostridium (0.5–3.2%), and Desulfobulbus (0.2–5.6%) presented comparatively high relative abundances as well. Among these, unclassified Anaerolineaceae, Longilinea, and Leptolinea were always dominant (Fig. 4 and Table 2). Anaerolinea was the dominant genus capable of remediation of field contaminated by organic pollutants such as dichlorodiphenyltrichloroethane (DDT), hexachlorocyclohexane (HCH), and atrazine (ATZ) (Fang et al. 2014). Sequences of Leptolinea showed high similarity (97–99%) with those of reductive dechlorinating bacteria enriched by chlorinated organic compounds (Praveckova et al. 2015). The genus vadinBC27 is important in degrading polybrominated diphenyl ethers (PBDE) enhanced with electron donors and this group has played a pivotal role in the treatment of recalcitrant organic contaminants in landfill leachate (Xie et al. 2014). Compared with other samples at 160 days, Atopobium, unclassified Christensenellaceae, and Ignavibacterium possessed significant advantages under methanogenic conditions. Under sulfate reducing conditions, the abundances of Anaerolinea, Geobacter, Olsenella, unclassified Xanthomonadales, Desulfuromonas, and Sphingobium were reduced with sulfate addition, while the abundance of Azospira, Olsenella, and Sphingobium increased in electronic donor promoting conditions.
We identified 10 genera (Table 2) that likely contribute to the dechlorination of TCBPA. They belong to the four phyla: Chloroflexi, Firmicutes, Bacteroidetes, and Chlorobi. The most abundant groups within Chloroflexi were unclassified Anaerolineaceae, Longilinea, Leptolinea, Anaerolinea, and Bellilinea and they significantly increased from 0 to 160 days, followed by vadinBC27, unclassified Porphyromonadaceae, Clostridium, Fastidiosipila, and Ignavibacterium. Clostridium is an important member of a synthetic dehalogenation culture and can act as a hydrogen supplier via glucose fermentation. Chen et al. (2013) showed that Clostridium was the predominant bacteria involved with DDT degradation in paddy soils and Clostridium sp. T-RF 521 was very similar to dechlorinating bacteria. Ignavibacterium had an advantage at 160 days under the methanogenic condition compared with the other treatments. Form an initial level of 12% at 0 day, the total relative abundances of these potential dechlorinating bacteria increased to 37% (electron donor), 43% (methanogenic condition), 38% (TCBPA + electron donor), and 29–40% (sulfate reducing condition). The TCBPA degradation efficiency order (high to low) was methanogenic condition > electron donor > sulfate reducing condition. These dechlorinators were more abundant in sulfate reducing conditions of 1 mM initial sulfate than in 20 mM sulfate. Relative abundance of genera Desulfobulbus, Geobacter, Desulfuromonas of Deltaproteobacteria increased with the addition of sulfate. Geobacter use organohalide respiration allowing for energy conservation (Richardson 2013). Geobacter lovleyi causes reductive dechlorination of tetrachloroethene (PCE) and trichloroethene (TCE) and combines this with energy conservation and growth (Philips et al. 2013; Wagner et al. 2012). The presence of Geobacter supported Dehalococoides dechlorination activity (Yan et al. 2012). Desulfobulbus and Desulfuromonas were formerly known as SRB (Richardson 2013). Several species belonging to SRBs are dehalogenating anaerobes like Desulfomonile tiedjei DCB-1 and dehalogenation of halogenated aromatic compounds like chlorophenols by SRBs has been reported (DeWeerd et al. 1991; Rasool et al. 2015). The role of SRBs in dehalogenation due to the inhibition of dehalogenases by sulfate, sulfite, or hydrogen sulfide, and competition for electron donor between SRBs and dehalogenators has been controverial (Aulenta et al. 2008; Liu et al. 2014; Panagiotakis et al. 2014; Voordeckers et al. 2002). In this study, we found that less sulfate in the system produced more efficient dechlorination of TCBPA. This indicated that negative influence of sulfate reduction was greater than the positive affect of SRBs on dechlorination. Therefore, the dechlorination efficiency of TCBPA with high sulfate concentration was not optimal due to inhibition of dechlorinating bacteria performance by high sulfate concentration (Panagiotakis et al. 2014).
The rate and extent of anaerobic TCBPA loss varied under different reducing conditions. The most effective anaerobic system for dechlorination of 200 μM TCBPA over 160 days was methanogenic followed by electron donor enhanced and sulfate reducing conditions with 1 mM SO4 2−, 10 mM SO4 2−, and 20 mM SO4 2−. Analysis by high-throughput sequencing of the 16S rRNA gene showed that the addition of TCBPA to reductive dechlorinating conditions decreased of bacterial community diversity. Analysis of the bacterial community at phylum, class, and genus levels demonstrated that an increase of putative dechlorinating bacteria in the microcosm is the key factor for effective TCBPA degradation. Efficient degradation of TCBPA under anaerobic conditions depended on abundant dechlorinating bacteria whose performance would be affected by high sulfate concentrations lowering the degradation rate of TCBPA. Although TCBPA could be dehalogented to BPA under both methanofenic and sulfate-reducing conditions, further investigation on eliminate BPA persistent in anerobic condition is required.
References
Aulenta, F., Beccari, M., Majone, M., Papini, M. P., & Tandoi, V. (2008). Competition for H2 between sulfate reduction and dechlorination in butyrate-fed anaerobic cultures. Process Biochemistry, 43(2), 161–168.
Bao, P., Hu, Z., Wang, X., Chen, J., Ba, Y., Hua, J., Zhu, C., Zhong, M., & Wu, C. (2012). Dechlorination of p, p'-DDTs coupled with sulfate reduction by novel sulfate-reducing bacterium Clostridium sp. BXM. Environmental Pollution, 162, 303–310.
Bastos, P. M., Eriksson, J., Green, N., & Bergman, K. (2008). A standardized method for assessment of oxidative transformations of brominated phenols in water. Chemosphere, 70(7), 1196–1202.
Blanco, E., Casais, M. C., Mejuto, M. C., & Cela, R. (2005). Analysis of tetrabromobisphenol A and other phenolic compounds in water samples by non-aqueous capillary electrophoresis coupled to photodiode array ultraviolet detection. Journal of Chromatography A, 1071(1–2), 205–211.
Bunge, M., Adrian, L., Kraus, A., Opel, M., Lorenz, W. G., Andreesen, J. R., Gorisch, H., & Lechner, U. (2003). Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature, 421(6921), 357–360.
Casatta, N., Mascolo, G., Roscioli, C., & Viganò, L. (2015). Tracing endocrine disrupting chemicals in a coastal lagoon (Sacca di Goro, Italy): sediment contamination and bioaccumulation in Manila clams. Science of the Total Environment, 511, 214–222.
Chang, B., Yuan, S., & Ren, Y. (2012). Anaerobic degradation of tetrabromobisphenol-A in river sediment. Ecological Engineering, 49, 73–76.
Chen, M., Cao, F., Li, F., Liu, C., Tong, H., Wu, W., & Hu, M. (2013). Anaerobic transformation of DDT related to iron(III) reduction and microbial community structure in paddy soils. Journal of Agricultural and Food Chemistry, 61(9), 2224–2233.
Chu, S., Haffner, G. D., & Letcher, R. J. (2005). Simultaneous determination of tetrabromobisphenol A, tetrachlorobisphenol A, bisphenol A and other halogenated analogues in sediment and sludge by high performance liquid chromatography-electrospray tandem mass spectrometry. Journal of Chromatography A, 1097(1–2), 25–32.
DeWeerd, K. A., Concannon, F., & Suflita, J. M. (1991). Relationship between hydrogen consumption, dehalogenation, and the reduction of sulfur oxyanions by Desulfomonile tiedjei. Applied and Environmental Microbiology, 57(7), 1929–1934.
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., & Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 27(16), 2194–2200.
Eriksson, J., Rahm, S., Green, N., Bergman, K., & Jakobsson, E. (2004). Photochemical transformations of tetrabromobisphenol A and related phenols in water. Chemosphere, 54(1), 117–126.
Fan, Z., Hu, J., An, W., & Yang, M. (2013). Detection and occurrence of chlorinated byproducts of bisphenol A, nonylphenol, and estrogens in drinking water of China: comparison to the parent compounds. Environmental Science & Technology, 47(19), 10841–10850.
Fang, H., Cai, L., Yang, Y., Ju, F., Li, X., Yu, Y., & Zhang, T. (2014). Metagenomic analysis reveals potential biodegradation pathways of persistent pesticides in freshwater and marine sediments. Science of the Total Environment, 470–471, 983–992.
Fennell, D. E., Nijenhuis, I., Wilson, S. F., Zinder, S. H., & Häggblom, M. M. (2004). Dehalococcoides ethenogenes strain 195 reductively dechlorinates diverse chlorinated aromatic pollutants. Environmental Science & Technology, 38(7), 2075–2081.
Fukazawa, H., Hoshino, K., Shiozawa, T., Matsushita, H., & Terao, Y. (2001). Identification and quantification of chlorinated bisphenol A in wastewater from wastepaper recycling plants. Chemosphere, 44(5), 973–979.
Gallart-Ayala, H., Moyano, E., & Galceran, M. T. (2010). On-line solid phase extraction fast liquid chromatography-tandem mass spectrometry for the analysis of bisphenol A and its chlorinated derivatives in water samples. Journal of Chromatography A, 1217(21), 3511–3518.
Gomes, B. C., Adorno, M. A. T., Okada, D. Y., Delforno, T. P., Lima Gomes, P. C. F., Sakamoto, I. K., & Varesche, M. B. A. (2014). Analysis of a microbial community associated with polychlorinated biphenyl degradation in anaerobic batch reactors. Biodegradation, 25(6), 797–810.
Horikoshi, S., Miura, T., Kajitani, M., Horikoshi, N., & Serpone, N. (2008). Photodegradation of tetrahalobisphenol-A (X = Cl, Br) flame retardants and delineation of factors affecting the process. Applied Catalysis, B: Environmental, 84(3–4), 797–802.
Herlemann, D. P., Labrenz, M., Jurgens, K., Bertilsson, S., Waniek, J. J., & Andersson, A. F. (2011). Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME Journal, 5(10), 1571–1579.
Ismail, Z. Z., & Pavlostathis, S. G. (2010). Influence of sulfate reduction on the microbial dechlorination of pentachloroaniline in a mixed anaerobic culture. Biodegradation, 21(1), 43–57.
Jayachandran, G., Görisch, H., & Adrian, L. (2003). Dehalorespiration with hexachlorobenzene and pentachlorobenzene by Dehalococcoides sp. strain CBDB1. Archives of Microbiology, 180(6), 411–416.
Jayamani, I., & Cupples, A. M. (2015). Stable isotope probing reveals the importance of Comamonas and Pseudomonadaceae in RDX degradation in samples from a Navy detonation site. Environmental Science and Pollution Research, 22(13), 10340–10350.
Kalasekar, S. M., Zacharia, E., Kessler, N., Ducharme, N. A., Gustafsson, J. K., Kakadiaris, I. A., & Bondesson, M. (2015). Identification of environmental chemicals that induce yolk malabsorption in zebrafish using automated image segmentation. Reproductive Toxicology, 55, 20–29.
Leung, A. O. W., Luksemburg, W. J., Wong, A. S., & Wong, M. H. (2007). Spatial distribution of polybrominated diphenyl ethers and polychlorinated dibenzo-p-dioxins and dibenzofurans in soil and combusted residue at guiyu, an electronic waste recycling site in southeast China. Environmental Science & Technology, 41(8), 2730–2737.
Li, L. (2014). Transformation of tetrabromobisphenol A and tetrachlorobisphenol A in river sediments. Guang Zhou: South China University of Technology (in Chinese).
Lin, N., Shan, Z., Zhu, C., & Ren, Y. (2015). Heavy metal and PCB contamination in river sediments of an E-waste recycling site in Qingyuan. Environmental Chemistry, 34(9), 1685–1693. in Chinese.
Liu, D., Lei, L., Yang, B., Yu, Q., & Li, Z. (2013). Direct electron transfer from electrode to electrochemically active bacteria in a bioelectrochemical dechlorination system. Bioresource Technology, 148, 9–14.
Liu, S., Li, L., & Ren, Y. (2014). Anaerobic biodegradation of TCBPA in river sediment and the role of sulfate reducing bacteria (SRB) in TCBPA’s degradation. Environmental Chemistry, 33(06), 915–922. in Chinese.
Loffler, F. E., Yan, J., Ritalahti, K. M., Adrian, L., Edwards, E. A., Konstantinidis, K. T., Muller, J. A., Fullerton, H., Zinder, S. H., & Spormann, A. M. (2013). Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. International Journal of Systematic and Evolutionary Microbiology, 63(Pt 2), 625–635.
Magoc, T., & Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics, 27(21), 2957–2963.
Men, Y., Feil, H., Verberkmoes, N. C., Shah, M. B., Johnson, D. R., Lee, P. K., West, K. A., Zinder, S. H., Andersen, G. L., & Alvarez-Cohen, L. (2012). Sustainable syntrophic growth of Dehalococcoides ethenogenes strain 195 with Desulfovibrio vulgaris Hildenborough and Methanobacterium congolense: global transcriptomic and proteomic analyses. ISME Journal, 6(2), 410–421.
Miura, T., Yamazoe, A., Ito, M., Ohji, S., Hosoyama, A., Takahata, Y., & Fujita, N. (2015). The impact of injections of different nutrients on the bacterial community and its dechlorination activity in chloroethene-contaminated groundwater. Microbes and Environments, 30(2), 164–171.
Nies, L., & Vogel, T. M. (1990). Effects of organic substrates on dechlorination of Aroclor 1242 in anaerobic sediments. Applied and Environmental Microbiology, 56(9), 2612–2617.
Panagiotakis, I., Mamais, D., Pantazidou, M., Rossetti, S., Aulenta, F., & Tandoi, V. (2014). Predominance of Dehalococcoides in the presence of different sulfate concentrations. Water, Air, & Soil Pollution, 225, 1.
Pérez-de-Mora, A., Zila, A., McMaster, M. L., & Edwards, E. A. (2014). Bioremediation of chlorinated ethenes in fractured bedrock and associated changes in dechlorinating and nondechlorinating microbial populations. Environmental Science & Technology, 48(10), 5770–5779.
Philips, J., Haest, P. J., Springael, D., & Smolders, E. (2013). Inhibition of geobacter dechlorinators at elevated trichloroethene concentrations is explained by a reduced activity rather than by an enhanced cell decay. Environmental Science & Technology, 47(3), 1510–1517.
Podosokorskaya, O. A., Kadnikov, V. V., Gavrilov, S. N., Mardanov, A. V., Merkel, A. Y., Karnachuk, O. V., Ravin, N. V., Bonch-Osmolovskaya, E. A., & Kublanov, I. V. (2013). Characterization of Melioribacter roseus gen. nov., sp. nov., A novel facultatively anaerobic thermophilic cellulolytic bacterium from the class Ignavibacteria, and a proposal of a novel bacterial phylum Ignavibacteriae. Environmental Microbiology, 15(6), 1759–1771.
Praveckova, M., Brennerova, M. V., Cvancarova, M., De Alencastro, L. F., Holliger, C., & Rossi, P. (2015). Divergent PCB organohalide-respiring consortia enriched from the efflux channel of a former Delor manufacturer in Eastern Europe. Ecotoxicology and Environmental Safety, 120, 223–234.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., Peplies, J., & Gloeckner, F. O. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research, 41(D1), D590–D596.
Rasool, K., Mahmoud, K. A., & Lee, D. S. (2015). Influence of co-substrate on textile wastewater treatment and microbial community changes in the anaerobic biological sulfate reduction process. Journal of Hazardous Materials, 299, 453–461.
Richardson, R. E. (2013). Genomic insights into organohalide respiration. Current Opinion in Biotechnology, 24(3), 498–505.
Ruan, T., Liang, D., Song, S., Song, M., Wang, H., & Jiang, G. (2015). Evaluation of the in vitro estrogenicity of emerging bisphenol analogs and their respective estrogenic contributions in municipal sewage sludge in China. Chemosphere, 124, 150–155.
Sánchez-Brunete, C., Miguel, E., & Tadeo, J. L. (2009). Determination of tetrabromobisphenol-A, tetrachlorobisphenol-A and bisphenol-A in soil by ultrasonic assisted extraction and gas chromatography-mass spectrometry. Journal of Chromatography A, 1216(29), 5497–5503.
Schmieder, R., & Edwards, R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics, 27(6), 863–864.
Song, S., Song, M., Zeng, L., Wang, T., Liu, R., Ruan, T., & Jiang, G. (2014). Occurrence and profiles of bisphenol analogues in municipal sewage sludge in China. Environmental Pollution, 186, 14–19.
Suzuki, D., Baba, D., Satheeja Santhi, V., Jebakumar Solomon, R. D., & Katayama, A. (2013). Use of a glass bead-containing liquid medium for efficient production of a soil-free culture with polychlorinated biphenyl-dechlorination activity. World Journal of Microbiology and Biotechnology, 29(8), 1461–1471.
Terasaki, M., Kosaka, K., Kunikane, S., Makino, M., & Shiraishi, F. (2011). Assessment of thyroid hormone activity of halogenated bisphenol A using a yeast two-hybrid assay. Chemosphere, 84(10), 1527–1530.
Tsydenova, O., & Bengtsson, M. (2011). Chemical hazards associated with treatment of waste electrical and electronic equipment. Waste Management, 31(1), 45–58.
Voordeckers, J. W., Fennell, D. E., Jones, K., & Haggblom, M. M. (2002). Anaerobic biotransformation of tetrabromobisphenol A, tetrachlorobisphenol A, and bisphenol A in estuarine sediments. Environmental Science & Technology, 36(4), 696–701.
Wagner, D. D., Hug, L. A., Hatt, J. K., Spitzmiller, M. R., Padilla-Crespo, E., Ritalahti, K. M., Edwards, E. A., Konstantinidis, K. T., & Loffler, F. E. (2012). Genomic determinants of organohalide-respiration in Geobacter lovleyi, an unusual member of the Geobacteraceae. BMC Genomics, 13, 200.
Xie, Z., Wang, Z., Wang, Q., Zhu, C., & Wu, Z. (2014). An anaerobic dynamic membrane bioreactor (AnDMBR) for landfill leachate treatment: performance and microbial community identification. Bioresource Technology, 161, 29–39.
Yamada, T., & Sekiguchi, Y. (2009). Cultivation of uncultured Chloroflexi subphyla: significance and ecophysiology of formerly uncultured chloroflexi ‘Subphylum I’ with natural and biotechnological relevance. Microbes and Environments, 24(3), 205–216.
Yan, J., Ritalahti, K. M., Wagner, D. D., & Loffler, F. E. (2012). Unexpected specificity of interspecies cobamide transfer from Geobacter spp. to organohalide-respiring Dehalococcoides mccartyi strains. Applied and Environmental Microbiology, 78(18), 6630–6636.
Yang, Y., Lu, L., Zhang, J., Yang, Y., Wu, Y., & Shao, B. (2014). Simultaneous determination of seven bisphenols in environmental water and solid samples by liquid chromatography-electrospray tandem mass spectrometry. Journal of Chromatography A, 1328, 26–34.
Yatsunenko, T., Rey, F. E., Manary, M. J., Trehan, I., Dominguez-Bello, M. G., Contreras, M., Magris, M., Hidalgo, G., Baldassano, R. N., Anokhin, A. P., Heath, A. C., Warner, B., Reeder, J., Kuczynski, J., Caporaso, J. G., Lozupone, C. A., Lauber, C., Clemente, J. C., Knights, D., Knight, R., & Gordon, J. I. (2012). Human gut microbiome viewed across age and geography. Nature.
Yuan, S. Y., Chen, S. J., & Chang, B. V. (2011). Anaerobic degradation of tetrachlorobisphenol-A in river sediment. International Biodeterioration & Biodegradation, 65(1), 185–190.
Yuan, S. Y., Li, H. T., Huang, W., & Chang, B. V. (2010). Biodegradation of tetrachlorobisphenol-A in river sediment and the microbial community changes. Journal of Environmental Science and Health Part B-Pesticides Food Contaminants and Agricultural Wastes, 45(PII 9225884225), 360–365.
Zhang, Y., Wang, X., Hu, M., & Li, P. (2015). Effect of hydraulic retention time (HRT) on the biodegradation of trichloroethylene wastewater and anaerobic bacterial community in the UASB reactor. Applied Microbiology and Biotechnology, 99(4), 1977–1987.
Zhou, J., Zhou, X., Li, Y., & Xing, J. (2015). Bacterial communities in haloalkaliphilic sulfate-reducing bioreactors under different electron donors revealed by 16S rRNA MiSeq sequencing. Journal of Hazardous Materials, 295, 176–184.
Ziv-El, M., Delgado, A. G., Yao, Y., Kang, D., Nelson, K. G., Halden, R. U., & Krajmalnik-Brown, R. (2011). Development and characterization of DehaloR^2, a novel anaerobic microbial consortium performing rapid dechlorination of TCE to ethene. Applied Microbiology and Biotechnology, 92(5), 1063–1071.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 51178190), Special Funds for the Construction of Key Disciplines Funded Projects in Guangdong Province (Grant No. 2013CXZDA004), and the Research Fund of SIT of Guangzhou (Grant No. 2013J4100107). LetPub (www.letpub.com) provided linguistic assistance during manuscript preparation.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 20 kb)
Rights and permissions
About this article
Cite this article
Zhang, J., Liu, S., Li, L. et al. Anaerobic Dechlorination of Tetrachlorobisphenol A in River Sediment and Associated Changes in Bacterial Communities. Water Air Soil Pollut 228, 78 (2017). https://doi.org/10.1007/s11270-017-3254-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11270-017-3254-3