Abstract
The spread of antimicrobial resistance (AMR) is a global concern, high research priority being given to the environmental contamination, as the prevalence of organisms exhibiting AMR continues to increase. Multiresistant bacteria carrying different mobile genetic elements have been detected in sites with different degrees of urbanization, surface waters receiving insufficiently treated effluents being at high risk. The aim of the present study was to investigate the loads, antibiotic susceptibility, and class 1 integron carriage of Enterobacteriaceae isolated from surface waters and wastewaters around a large Romanian city. Searching for a valuable genetic marker of the displayed antibiotic resistance, the link between the AMR and the presence of int1I gene was explored in a total of 166 waterborne strains. Overall, amoxicillin-clavulanate resistance displayed the highest frequency (71.1 %), followed by ampicillin (63.9 %), cefuroxime (21.1 %), ciprofloxacin (17.5 %), cefotaxime (15.7 %), ceftriaxone (10.8 %), and gentamicin (6.6 %). The frequencies of isolates resistant to ampicillin, amoxicillin-clavulanate, ciprofloxacin, and gentamicin and also the prevalence of multiresistant strains were greater in surface waters, compared to wastewaters. The Int1I gene was detected in 21.7 % waterborne Enterobacteriaceae. A decrease in coliform counts and intI1-bearing cells, but a general increase in AMR and multiresistant bacteria, occurred during the wastewater treatment. A weak positive correlation was found between multidrug resistance int1I carriage in wastewater effluent but no sufficient evidence of a linkage between phenotypic AMR and int1I, overall. The presence of class 1 integron can be associated with anthropogenic influence, but the simple detection of intI1 gene cannot explain the complex antibiotic resistance phenotype.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Antimicrobial resistance (AMR) may seriously impair the prevention and treatment of an ever-increasing range of infections. The problem has been identified as a major threat to public health at least 20 years ago (Wise et al. 1998), and it continues to expand. In the future, the emergence and dispersion of non-susceptible microorganisms carrying newly acquired virulence determinants threaten to lead to a post-antibiotic era, in which common infections and minor injuries will be fatal (WHO 2014). Today, worldwide authorities warn on the factors contributing to the emergence and spread of AMR. The increasing anthropogenic pressure, consisting in chemical and microbiological pollution, the uncontrolled use of antimicrobial agents, and increased antibiotic consumption as well as the free movement of population and goods are the main factors facilitating the global invasion of bacteria with extremely high resistance to antibiotics.
The prevalence of organisms exhibiting AMR, especially resistance to multiple antibiotics, continues to increase across Europe. Data from the European Antimicrobial Resistance Surveillance Network (EARS-Net) show large variations in percentages of AMR, depending on the microorganism, antimicrobial agent, and geographical region (ECDC 2015). Initially, the AMR surveillance was focused on healthcare-associated infections, limited information being available toward the environmental contamination with antimicrobials. Today, the increasing trend of AMR awareness leads to extensive investigations in various ecosystems. The presence of multiresistant strains, bacteria carrying mobile genetic elements, and gene cassettes encoding resistance to antibiotics or biocides was demonstrated in sites with different degrees of urbanization, and also in natural environments. The core findings of the case studies demonstrate that increased levels of multidrug-resistant bacteria in urban water cycling are mainly due to wastewater discharge effluents (Laroche et al. 2009; Figueira et al. 2012; Czekalski et al. 2012; Koczura et al. 2012).
Although water availability is sufficient in terms of quantity, numerous studies warn upon the chemical and microbiological contamination of Romanian water resources (Farkas et al. 2010a, b; Mureşan et al. 2010; Muntean et al. 2010). Furthermore, opportunistic pathogens and also emerging pathogens bearing integrons and genetic elements encoding AMR have been identified to enter a drinking water treatment plant, as a consequence of fecal pollution (Farkas et al. 2012, 2013). Limited information is available regarding the antibiotic susceptibility of environmental bacteria (Bodoczi Florea 2011; Marinescu et al. 2015), but none in the area of Cluj-Napoca, the second largest city in Romania.
Near large cities, increased anthropogenic pressure leads to enhanced occurrence of pathogens and bacteria possessing considerable antibiotic resistances in rivers (Abraham 2011). The human gut is recognized as a reservoir of potentially pathogenic microorganisms. A comprehensive environmental monitoring of the pathogenic risk would be virtually impossible, due to the wide variety of hazardous microbes. Therefore, fecal indicators are a general tool currently used to assess water quality and also for microbial source tracking. More recently, the gastrointestinal tract was found to be also a hotspot for the selection of AMR bacteria and resistance genes (Penders et al. 2013; Huddleston 2014). Once released into aquatic habitats, the potential of resistant bacteria remains largely unknown, due to a variety of genetic mechanisms enabling the emergence and transfer of new resistance mechanisms (Rolain 2013). The improved management of water resources needs new tools to quickly assess and eventually predict dynamics of the complex AMR phenomenon. Low antimicrobial susceptibility is frequently linked to the carriage of specific resistance genes. Since several hundreds of resistance genes have been characterized up to date (Amábile-Cuevas 2016) and their emergence and variability are continuously expanding, it would be difficult to select which to be screened. Reliable genetic markers are still to be found. Recently, the intI1 gene was recognized as a generic marker of anthropogenic influence, due to a series of key advantages, such as the following: universal presence and high abundance in the commensal bacteria of humans and domestic animals, a consequently high representation in waste streams, low abundance in less affected environments, and a uniform and highly conserved DNA sequence (Gillings et al. 2014).
The scope of the present study was to assess the levels of microbial burden, antimicrobial susceptibility, and class 1 integron carriage in waterborne Enterobacteriacee isolated from Someşul Mic River and from a wastewater treatment plant. In addition, by exploring the links between the AMR and the presence of the int1I gene, the hypothesis of a new genetic marker for the displayed antibiotic resistance was tested.
2 Materials and Methods
2.1 Sampling Site
Belonging to the Someş-Tisa river basin, Someşul Mic River is the most important water resource of Cluj County. With a catching area of 3773 km2 and a total length of 178 km, Someşul Mic River results by the confluence of Someşul Cald with Someşul Rece Rivers. Crossing the Cluj city, the river receives many tributaries. Recently, Vigh and Pandi (2013) monitored few physico-chemical parameters in Someşul Mic tributaries, but no data are available regarding the microbial load, nor the environmental impact of the wastewater treatment plant (WWTP). As the area is becoming increasingly more populated, anthropogenic effects could lead to a severe biological contamination of surface waters. Today, the main sources of river pollution are domestic sewage, wastewater plant effluents, and animal manure, and less substantial are the industrial waters. The WWTP of Cluj-Napoca, although recently renovated, could still be a source of contamination with micropollutants such as antimicrobials and microorganisms surviving the treatment.
2.2 Water Sampling and Analysis
During March 2015, water samples were collected in two sampling campaigns, from four sampling points: river water upstream the city (RWU), municipal wastewater inlet (WWI), treated effluent (WWE), and river water downstream the WWTP (RWD) (Fig. 1). Maps in this article were created using ArcGIS Web Map by Esri. The base map sources were Earthstar Geographics, CNES/Airbus DS, Esri Romania, Esri HERE, DeLorme, NGA, USGS, and METI/NASA, with the overlay World Hydro Reference Map.
To restrict microbial regrowth, recipients were transported under specific conditions (4–8 °C) to the laboratory, where analyses were performed within 4 h after sampling. The Enterobacteriaceae colony-forming units (CFUs) were estimated by inoculation of water samples and subsequent dilutions into specific culture media: Tergitol-7 TTC (T7-TTC) Agar, Eosin Methylene Blue (EMB) Agar, MacConkey Agar, and Violet Red Bile Glucose (VRBG) Agar. Presumptive Enterobacteriacee were selected from each culture medium: yellow colonies able to decrease pH in T7-TTC Agar, dark colonies with or without metallic sheen from EMB Agar, red colonies from MacConkey Agar, and purple/pink colonies from VRBG Agar. From each plate, up to ten colonies with characteristic colony morphology were isolated on Tryptic Soy Agar and subjected to biochemical testing (oxidase production) in order to be confirmed as Enterobacteriaceae. The aim of the assay is to select the most adequate culture medium for detection and isolation environmental Enterobacteriaceae in waters with high microbial load, such as wastewaters.
Quality and reproducibility controls were realized with MicroBioLogics strains Escherichia coli ATCC 25922, Enterococcus faecalis ATCC 19433 29212, and Pseudomonas aeruginosa ATCC 27853. Culture media and reagents were purchased from Liofilchem, Merck Millipore, Oxoid, Sartorius Biotech, Scharlau, and bioMérieux.
2.3 Selection of Waterborne Enterobacteriaceae
In order to select the study strains, the ability of culture-based methods to detect waterborne Enterobacteriaceae was verified using molecular methods. PCR amplifications were performed using two sets of primers (Table 1), targeting the lacZ gene and the 16s rRNA gene (Maheux et al. 2014). PCR reaction mix contained in 25-μl total volume: 12.5 μl DreamTaq Green PCR master mix (2×), 10.25 μl nuclease-free water, 0.125 μl each primer to a final concentration of 0.5 μM, and 2 μl bacterial suspension adjusted to a 0.5 McFarland standard. The following reaction conditions were set up using a Biometra Trio thermocycler: initial denaturation 94 °C for 5 min followed by 35 cycles of 94 °C for 30 s, 58 °C for 30 s, 72 °C for 45 s, and a final extension at 72 °C for 5 min. Amplified PCR products were separated in 1.5 % agarose gel in 1× TBE buffer and stained with ethidium bromide 0.5 μg/ml. PCR primers and molecular reagents were purchased from Cleaver Scientific, Eurogentec, Lonza, and Thermo Fisher Scientific.
2.4 Antimicrobial Susceptibility Testing
Since large cities and their WWTPs are known to be major contributors to the microbial pollution in the environment, the spread of AMR waterborne bacteria was further investigated on a total number of 166 bacterial strains (30 RWU, 48 WWI, 48 WWE, and 40 RWD isolates).
Antibiotic susceptibility phenotypes were assessed using the disk-diffusion method, according to EUCAST manual (EUCAST 2014). Seven antibiotics were included in AMR testing: ampicillin 10 μg (AMP), amoxicillin-clavulanate 10–2 μg (AMC), cefotaxime 5 μg (CTX), ceftriaxone 30 μg (CRO), cefuroxime 30 μg (CXM), ciprofloxacin 5 μg (CIP), and gentamicin 10 μg (CN). They belong to four categories of antibiotics (penicillins, cephalosporins, quinolones, and aminoglycosides).
Overnight, bacterial cultures were diluted in saline solution to prepare cell suspensions yielding approximately 1.5 × 108 cells/ml, using a 0.5 McFarland standard as a visual comparison. After inoculation and antibiotic disk placement, the Muller-Hinton Agar plates were incubated for 16 to 20 h at 35 ± 1 °C, and the inhibition zones were measured. The breakpoints used to define susceptible and resistant categories for each antimicrobial agent were those recommended by the EUCAST guidelines, version 5 (EUCAST 2015).
2.5 Screening for Class 1 Integron
The Enterobacteriaceae isolates under the study were screened for the presence of class 1 integrons, by PCR detection of the integrase gene intI1 (Márquez et al. 2008), as detailed in Table 1. The PCR reaction mixtures were prepared as previously described (Section 2.3) and subjected to thermal cycling: initial denaturation 94 °C for 5 min followed by 35 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 45 s, and a final extension at 72 °C for 5 min. Positive controls, established based on sequencing analysis, were included in each PCR experimental set. An agarose-gel analysis of the amplified PCR products was performed.
2.6 Statistical Analysis
Frequencies and proportions of AMR isolates, the abundances of multidrug-resistant (MDR), and intI1 bearing strains were calculated for each sampling point and overall. MDR was defined as non-susceptibility to at least one agent in three or more antimicrobial categories (Magiorakos et al. 2012). The results were statistically assessed using the analysis of variance (ANOVA with Bonferroni correction) to find out any significant differences between the phenotypic resistance to antibiotics of Enterobacteriaceae in the four types of waters. Additional comparison was performed to assess the degree of statistical difference between the inhibition zone diameters of the isolates with different origins to each of the tested antimicrobial agents.
Gini-Simpson index of diversity took into account the relative abundance of each phenotype (Guiasu and Guiasu 2012). Resistant and susceptible phenotypes were converted to a binary matrix (1-0), where the intermediate phenotype was considered susceptible. PCoA implemented with the GenAlEx version 6.5 program (Peakall and Smouse 2006, 2012) was carried out to spatially represent the community relationships based on the distance matrix and to detect the differentiation consistency between Enterobacteriaceae populations defined by the cluster analysis.
Inferential statistics by co-dependency was analyzed for strains isolated from each sampling point and also overall. First, it was verified whether occurrences of phenotypic resistance were correlated with int1I carriage. Then, the analysis of covariance (ANCOVA) was used to compare the four clusters of Enterobacteriaceae, with antibiotic susceptibility and the integron carriage as covariates. Two-sided 5 % significance levels were used to identify statistically significant results. Statistical analyses were performed using the Real Statistics Resource Pack software for Microsoft Excel (Zaiontz 2015), with a significance level of p = 0.05.
3 Results and Discussion
3.1 Microbial Loads in River Water and Wastewater Treatment Plant
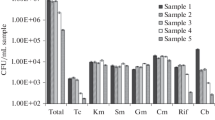
As expected, the highest values for Enterobacteriaceae counts were obtained in raw wastewaters, their number decreasing into the final effluent. As a consequence, the observed log-reduction during the treatment process reached an average value of 2.78 (99.83 %). In surface waters, microbial pollution increased along the Someşul Mic River (Table 2). According to the European Surface Water Abstraction Directive 75/440/EEC, the river water meets the requirements for A2 quality category upstream the city, while for A3 quality category downstream, with respect to coliform counts.
Based on the growth rates and proportions of biochemically confirmed CFUs, T7-TTC Agar and EMB Agar had the highest sensitivity to detect waterborne Enterobacteriaceae in samples with high microbial loads. These culture media, as well as MacConkey Agar, are commonly used for detection of lactose-fermenting coliforms in water. Since not all enterobacteria, as per example Salmonella spp. and Shigella spp., are able to use lactose but glucose (Brenner et al. 2005), VRBG Agar was supposed to yield higher bacterial counts. Instead, the CFU number was lower on VRBG Agar, comparing to the growth rates obtained on lactose-containing media. Also, increased percentages of presumptive colonies on VRBG agar (pink with or without red precipitate) were non-enterobacteria, giving a positive reaction to oxidase testing. Selection of the most adequate methods for detection and isolation environmental Enterobacteriaceae is particularly important, especially in waters with low levels of fecal contamination. Due to the wide diversity of microbial consortia in aquatic ecosystems, a poor selectivity of the culture media may lead to the excessive growth of competing bacteria, creating difficulties for the recovery and discrimination of target colonies. Meanwhile, an intensive selective pressure has the risk of growth inhibition, including for bacteria of interest.
A total number of 187 oxidase-positive strains were selected from T7-TTC agar and subjected to molecular confirmation. Of these, 166 isolates yielded a positive PCR signal for at least one of the Enterobacteriaceae specific genes, lacZ (89.8 %) or 16 s rRNA gene (85.5 %), being further defined as the study strains.
3.2 Antimicrobial Susceptibility of Waterborne Enterobacteriaceae
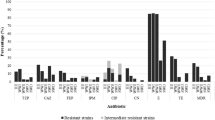
Assessment of antimicrobial susceptibility was performed on 166 strains (30 RWU, 48 WWI, 48 WWE, and 40 RWD isolates). A percentage of 13 % Enterobacteriaceae isolated from Someşul Mic River upstream the city, 21 % from raw wastewaters, 16 % from treated effluents, and only 3 % from river waters downstream the city were entirely susceptible. Most of the isolates were resistant to at least one antibiotic. The maximum number of drugs with observed phenotypic resistance per strain decreased from six antibiotics in RWU and WWI to five antibiotics in WWE and four antibiotics in RWD (Fig. 2). Overall, amoxicillin-clavulanate resistance displayed the highest frequency (71.1 %), followed by ampicillin (63.9 %), cefuroxime (21.1 %), ciprofloxacin (17.5 %), cefotaxime (15.7 %), ceftriaxone (10.8 %), and gentamicin (6.6 %) (Table 3). Unexpectedly, the percentage of strains resistant to ampicillin, ciprofloxacin, and gentamicin was higher in surface waters, comparing to wastewaters.
Lower antibiotic susceptibility was observed in enterobacteria isolated from Someşul Mic River, when compared to European surface waters. Environmental isolates without antibiotic resistance encountered up to 90 % of the isolates in Ardas and Evros rivers, Greece (Alexopoulos et al. 2006), 87 % in Vistula River, Poland (Kotlarska et al. 2015), 69.8 % in Seine estuary, France (Laroche et al. 2009), 34.84 % in Tagus estuary, Portugal (Pereira et al. 2013), and 23 % in Nowohucki Reservoir, Poland (Wolny-Koładka and Lenart-Boroń 2016).
The greatest frequency of multiresistant Enterobacteriaceae was observed in river waters, with 23 % MDR strains in RWU and 30 % in RWD samples. In wastewaters, 10 % of WWI and 9 % of WWE strains were multidrug-resistant (Table 3). Increased frequency of phenotypes intermediate resistant to cefotaxime (17 %), ciprofloxacin (13 %), ceftriaxone (7 %) in RWU and gentamycin (50 %), ciprofloxacin (10 %), cefotaxime (5 %), ceftriaxone (5 %) in RWD, respectively, contributed to the elevated rates of MDR in surface waters. Lower levels of intermediate resistance were detected in WWI isolates (4 % intermediate resistant to ceftriaxone) and WWE strains (6 % intermediate resistance to ceftriaxone, while 2 % intermediate resistant to ciprofloxacin).
Conclusions reported by other authors describe increased frequencies of AMR and MDR bacteria in water samples collected downstream the WWTPs in comparison to upstream river samples, being related to effluent discharge, where the highest percentages were usually found (Ferreira da Silva et al. 2007; Koczura et al. 2012). The findings of this study indicate important proportions of resistant (87 %) and multiresistant (23 %) enterobacteria in upstream river water and need to be further investigated. The sources of environmental resistance may reside in the contaminant resistome, natural resistome, or both. Increased levels of fecal pollution have been previously reported to occur in the upper basin of Someşul Mic River (Farkas et al. 2010a, b; Muntean et al. 2010). At the same time, contamination with antibiotic resistance genes was recently shown to be widely spread and persistent, even in the absence of a positive selection pressure (Martínez 2012). Bacterial dynamics through the urban water cycle as well as genome variations triggered by the acquired, intrinsic, and silent resistance may have wide implications on the ecology of antibiotic-resistant bacteria (Vaz-Moreira et al. 2014).
The most prevalent AMR patterns in waterborne Enterobacteriaceae were AMP-AMC, followed by AMC for all sites (Fig. 3). Surveillance of antibiotic resistance in wastewater was suggested to be a valuable tool for the screening of resistance trends in human population (Kwak et al. 2015). Indeed, the results of the present study indicated high similarities between the AMR profiles of waterborne bacteria and the antibiotic usage in humans. The high proportion of resistant waterborne strains correlates with integrated data indicating the antibiotic overuse in human population, at country level. Romania holds the second place in Europe regarding the consumption of antibacterials. Penicillins, cephalosporins, and other beta-lactam antibiotics, followed by quinolones, are the most frequently used in the therapy of infections (Popescu et al. 2013; ECDC 2014). Two other studies assessing AMR in E. coli strains isolated from Romanian surface waters also indicated high levels of resistance to beta-lactams, in Arieş River (Bodoczi Florea 2011) and in Dâmboviţa River (Marinescu et al. 2015). On the other hand, antibiotic resistance mediated by beta-lactam resistance encoding genes is the most widespread resistance mechanism, with more than 1000 enzymes identified. Even though the beta-lactamases have been present for millions of years, their extent diversity seems to be the consequence of both the recent evolution in response to the clinical use of antibiotics and also generated by the environmental resistome (Galán et al. 2013).
To deeply assess the degree of diversity in AMR patterns into the study strains, the Gini–Simpson was used. It accounted for the relative abundance of each antibiotic resistance phenotype in each type of water, as detailed in Table 4. The greatest value of Gini–Simpson coefficient (0.80) indicated the highest diversity of antibiotic resistance phenotypes in Enterobacteriaceae cells recovered from river waters upstream the city. Less diversity was detected in wastewater inlet (0.75) and outlet (0.76), as well as in river water downstream the city (0.77). Additionally, the principal coordinate analysis (PCoA) showed a wide variability of AMR patterns in RWU and WWI enterobacteria. The isolates in the four groups did not cluster independently, meaning that the resistance phenotype profiles in waterborne Enterobacteriaceae were similar, with the evidence that RWD patterns resulted as a subgroup of the resistance phenotypes deriving from the river upstream and from the wastewater treatment plant (Fig. 4).
Comparing the four clusters of Enterobacteriaceae (ANOVA with Bonferroni correction), it seemed that no significant difference occurred with respect to the phenotypic resistance, when accounting the total number of antibiotics displaying resistance per strain (p = 0.206). Also, no statistical significant difference between the four groups was observed (p = 0.119) when considering the resistance to each drug. Therefore, a more detailed statistical assay was performed to compare the seven antibiotic susceptibilities apart. The analysis of variance for the four clusters of Enterobacteriaceae indicated significant differences between the inhibition zone diameters in the testing of AMP (p = 0.018), AMC (p = 0.0007), CIP (p = 0.001), and CN (p = 1.2 × 10−20). No significant differences were observed in the testing of cephalosporins: CTX (p = 0.185), CRO (p = 0.058), and CXM (p = 0.126).
3.3 Association Between Antibiotic Resistance and Class 1 Integron Carriage
For further analysis, the observed phenotypic resistance to antibiotics was compared with class 1 integron-integrase gene carriage into the study strains. Int1I was detected in 21.7 % of the isolates, with the highest frequency in WWI (43.75 %) and the lowest in WWE (9 %) strains. The spread of Int1I-bearing enterobacteria was lower in river waters upstream (13 %) than downstream (16 %) the city (Fig. 2). Comparing to other studies, the proportions of class 1 integron-positive Enterobacteriaceae were higher in wastewater affluent and also in river water. Among the Escherichia spp., Ferreira da Silva et al. (2007) found a similar frequency of class 1 integrons in the raw (10.0 %) and treated (9.6 %) wastewater. The integron-positive isolates made up 11 % of E. coli isolates recovered from the WWTP, while 6 and 14 % of isolates cultured from upstream and downstream river water in a study conducted by Koczura et al. (2012). The incidence of the intI1 gene found by Mocraka et al. (2012) among Enterobacteriaceae originated from sewage was 22.5 %, while 9.1 % in the final effluent. In surface waters, the intI1 gene was detected in 11.36 % E. coli isolates from Tagus estuary (Pereira et al. 2013) and up to 14.7 % in Seine estuary (Laroche et al. 2009). Assessing for the presence of class 1 integrons only in resistant E. coli, up to 38.3 % positive strains were detected in raw wastewater, whereas up to 37.1 % in treated wastewater, and 27.6 % in Vistula River, respectively (Kotlarska et al. 2015).
The observed significant reduction of the intI1-bearing strains during wastewater treatment needs special attention. Wastewater treatment procedures were described as inefficient in the removal of AMR carrying resistance genes, which were released to the receiving surface waters (Czekalski et al. 2012; Munir et al. 2011; Mocraka et al. 2012). The WWTP investigated in this study was recently upgraded with a tertiary stage for nitrogen and phosphorus removal, but the process does not include disinfection and was not designed to remove emergent contaminants such as DNA elements. The present evaluation indicates good performances for the reduction of integron-bearing Enterobacteriaceae (log-reduction value 0.68, meaning 79.22 %).
The association between multiresistance and prevalence of class 1 integron was previously described (Koczura et al. 2012; Pereira et al. 2013), but not all authors did find a significant correlation (Laroche et al. 2009). In this investigation, we only found a weak positive correlation between MDR and the presence of intI1 in wastewater effluent (r = 0.39; p = 0.06), but no sufficient evidence of a linkage between phenotypic AMR and int1I gene carriage, overall, although the comparison of the four clusters of Enterobacteriaceae revealed no significant difference between groups, with respect to the emergence of resistant strains bearing the class 1 integron integrase (p = 0.166, ANCOVA). Also, the Int1I gene presence in 12 % of the entirely susceptible strains is worth mentioning. Class 1 integrons have been widely associated with anthropogenic pressure, as well as with acquisition and dissemination of antibiotic resistance genes, but our findings demonstrate that it is not possible to explain the antibiotic-resistance phenotype based on int1I carriage only.
4 Conclusions
A high proportion of waterborne Enterobacteriaceae displayed antibiotic resistance, especially to penicillins, which are the most frequently used antibiotics in Romania. A decrease in coliform counts but a general increase in AMR and multiresistant bacteria occurred during the wastewater treatment. The most important, a significant reduction of the intI1-bearing cells occurred. Performances for the reduction of key DNA elements need special attention, as valuable outcomes could be driven for new strategies in order to reduce the incidence and spread of emergent contaminants in the environment.
The frequencies of MDR strains in Someşul Mic River, both upstream and downstream the Cluj-Napoca city, were greater compared to those found in wastewaters. Further investigations are needed to deeply analyze if the natural resistome or other sources of contamination are involved.
A weak positive correlation between MDR and the presence of intI1 was only driven in wastewater effluent, but no sufficient evidence was found on the linkage between phenotypic AMR and int1I gene carriage. The presence of class 1 integron can be merely associated with anthropogenic pressure, but the simple detection of intI1 gene cannot explain the complex antibiotic resistance phenotype.
References
Abraham, W. R. (2011). Megacities as sources for pathogenic bacteria in rivers and their fate downstream. International Journal of Microbiology, 2011, 798292.
Alexopoulos, A., Voidarou, C., Stefanis, C., Papadopoulos, I., Vavias, S., Tsiotsias, A., Kalkani, E., Charvalos, E., & Bezirtzoglou, E. (2006). Antibiotic resistance profiles and integrons in Enterobacteriaceae from the riverside of Evros-Ardas with respect to chemical and waste pollution. Microbial Ecology in Health and Disease, 18(3–4), 170–176.
Amábile-Cuevas, C. F. (2016). Antibiotics and antibiotic resistance in the environment. Boca Raton: CRC Press.
Bodoczi Florea, A. (2011). Antimicrobial susceptibility of Escherichia coli isolated from Arieş River (Romania). Analele Universităţii din Oradea - Fascicula Biologie, 18(1), 34–38.
Brenner, D. J., Farmer, J. J., III, & Family, I. (2005). Enterobacteriaceae. In G. M. Garrity, D. J. Brenner, N. R. Krieg, & J. T. Staley (Eds.), Bergey’s manual of systematic bacteriology, Vol 2, Part B (pp. 587–850). New York: Springer.
Czekalski, N., Berthold, T., Caucci, S., Egli, A., & Bürgmann, H. (2012). Increased levels of multiresistant bacteria and resistant genes after wastewater treatment and their dissemination into Lake Geneva, Switzerland. Frontiers in Microbiology, 3, 106.
ECDC. (2014). Summary of the latest data on antibiotic consumption in the European Union. Stockholm: European Centre for Disease Prevention and Control.
ECDC. (2015). Antimicrobial resistance (AMR) reporting protocol. European Antimicrobial Resistance Surveillance Network (EARS-Net) surveillance data for 2014. Stockholm: European Centre for Disease Prevention and Control.
EUCAST. (2014). Antimicrobial susceptibility testing. EUCAST disk diffusion method. Stockholm: European Committee on Antimicrobial Susceptibility Testing.
EUCAST. (2015). Breakpoint tables for interpretation of MICs and zone diameters, Version 5. Stockholm: European Committee on Antimicrobial Susceptibility Testing.
Farkas, A., Bocoş, B., Ţigan, Ş., Ciatarâş, D., Drăgan-Bularda, M., & Carpa, R. (2010a). Surveillance of two dam reservoirs serving as drinking water sources in Cluj, Romania. In M. A. Dimkic (Ed.), Balkans Regional Young Water Professionals Conference Proceedings (pp. 91–97). Belgrade: Jaroslav Černi Institute for the Development of Water Resources.
Farkas, A., Ciatarâş, D., Bocoş, B., & Ţigan, Ş. (2010b). Monitoring of water source Gilău and its affluent Someşul Rece during 2005–2009. Applied Medical Informatics, 26(1–2), 27–34.
Farkas, A., Drăgan-Bularda, M., Ciatarâş, D., Bocoş, B., & Ţigan, Ş. (2012). Opportunistic pathogens and faecal indicators in drinking water associated biofilms in Cluj, Romania. Journal of Water and Health, 10, 471–483.
Farkas, A., Butiuc-Keul, A., Ciatarâş, D., Neamţu, C., Crăciunaş, C., Podar, D., & Drăgan-Bularda, M. (2013). Microbiological contamination and resistance genes in biofilms occurring during the drinking water treatment process. Science of the Total Environment, 443, 932–938.
Ferreira da Silva, M., Vaz-Moreira, I., Gonzalez-Pajuelo, M., Nunes, O. C., & Manaia, C. M. (2007). Antimicrobial resistance patterns in Enterobacteriaceae isolated from an urban wastewater treatment plant. FEMS Microbiology Ecology, 60(1), 166–176.
Figueira, V., Serra, E. A., Vaz-Moreira, I., Brandão, T. R., & Manaia, C. M. (2012). Comparison of ubiquitous antibiotic-resistant Enterobacteriaceae populations isolated from wastewaters, surface waters and drinking waters. Journal of Water and Health, 10(1), 1–10.
Galán, J. C., González-Candelas, F., Rolain, J. M., & Cantón, R. (2013). Antibiotics as selectors and accelerators of diversity in the mechanisms of resistance: from the resistome to genetic plasticity in the β-lactamases world. Frontiers in Microbiology, 4, 9.
Gillings, M. R., Gaze, W. H., Pruden, A., Smalla, K., Tiedje, J. M., & Zhu, Y. G. (2014). Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. The ISME Journal, 9, 1269–1279.
Guiasu, R. C., & Guiasu, S. (2012). The weighted Gini-Simpson index: revitalizing an old index of biodiversity. International Journal of Ecology, 2012, 10.
Huddleston, J. R. (2014). Horizontal gene transfer in the human gastrointestinal tract: potential spread of antibiotic resistance genes. Infection and Drug Resistance, 7, 167–176.
Koczura, R., Mokracka, J., Jabłońska, L., Gozdecka, E., Kubek, M., & Kaznowski, A. (2012). Antimicrobial resistance of integron-harboring Escherichia coli isolates from clinical samples, wastewater treatment plant and river water. Science of the Total Environment, 414, 680–685.
Kotlarska, E., Łuczkiewicz, A., Pisowacka, M., & Burzyński, A. (2015). Antibiotic resistance and prevalence of class 1 and 2 integrons in Escherichia coli isolated from two wastewater treatment plants, and their receiving waters (Gulf of Gdansk, Baltic Sea, Poland). Environmental Science and Pollution Research, 22, 2018–2030.
Kwak, Y. K., Colque, P., Byfors, S., Giske, C. G., Möllby, R., & Kühn, I. (2015). Surveillance of antimicrobial resistance among Escherichia coli in wastewater in Stockholm during 1 year: does it reflect the resistance trends in the society? International Journal of Antimicrobial Agents, 45(1), 25–32.
Laroche, E., Pawlak, B., Berthe, T., Skurnik, D., & Petit, F. (2009). Occurrence of antibiotic resistance and class1, 2 and 3 integrons in Escherichia coli isolated from a densely populated estuary (Seine, France). FEMS Microbiology Ecology, 68(1), 118–130.
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., Harbarth, S., Hindler, J. F., Kahlmeter, G., Olsson-Liljequist, B., Paterson, D. L., Rice, L. B., Stelling, J., Struelens, M. J., Vatopoulos, A., Weber, J. T., & Monnet, D. L. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection, 18, 268–281.
Maheux, A. F., Boudreau, D. C., Bisson, M. A., Dion-Dupont, V., Bouchard, S., Nkurang, M., Bergeron, M. G., & Rodriguez, M. J. (2014). Molecular method for detection of total coliforms in drinking water samples. Applied and Environmental Microbiology, 80(14), 4074–4084.
Marinescu, F., Marutescu, L., Savin, I., & Lazar, V. (2015). Antibiotic resistance markers among Gram-negative isolates from wastewater and receiving rivers in South Romania. Romanian Biotechnological Letters, 20(1), 10055–10069.
Márquez, C., Labbate, M., Raymondo, C., Fernández, J., Gestal, A. M., Holley, M., Borthagaray, G., & Stokes, H. W. (2008). Urinary tract infections in South American population: dynamic spread of class 1 integrons and multidrug resistance by homologous and site-specific recombination. Journal of Clinical Microbiology, 46(10), 3417–3425.
Martínez, J. L. (2012). Natural antibiotic resistance and contamination by antibiotic resistance determinants: The two ages in the evolution of resistance to antimicrobials. Frontiers in Microbiology, 3, 1.
Mocraka, J., Koczura, R., & Kaznowski, A. (2012). Multiresistant Enterobacteriaceae with class 1 and class 2 integrons in a municipal wastewater treatment plant. Water Research, 46, 3353–3363.
Munir, M., Wong, K., & Xagoraraki, I. (2011). Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Research, 45(2), 681–693.
Muntean, V., Maier, C. G., Carpa, R., & Farkas, A. (2010). Microbiological and enzymological study on sediments and water of the river Someşul Mic upstream the Gilău (Cluj County) treatment plant. Studia Universitatis Babes-Bolyai, Biologia, 55(1), 131–138.
Mureşan, C., Farkas, A., Vele, D., & Chakirou, C. (2010). Study on private water sources (dug wells and springs) in Cluj County. Romaqua, 73, 38–42.
Peakall, R., & Smouse, P. E. (2006). GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6, 288–295.
Peakall, R., & Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics, 28, 2537–2539.
Penders, J., Stobberingh, E. E., Savelkoul, P. H., & Wolffs, P. F. G. (2013). The human microbiome as a reservoir of antimicrobial resistance. Frontiers in Microbiology, 4, 87.
Pereira, A., Santos, A., Tacão, M., Alves, A., Henriques, I., & Correia, A. (2013). Genetic diversity and antimicrobial resistance of Escherichia coli from Tagus estuary (Portugal). Science of the Total Environment, 461–462, 65–71.
Popescu, G. A., Mathyas, L., Ciolan, C., Serban, R., & Pistol, A. (2013). Antibacterial consumption in Romania in 2012: specific features and quality indicators for community usage. l. BMC Infectious Diseases, 13(Suppl 1), O17.
Rolain, J. M. (2013). Food and human gut as reservoirs of transferable antibiotic resistance encoding genes. Frontiers in Microbiology, 4, 173.
Vaz-Moreira, I., Nunes, O.C., Manaia, C.M. (2014). Bacterial diversity and antibiotic resistance in water habitats: searching the links with the human microbiome. FEMS Microbiology Reviews, 38(4), 761–778.
Vigh, M., & Pandi, G. (2013). Quality parameters for Someşul Mic River tributaries in Cluj-Napoca. Present Environment and Sustainable Development, 7(2), 123–131.
WHO. (2014). Antimicrobial Resistance. Global Report on Surveillance. Geneva, Switzerland: WHO Press, World Health Organization.
Wise, R., Hart, T., Cars, O., Streulens, M., Helmuth, R., Huovinen, P., & Sprenger, M. (1998). Antimicrobial resistance. Is a major threat to public health. British Medical Journal, 317(7159), 609–610.
Wolny-Koładka, K., & Lenart-Boroń, A. (2016). Phenotypic and molecular assessment of drug resistance profile and genetic diversity of waterborne Escherichia coli. Water Air Soil Pollution, 227, 146.
Zaiontz, C. (2015). Real statistics using Excel. www.real-statistics.com.
Acknowledgments
This work has been partially supported by the grant RO-04 66230, Methodological guide for monitoring antibiotic residues and antibiotic resistance in the environment as a supporting instrument for an enhanced quality management of surface waters and groundwater (EnviroAMR), financed by Iceland, Liechtenstain and Norway through the EEA Financial Mechanism.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Farkas, A., Bocoş, B. & Butiuc-Keul, A. Antibiotic Resistance and intI1 Carriage in Waterborne Enterobacteriaceae . Water Air Soil Pollut 227, 251 (2016). https://doi.org/10.1007/s11270-016-2944-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11270-016-2944-6