Abstract
Bovine leukemia virus is a retrovirus that causes enzootic bovine leukosis and is associated with global economic losses in the livestock industry. The aim of this study was to investigate the genotype determination of BLVs from cattle housed in 6 different farms in Türkiye and the characterization of their LTR and pX (tax, rex, R3, and G4 gene) regions. For this purpose, blood samples from 48 cattle infected with BLV were used. The phylogenetic analysis based on the env gene sequences revealed that all BLVs were clustered in genotype 1 (G1), and the sequences of the LTR (n = 48) and the pX region (n = 33) of BLVs were obtained. Also, analysis of these nucleic acid and amino acid sequences allowed assessments similar to those reported in earlier studies to be relevant to transactivation and pathogenesis. This study reports the molecular analysis of the LTR and pX region of BLVs in Türkiye for the first time.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bovine Leukemia Virus (BLV) is a retrovirus that causes "Enzootic Bovine Leukosis" (EBL), which causes tumor formation in almost all organs of cattle older than two years and an increase in lymphocyte counts, resulting in significant yield loss in cattle [1]. BLV infection may be clinically quiet in cattle in the aleukemic (AL) state or manifest as persistent lymphocytosis (PL), defined by an increase in the number of B lymphocytes, and, less frequently, as B-cell lymphomas in lymph nodes after such a long latency period [2].
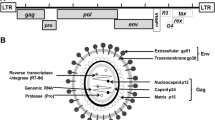
BLV is a member of the Ortervirales order, Retroviridae family, Orthoretrovirinae subfamily, and the Deltaretrovirus genus [3]. The genome consists of a total of 8714 nucleotides containing the structural and enzymatic proteins, the pX region (tax, rex, R3, and G4), and two 'Long Terminal Repeat' (LTR) [4, 5]. In addition, the presence of two transcripts, AS1 (Antisense 1) and AS2 (Antisense 2), [6] and five microRNAs (miRNAs) encoded between the gp51 (env) and R3 genes were detected in BLV genome [7, 8]. LTR regions flank the 5' and 3' ends of the BLV genome. Each LTR has three regions named U3, R, and U5. U3 contains several regulatory elements essential to the regulation of viral promoter and promoter activity and plays a crucial role in triggering BLV transcription [9, 10]. It has been shown that sequence variations in the BLV-LTR region modify interactions with transcription factors, leading to changes in viral gene expression and replication [11]. The BLV contains a pX region containing 3' segments of genes encoding four regulatory proteins in overlapping reading frames. This region contains Tax, an activator of viral and cellular transcription that is responsible for tumor progression; Rex, which posttranscriptionally binds and maintains viral RNAs and enables their translation; and gene regions encoding the R3 and G4 helper proteins, which are likely responsible for sustaining viral loads during persistent infection [5, 12].
The gene region encoding BLV gp51 (env) is used in BLV genotyping studies, and these studies have demonstrated that BLV can be grouped into a minimum of 12 genotypes (from genotype 1 to genotype 12) [13,14,15]. BLV infections have been studied in Türkiye for a long time, with prevalence rates in earlier studies [16,17,18,19], the studies on the genotyping of BLV circulating in cattle are very limited [20,21,22].
In this study, we have fulfilled i) to determine the genotype(s) of BLVs from animals on six farms in different years (2012–2017), ii) to analyze the sequences of the other two gene regions of BLVs detected, the LTR and the pX regions, and iii) to evaluate these sequences in the light of knowledge about the occurrence of various infection patterns (as AL and PL) and possible pathogenetic mechanisms.
Materials and methods
History of study and selection of the samples
In this study, retrospective blood samples from 48 cattle that were found to be positive for BLV infection by antibody ELISA were used. These cattle were from six farms located in different provinces (Kırklareli, Balıkesir, Bursa, Ankara, Şanlıurfa, and Adana) (Table 1). The env gene sequences of BLVs from 11 samples used in this study were reported in a previous study [20]. Besides the env gene sequences of the remaining BLV strains (n = 37), this study aimed to analyze the sequences related to the LTR and pX regions of BLVs in all samples.
Isolation of viral nucleic acid and PCRs
After the centrifugation of blood samples, total DNA was extracted using the phenol–chloroform method [23] and/or the commercial extraction kit (QIAGEN Qiamp DNA Mini Kit (Cat. No.:51304) following the manufacturer’s protocol. Nested PCRs and PCRs for the amplification of the target gene region were performed with the primers [24,25,26,27] and methods previously described (Supplement 1), using Taq DNA Polymerase (#EP0401; Thermo Scientific, USA) for the env gene and DreamTaq DNA Polymerase (#EP0702; Thermo Scientific, USA) for the LTR and pX regions. The reaction mixture consisted of 19.85 μl nuclease-free water, 2.4 μl MgCl2 (25 mM), 3 μl 10 × Taq buffer, 0.5 μl of each primer, 0.5 μl dNTP (10 mM each) (R0192; Thermo Scientific, USA), 0.25 μl Taq polymerase (500 U/μl), and 3 μl DNA for the env gene. For the LTR and pX regions, 2.5 µl of DNA was added to a mixture of 2.5 µl of 10X DreamTaq buffer, 0.5 µl of dNTP (10 mM each) (#R0192; Thermo Scientific, USA), 1 µl of each primer, 0.25 µl of DreamTaq DNA polymerase (5 U/µl), and 17.25 µl nuclease-free water.
For the BLV env gene, the procedure was as follows: after the appropriate PCR mixtures, initial denaturation at 95 °C for 5 min, and finally, 35 amplification cycles of amplification [at 95 °C for 30 s, 62 °C for the 30 s (for the first round; 598 bp), 58 °C for 30 s (for the second round; 444 bp), 72 °C for one min] and a final extension step at 72 °C for 10 min.
The nested PCR program for the LTR gene amplification was as follows for both rounds: an initial denaturation phase at 95 °C for 4 min, the cycles of amplifications [ at 95 °C for 30 s, 52 °C for the 30 s (for both rounds), 72 °C for one min] and a final extension step at 72 °C for 10 min. Each PCR cycle consisted of 35 cycles.
For the detection of the pX region, three primer pairs overlapping each other were used, and the corresponding products were obtained. The procedure for PCRs is as follows: initial denaturation at 95 °C for 4 min, and finally, 35 amplification cycles of amplification [at 95 °C for the 30 s, at annealing steps 59 °C (for pXBLV1F/pXBLV1R) and 56 °C (for pXBLV2F/pXBLV2R and pXBLV3F/pXBLV3R) for the 30 s, 72 °C for one min] and a final extension step at 72 °C for 10 min.
Genome sequencing and phylogenetic analysis
After the PCRs, amplicons were visualized using gel electrophoresis. Nucleotide sequencing of the amplicons with expected sizes (∼ 444 bp for env gene, 571 bp for LTR gene, 609 bp for pX1, 669 bp for pX2, and 478 bp for pX3 (for pX region, three primer pairs overlapping each other were used)) were obtained. Products of expected sizes obtained from PCR were sequenced by the Sanger sequencing method in both directions with the same primers used for amplification. It was conducted by a commercial company. The BLV genome sequence data acquired in this work were validated using the NCBI (National Center for Biotechnology Information)-provided BLAST (Basic Local Alignment Search Tool) website. The obtained sequences were evaluated using the multiple alignment function of the Aliview program [28] and compared with the BLV genome sequences found in GenBank.
MEGA X (Molecular Evolutionary Genetics Analysis across Computing Platforms) was used to build phylogenetic trees of all gene regions studied [29]. The best-fitting models were identified using the "Find Best Model" function in the program. Phylogenetic analysis were conducted using the maximum likelihood method with the Kimura-2 parameter plus gamma distribution, the Tamura-3 parameter plus gamma distribution, and the Tamura-Nei parameter plus gamma distribution for the env, LTR, and pX regions, respectively. Phylogenetic distances were statistically supported by bootstrapping with 1,000 replicates. The nucleotide (nt) and amino acid (aa) identities were calculated by using the SIAS online tool [30].
Results
As a result of PCRs and sequence analysis for the gene regions studied, nucleotide sequences of the env gene for 32 BLVs could be obtained; also, 48 and 33 strains provided the nucleotide sequences for the LTR and pX regions, respectively (Table 1). The sequences were deposited in the GenBank database, and the accession numbers are presented in Supplement 2.
Analysis of sequences and phylogenetic trees
In this part, phylogenetic analysis and pairwise comparison of nucleotide sequences of our strains and strains deposited in Genbank and the nucleotide sequences of some regions in the LTR, env, and Tax gene regions were examined in detail.
Phylogenetic analysis and pairwise comparison
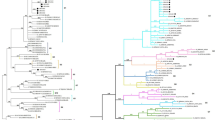
The phylogenetic analysis based on nucleotide sequences of the env gene of the strains revealed that all strains clustered in Genotype 1 (Fig. 1). When the phylogenetic analyses performed with the samples obtained in the study are examined, it is observed that most of the sequences obtained from the same farms did not cluster together and are not specific to a particular geographic region. Similarly, our strains were also included with genotype 1 viruses in the phylogenetic trees for LTR (n = 48) and tax gene region (n = 33) (Supplement 3).
Phylogenetic analysis of BLV partial env gene sequences of local viruses in this study and BLV env gene sequences from other countries (genotypes 1–12) in GenBank. The phylogenetic tree was generated using the maximum likelihood method provided by MEGAX software. Phylogenetic distances were statistically supported by bootstrapping with 1,000 replicates. The naming of local BLVs in the study is “sample code (Farm code-Sample No)/Sampling Year/Country”. The positions of BLV strains identified in this study are indicated by the icons showing every farms ( ■: TG, □: TH, ▲: KRC, ●: P, ♦: CP, ○: CKR). The scale bar indicates nucleotide substitutions per site. The genotypes are indicated at the right-hand side of each phylogenetic tree
The nucleotide (LTR) and amino acid (env, tax, rex, R3, and G4) identities of strains in this study were calculated by the online tool SIAS [30]. The identities of the partial env, LTR, and tax gene sequences from BLVs studied with the reference strain K02120 were 98.59–100%, 98.84–99.62%, and 98.7–99.67%, respectively. Also, according to the analysis of the partial env, LTR, and tax gene sequences, the local BLVs shared 97.18–100%, 98.69–100%, and 98.05–100% sequence identity to each other, 90.84–100%, 96.77–100%, and 92.53–100% sequence identity to other countries sequences used in the trees (Supplement 2), respectively.
The amino acid identities of the rex, R3, and G4 gene region sequences of strains obtained in the study with the reference strain K02120 were 98.64–100%, 96.29–100%, respectively. The sequences mentioned (the rex, R3, and G4 gene regions, respectively) of the local viruses showed 95.28–100%, 97.29–100%, 92.59–100%, and 92.45–100% identities with each other, and also 89.86–100%, 77.77–100%, and 92.45–100% identities with the sequences from other countries (Supplement 2).
Analysis of the amino acid sequences
Env gene sequences
Amino acid sequences of 32 partial env gene were aligned to obtain information on the genetic diversity of the BLVs detected in this study. 11 different amino acid substitutions were observed in 12 different local viruses, whereas the same aa sequence as in the reference strain K02120 was found in the remaining 20 BLVs. When these aa substitutions are summarized by functional regions; In the "zinc-binding peptide" and "2. neutralizing domain" regions H148Y replacement in three samples (KRC-S11, KRC-9, KRC-K10); H148L substitution in three samples (TG-5, TG-9, TG-8430) were observed. In the E epitope, S189G substitution was observed in TG-8304. In the CD4 + epitope, P112A substitution was identified in TH-37 (Supplement 4).
LTR region sequences
The LTR region of BLV comprises three regions called U3, R, and U5 which plays a crucial role in virus replication and infection pathogenesis. Analysis of the nucleotide sequences of the U3, R, and U5 regions with those of K02120 revealed some nucleotide substitutions. These are A69T in three samples (TH-37, TH-39, P-6), G148A in seven samples (TG-5, TG-9, TG-8430, KRC-52, KRC-55, KRC-S12, KRC-K13), T175C in 15 samples (TG-10, TG-18, TG-8304, KRC-70A, KRC-S5, KRC-S11, KRC-S15, KRC-5, KRC-9, KRC-2, KRC-10, P-8, P-14, P-31, P-34 (Table 2).
pX region sequences
The BLV contains a pX region containing 3' segments of genes encoding four regulatory proteins (Tax, Rex, R3, and G4) in the form of overlapping reading frames. A detailed analysis of amino acid sequences in the functional regions of every gene region coding these proteins revealed some aa substitutions at the local viruses in this study. All analyses were performed under the guidance of the K02120 strain.
In the amino acid analysis of the Tax gene; Q40R (for KRC-56), E42K (for TG-5, TG-8, TG-11, TG-8430, CKR-6, CKR-7, CKR-8, CKR-9, and CKR-11) and R43K (for KRC-S15 and CKR-7) were detected in the region "Zinc finger". In the "T-cell epitope" region, three different mutations were detected. These are; C118Y for CP-2, a V146L aa for TG-8430, and a V146I for KRC-55. In the "CD8 + CTL epitope" region, an I186V aa substitution was detected in two samples (P-5, P-6) and a C196R aa substitution in TG-11 (Table 3).
The analysis of the rex gene also indicated the presence of some substitutions. In the NES region, which is thought to be essential for Rex protein function, an S74L substitution was detected in seven samples (TH-29A, TH-34, TH-39, CP-1, CP-64, KRC-S12, KRC-K13), an R77C substitution in two samples (TG-11, KRC-S8), a D81N substitution in two samples (TG-9, TG-8430) were detected (Fig. 2a).
pX region contains several open reading frames for Tax, Rex, R3, and G4 regulatory proteins. The analysis of the G4 gene region revealed an F8L change in KRC-52 and an F22S change in KRC-56 in the "leader peptide" region, which is encoded by the overlapping part with the LTR region and is defined as the functional region encoded in the sequences of the BLV pX region, and the A65V aa change in KRC-S15 was observed. In addition, three aa insertions (LLP) were detected between the 50th and 51st aa in KRC-56. In samples TG-8 and TG-8090, no aa was synthesized from these codons due to the deletion in the 49th and 50th position nucleotides. Although there was no functional region in 16 of the 33 samples from which the pX region/G4 gene sequence information was obtained, an H > L change was persistently observed at the 52nd amino acid. In addition, it was found that the mentioned H > L substitution was found in four other strains from other countries (Fig. 2b).
There is no functional area identified in the R3 gene region yet. The analysis of the aa sequences of the R3 protein of local strains under the guidance of reference strain K02120, R31S change in five samples (TH-29A, TH-34, TH-39, CP-1, CP-64) identified (Fig. 2c). In summary from the data, mutations in some regions of our BLVs, which were important for the pathogenesis of the BLV infection reported by other researchers, are presented in Table 4.
Discussion
BLV is common in many countries, with prevalence rates varying from country to country, and spreads through trade between countries, causing significant economic losses. The literature shows an interaction between some geographical regions and genotypes of BLV. Thus, genotypes 2, 5, 6, and 9 were found mostly in South American countries, genotypes 7 and 8 in Russia and Eastern European countries, genotype 10 in Thailand, China, and Myanmar, and genotypes 1, 3, and 4 on almost all continents [13, 31,32,33,34,35,36,37]. Genotype 11 and 12, most recently identified, was detected in China and Kazakhstan, respectively [14, 15]. While BLV infections have been detected in Türkiye, with prevalence rates in earlier studies [16,17,18,19], the studies limited on the genotyping of BLV circulating in cattle [20,21,22] have revealed the existence of two distinct genotypes (G1 and G4). This study included animal materials (n = 48) in 6 organized farms located in different provinces between 2012 and 2017. Similar to those presented in the previous study [20], which reported the genotypes of BLVs (n = 16) from the same farms (coded as TG, TH, KRC, P, CP, CKR in Table 1) sampled in this study, the results from our BLVs (n = 32) also revealed all strains belonged to genotype 1 (Fig. 1). When the phylogenetic analyses performed with the samples obtained in the study are examined, it is observed that most of the sequences obtained from the same farms did not cluster together and are not specific to a particular geographic region. This may be caused by the mentioned farms' intense animal mobility, including the entry of imported animals. In this study, in addition to the env gene sequences of different animals (n = 32) obtained from the same herds, the LTR and pX (tax, rex, R3, and G4) gene region sequences, which were not performed in the previous study [20], were obtained and analyzed for nucleic acid and amino acid sequences. Also, phylogenetic analyses for the LTR gene region and tax gene were performed. Generally, env gene sequences are used in genotyping studies of BLVs. In the phylogenetic trees for LTR (Supplement 3a) and tax (Supplement 3b), local viruses are also located in G1, such as Japan, the USA, Vietnam, and Uruguay.
In addition to identifying the genotype(s) circulating in a herd or nation, providing information about genetic sequence variation of the parts of the BLV genome and their impact on disease outcome is an important subject of this research. Therefore, an objective of this study was to assess the possible relationship between disease progression and sequence variation of the BLV Env, Tax, and LTR regions in infected animals.
The glycoprotein Env gp51 promotes the production of neutralizing antibodies and comprises seven epitopes, denoted G, H, F, E-E', B-B', D-D', and A, three of which (G, H, and F) are conformational epitopes that are significant determinants of viral neutralization and suppression of syncytium formation [38]. In this study, molecular characterization of Env gp51 of BLV viruses revealed some mutations in functional/nonfunctional regions. The aa changes that occur as a result of these mutations are D128E, N129H, H148Y/L, and S189G (Supplement 4). As a result of comparing the sequences in this study with the reference strain K02120, it was found that epitopes B, G, and F were conserved.
Three neutralization domains (ND1, ND2, and ND3) identified in BLV gp51 were reported to induce BLV-neutralizing antibodies [38]. While ND1 and ND3 were conserved from the neutralization domains in the native viruses in the study, two amino acid substitutions (H148Y and H148L) were detected at two different positions in ND2 (Supplement 4). Even though it is not clear how these mutations affect infectivity, it is thought that they may affect viral fusion and in vivo infectivity of BLV [38, 39].
The LTR region of the BLV genome consists of three regions called U3, R, and U5. The majority of BLV mutations have been found in the LTR region which contains the viral promoter and enhancer [10, 40]. The PU.1/Spi-B element, which serves as the binding site for the GRE and ETS that sensitize to dexamethasone in the presence of Tax, is one of several response elements found in the U3 region [10]. Zhao et al. [25] reported that the G148A change in the GRE region observed in their strains could potentially affect the glucocorticoid response of BLV. The 5' LTR genome sequences of three BLV lymphosarcoma isolates (LS1, LS2, and LS3) were compared with the complete BLV genome sequences available at the time by Moratorio et al. [26]. They reported that samples isolated from LS1 and other isolates had a change at the same location (G148A) in the GRE binding site. It is hypothesized that the base activity of LTR is greatly decreased by GRE mutation in the absence of Tax, which may result in enhanced viral transcription suppression in lymphosarcoma strains as a defense against host immune response. It is noteworthy that G148A mutations in the region GRE were detected in seven local viruses (TG-5, TG-9, TG-8430, KRC-52, KRC-55, KRC-S12, KRC-13) (Table 2).
The promoter boxes "CAT" and "TATA" are also in the U3 region. Using a reverse genetics technique, researchers [40] examined how different nucleotide alterations in the TATA box area affected BLV transactivation. They found that spontaneous mutations at nucleotide 175 in the TATA box region may affect viral productivity by changing transactivation. C175T decreases viral production dramatically, while T175C has the reverse effect. In this study, T175C change was observed in seven local viruses (TG-10, KRC-S11, KRC-S15, KRC-9, KRC-K10, P-8, P-14) while others (n = 41) had C175T (Table 2).
There are many different types of microorganisms whose genomes include overlapping reading frames, but viruses are the most prevalent. When the nucleotide sequences of two genes overlap, the mutation may influence the evolution of both genes. The BLV contains overlapping reading frames and a pX region that contains 3' segments of genes encoding four regulatory proteins: Tax, Rex, and two accessory proteins, R3 and G4 [12]. Although Tax protein has been identified as the first oncogenic transactivator that triggers disease progression, activation of Tax is not sufficient to trigger malignancy. Many host factors influence malignancy or disease progression, such as activation of cell oncogenes, cell survival pathways, and induction of growth factors [12, 41]. It was reported that Tax, the most conserved among the four regulatory proteins, showed purifying selection consistent with its importance in the viral life cycle [5]. According to two separate studies [42, 43]; the incubation period for the development of EBL with BLV carrying the 233P-Tax gene is significantly longer than the incubation period for 233L-Tax, and the risk of developing EBL may be higher in those infected with BLV carrying the 233L-Tax gene than in those infected with 233P-Tax It is interesting that the 233L-Tax protein variation was found to be encoded by all tax gene sequences obtained from local viruses in this study (Table 3).
Replication of BLV is inhibited in part by cellular immunity. Antibodies against viral proteins have been shown to remain in BLV-infected cattle, which is indicative of sustained, although suppressed, viral gene expression. A robust response from CD8 + cytotoxic T lymphocytes (CTL) and CD4 + helper T cells is elicited in response to Tax, in addition to structural proteins (envelope protein gp51 and capsid protein p24) [44]. Niewiesk et al. [44] hypothesized that escape from the anti-Tax CTL response in vivo caused by mutations in HTLV-1 Tax CTL epitopes contributed to the pathogenesis of the illness. For these reasons, it has been reported that mutations in this region can promote the spread of the virus in the host [45]. Although C196R (for TG-11) and I186V changes (for P-5 and P-6) in the CTL epitope region were detected, it is not possible to evaluate their effects on infectivity (Table 3).
Viral RNA is transported by Rex proteins, which are encoded in the BLV pX area. The rex gene stabilizes viral RNA, facilitating its translation, and regulates viral replication post-transcriptionally [46, 47]. Rex proteins use nuclear localization signal (NLS) and nuclear export signal (NES) to mediate transport between the nucleus and the cytoplasm [48]. It has been reported that mutation of a cysteine residue in the NES region to alanine (C86A) partially impairs but does not entirely inhibit NES function [48, 49]. In the NES region, S74L (for TH-29A, TH-34, TH-39, KRC-S12, KRC-K13, CP-1, CP-64), R77C (for TG-11, KRC-S8) and D81N (for TG-9, TG-8430) substitutions was observed in BLVs in this study (Fig. 2a).
The G4 gene is mainly expressed in the leukemia phase in vivo and since it may immortalize primary embryonic fibroblasts, is notably related to leukogenesis [13, 27, 50].The amino acid sequence of the G4 protein encoded by BLV includes the "leader peptide" (1–24 aa), the "Myb-like motif" (MYB), and an arginine-rich region (ARR) in the middle of the protein [51]. In this study, an F8L change was detected in KRC-52, an F22S change in KRC-56 in the "leader peptide" region of G4 gene, and an A65V change in the arginine-rich region at KRC-S15. In the comprehensive genomic study of three bovine B-cell lymphosarcomas from different farms, Moratorio et al. [26] reported that the BLV LS1 sample had an A29V substitution in the G4 amino acid sequences. An A29T change was also detected in the G4 gene sequence from TG-8430. On the other hand, in two local BLVs, TG-8 and TG-8090, deletions in the codon nucleotides at the 49th and 50th positions were identified (Fig. 2b). Murakami et al. [52] reported that a BLV variant with a deletion mutation of amino acids 48–51 from the N-terminus of the G4 protein results in lower virus production in vitro. As a result, mutant viruses would not propagate effectively in vivo due to ineffective virus production.
To date, no functional regions have been reported in the R3 gene region. Although R31S substitution in the R3 gene region has been detected in five local viruses (TH-29A, TH-34, TH-39, CP-1, CP-64), it is not possible for us to know whether this substitution causes a change in R3 gene function (Fig. 2c).
Although the literature highlights an association between some substitutions in the parts of the BLV genome and clinical stages of infection [26, 27], Montero-Machuca et al. [53] reported that they could not establish a relationship between BLV-infected cattle and changes in the pX region at different clinical stages of lymphocytosis, lymphoma, asymptomatic state, or proviral DNA load. So, there needs to be more clarity on this issue. This study used a large sample population (n = 48) from six herds, in which two of the herds were sampled in two different years, and one was sampled in four different years (Table 1), and also the sequences of the env, LTR and pX regions were evaluated. Unfortunately, the samples are retrospective there are no data on the clinical stages of infection (PL, AL, or lymphosarcoma formation) or hematological findings in sampling cattle. However, the mutations detected local viruses allowed some evaluations in the light of literature data [10, 26, 40, 45, 52]. It was noteworthy that mutations were detected in cattle harboring viruses (two samples) with a deletion in amino acids 49–50 of the G4 gene region associated with low transcriptional activity, and a G148A substitution (seven samples), which has been reported to be able to significantly decrease LTR activity in lymphosarcoma strains (LS1, LS2, LS3). animals in which viruses with mutations in the LTR T175C mutation (seven samples), LTR "Nfkb like" region (four samples), and the Tax CTL region (three samples) associated with high transcriptional activity, were different (Table 4). We believe that further studies will allow the data to evaluate changes that clearly distinguish the clinical stages and pathogenesis of BLV infection.
While the majority of the BLV genome seems to be conserved, significant variation has been identified in several areas. To date, the prevailing view is that sequence variation is due to the geographic origin of the isolate rather than mutational pressure from BLV pathology. The few point mutations that separate the various isolates are interesting, but whether they are functional is yet to be known. Considering the complexity of the biology of retroviruses, the size of the economic losses due to BLV, and their zoonotic potential, it was concluded that the molecular epidemiology and pathogenesis mechanism of BLV infection should be investigated in further studies, including samples from cattle with different clinical stage, and also different geographic locations.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Maclachlan NJ, Dubovı EJ (2011) Fenner’s veterinary virology, 5th edn. Academic Press Elsevier, New York, pp 243–274
Tajıma S, Ikawa Y, Aıda Y (1998) Complete bovine leukemia virus (BLV) provirus is conserved in BLV-infected cattle throughout the course of B-cell lymphosarcoma development. J Vırol 72:7569–7576. https://doi.org/10.1128/JVI.72.9.7569-7576.1998
Coffin J, Blomberg J, Fan H, Gifford R, Hatziioannou T et al (2021) ICTV virus taxonomy profile: retroviridae. J Gen Virol 102:001712. https://doi.org/10.1099/jgv.0.001712
Schwartz I, Lévy DL (1994) Pathobiology of bovine leukemia virus. Vet Res 25:521–536
Zhao X, Mcgırr KM, Buehrıng GC (1994) Potential evolutionary influences on overlapping reading frames in the bovine leukemia virus pXBL region. Genomics 89:502–511. https://doi.org/10.1016/j.ygeno.2006.12.007
Durkın K, Rosewıck N, Artes M, Hahau V, Grıebel P et al (2016) Characterization of novel Bovine Leukemia Virus (BLV) antisense transcripts by deep sequencing reveals constitutive expression in tumors and transcriptional interaction with viral microRNAs. Retrovirology 13:1–16. https://doi.org/10.1186/s12977-016-0267-8
Kıncaıd RP, Burke JM, Sullıvan CS (2012) RNA virus microRNA that mimics a b-cell oncomiR. Proc Natl Acad Sci 109:3077–3082. https://doi.org/10.1073/pnas.1116107109
Rosewıck N, Momont M, Durkın K, Takeda H, Caıment F et al (2013) Deep sequencing reveals abundant noncanonical retroviral microRNAs in B-cell leukemia/ lymphoma. Proc Natl Acad Sci USA 110:2306–2311. https://doi.org/10.1073/pnas.1213842110
Gıllet N, Florıns A, Boxus M, Burteau C, Nıgro A et al (2007) Mechanisms of leukemogenesis induced by bovine leukemia virus: prospects for novel anti-retroviral therapies in human. Retrovirology 16:18. https://doi.org/10.1186/1742-4690-4-18
Pluta A, Wıllems L, Douvılle RN, Kuźmak J (2020) Effects of naturally occurring mutations in bovine leukemia virus 5′-ltr and tax gene on viral transcriptional activity. Pathogens 9:1–28. https://doi.org/10.3390/pathogens9100836
Pluta A, Rola-Łuszczak M, Douvılle RN, Kuźmak J (2018) Bovine leukemia virus long terminal repeat variability: identification of single nucleotide polymorphisms in regulatory sequences. Virol J 15:1–14. https://doi.org/10.1186/s12985-018-1062-z
Araınga M, Takeda E, Aıda Y (2012) Identification of bovine leukemia virus tax function associated with host cell transcription, signaling, stress response and immune response pathway by microarray-based gene expression analysis. BMC Genom 13:121. https://doi.org/10.1186/1471-2164-13-121
Polat M, Takeshıma SN, Hosomıchı K, Kım J, Mıyasaka T et al (2016) A new genotype of bovine leukemia virus in South America identified by NGS-based whole genome sequencing and molecular evolutionary genetic analysis. Retrovirology 13:1–23. https://doi.org/10.1186/s12977-016-0239-z
Sultanov A, Rola-Łuszczak M, Mamanova S, Ryło A, Osi’nski Z et al (2022) Molecular Characterization of Bovine Leukemia Virus with the evidence of a new genotype circulating in cattle from Kazakhstan. Pathogens 11:180. https://doi.org/10.3390/pathogens11020180
Yu C, Wang X, Zhou Y, Wang Y, Zhang X et al (2019) Genotyping bovine leukemia virus in dairy cattle of Heilongjiang, northeastern China. BMC Vet Res 15:1–9. https://doi.org/10.1186/s12917-019-1863-3
Burgu I, Akca Y, Alkan F, Kerim Urman H, Alcıgır G et al (1990) Türkiye’de Enzootik Sıgır Löykozu’nun Seroepidemiyolojisi Ve Patolojisi. Ankara Univ Vet Fak Derg 37:32–45
Akca Y, Alkan F, Bilge Dagalp S, Karaoglu T, Ozkul A et al (1996) The investigation of bovine leukosis virus (BLV) specific antibodies in milk and serum samples from dairy cattle using Enzyme-Linked Immunosorbent Assay (ELISA) and Agar Jel Immunodiffusion (AGID) test. Ankara Univ Vet Fak Derg 43:53–59
Bilge-Dagalp S, Can-Sahna K, Yıldırım Y, Karaoglu T, Alkan F et al (2008) Effects of bovine leucosis virus (BLV) infection on the bovine viral diarrhea virus (BVDV) and bovine herpes virus 1 (BHV1) seroprevalences in dairy herds in Türkiye. Rev Med Vet 159:385–390
Şevik M, Avcı O, İnce OB (2015) An 8-year longitudinal sero-epidemiological study of bovine leukaemia virus (BLV) infection in dairy cattle in Türkiye and analysis of risk factors associated with BLV seropositivity. Trop Anim Health Prod 47:715–720. https://doi.org/10.1007/s11250-015-0783-x
Alkan F, Karayel-Hacıoglu I, Duran Yelken S, Coskun N (2021) The genotype determination and molecular characterization of bovine leukemia virus in Türkiye. Vet Arh 91:237–247. https://doi.org/10.24099/vet.arhiv.1214
Değirmenci H (2011) Enzootıc Bovıne Leukosıs Vırus İnfeksiyonunun Marmara Bölgesinde Serolojik Ve Moleküler Yöntemler İle Araştırılması. Dissertation, University of Istanbul.
Doğan F, Bilge Dagalp S, Dik B, Farzani TA, Alkan F (2020) Detection of genotype 1 bovine leukemia virus from a C. schultzei pool: Do Culicoides spp. have a role on the transmission of bovine leukemia virus? Infect Genet Evol 85:104469. https://doi.org/10.1016/j.meegid.2020.104469
Sambrook J, Frıtsch EF, Manıatıs T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Fechner H, Kurg A, Geue L, Blankensteın P, Mewes G et al (1996) Evaluation of polymerase chain reaction (PCR) application in diagnosis of bovine leukaemia virus (BLV) infection in naturally infected cattle. Zentralbl Veterinarmed B 43:621–630. https://doi.org/10.1111/j.1439-0450.1996.tb00361.x
Zhao X, Jımenez C, Sentsuı H, Buehrıng GC (2007) Sequence polymorphisms in the long terminal repeat of bovine leukemia virus: evidence for selection pressures in regulatory sequences. Virus Res 124:113–124. https://doi.org/10.1016/j.virusres.2006.10.010
Moratorıo G, Fıscher S, Bıanchı S, Tomé L, Rama G et al (2013) A detailed molecular analysis of complete bovine leukemia virus genomes isolated from B-cell lymphosarcomas. Vet Res 44:1–11. https://doi.org/10.1186/1297-9716-44-19
Paneı CJ, Serena MS, Metz GE, Bravı ME, González ET et al (2013) Analysis of the pX region of bovine leukemia virus in different clinical stages of enzootic bovine leukemia in argentine holstein cattle. Virus Res 171:97–102. https://doi.org/10.1016/j.virusres.2012.08.001
Larsson A (2014) AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278. https://doi.org/10.1093/bioinformatics/btu531
Kumar S, Stecher G, Lı M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Bio Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
SIAS (2022) Sequence identity and similarity. Accessed 05 Jan 2022 http://imed.med.ucm.es/Tools/sias.html/.
Fechner H, Blankensteın P, Looman AC, Elwert J, Geue L et al (1997) Provirus variants of the bovine leukemia virus and their relation to the serological status of naturally infected cattle. Virology 237:261–269. https://doi.org/10.1006/viro.1997.8784
Moratorıo G, Obal G, Dubra A, Correa A, Bıanchı S et al (2010) Phylogenetic analysis of bovine leukemia viruses isolated in South America reveals diversification in seven distinct genotypes. Arch Virol 155:481–489
Ochırkhuu N, Konnaı S, Odbıleg R, Nıshımorı A, Okagawa T et al (2016) Detection of bovine leukemia virus and identification of its genotype in Mongolian cattle. Arch Virol 161:985–991. https://doi.org/10.1007/s00705-015-2676-8
Yang Y, Chen L, Dong M, Huang W, Hao X et al (2019) Molecular characterization of bovine leukemia virus reveals existence of genotype 4 in Chinese dairy cattle. Virol J 16:1–7. https://doi.org/10.1186/s12985-019-1207-8
Pandey GS, Sımulundu E, Mwıınga D, Samuı KL, Mweene AS et al (2017) Clinical and subclinical bovine leukemia virus infection in a dairy cattle herd in Zambia. Arch Virol 162:1051–1056. https://doi.org/10.1007/s00705-016-3205-0
Polat M, Moe HH, Shımogırı T, Moe KK, Takeshıma SN et al (2017) The molecular epidemiological study of bovine leukemia virus infection in Myanmar cattle. Arch Virol 162:425–437. https://doi.org/10.1007/s00705-016-3118-y
Polat MS, Takeshıma SN, Aıda Y (2017) Epidemiology and genetic diversity of bovine leukemia virus. Virol J 14:209. https://doi.org/10.1186/s12985-017-0876-4
Pluta A, Rola-Łuszczak M, Kubıś P, Balov S, Moskalık R et al (2017) Molecular characterization of bovine leukemia virus from Moldovan dairy cattle. Arch Virol 162:1563–1576. https://doi.org/10.1007/s00705-017-3241-4
Gatot JS, Callebaut I, Van Lınt C, Demonté D, Kerkhofs P et al (2002) Bovine leukemia virus SU protein interacts with zinc, and mutations within two interacting regions differently affect viral fusion and infectivity in vivo. J Virol 76:7956–7967. https://doi.org/10.1128/JVI.76.16.7956-7967.2002
Murakamı H, Todaka H, Uchıyama J, Sato R, Sogawa K et al (2019) A point mutation to the long terminal repeat of bovine leukemia virus related to viral productivity and transmissibility. Virology 537:45–52. https://doi.org/10.1016/j.virol.2019.08.015
Ashrafı F, Nassırı M, Javadmanesh A, Rahımı H, Rezaee SA (2020) Epigenetics evaluation of the oncogenic mechanisms of two closely related bovine and human deltaretroviruses: a system biology study. Microb Pathog 139:103845. https://doi.org/10.1016/j.micpath.2019.103845
Inoue E, Matsumura K, Soma N, Hırasawa S, Wakımoto M et al (2013) L233P mutation of the tax protein strongly correlated with leukemogenicity of bovine leukemia virus. Vet Microbiol 167:364–371. https://doi.org/10.1016/j.vetmic.2013.09.026
Tomiyasu T, Mori H, Okazaki K (2022) Epidemiological evidence for early-onset of enzootic bovine leukosis by L233-Tax-carrying bovine leukemia virus in Japanese black cattle. J Vet Med Sci 84:1216–1220. https://doi.org/10.1292/jvms.22-0169
Nıewıesk S, Daenke S, Parker CE, Taylor G, Weber J et al (1995) Naturally occurring variants of human T-cell leukemia virus type I Tax protein impair its recognition by cytotoxic T lymphocytes and the transactivation function of Tax. J Virol 69:2649–2653. https://doi.org/10.1128/jvi.69.4.2649-2653.1995
Pluta A, Blazhko NV, Ngırande C, Jorıs T, Wıllems L et al (2021) Analysis of nucleotide sequence of tax, miRNA and LTR of bovine leukemia virus in cattle with different levels of persistent lymphocytosis in Russia. Pathogens 10:246. https://doi.org/10.3390/pathogens10020246
Mcgırr KM, Buehrıng GC (2005) Tax and rex sequences of bovine leukaemia virus from globally diverse isolates: rex amino acid sequence more variable than tax. J Vet Med B Infect Dis Vet Public Health 52:8–16. https://doi.org/10.1111/j.1439-0450.2004.00815.x
Stott ML, Thurmond MC, Dunn SJ, Osburn BI, Stott JL (1991) Integrated bovine leukosis proviral DNA in T helper and T cytotoxic/suppressor lymphocytes. J Gen Virol 72:307–315. https://doi.org/10.1099/0022-1317-72-2-307
Choı EA, Hope TJ (2005) Mutational analysis of bovine leukemia virus rex: identification of a dominant-negative inhibitor. J Virol 79:7172–7181. https://doi.org/10.1128/JVI.79.11.7172-7181.2005
Henderson BR, Eleftherıou A (2000) A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp Cell Res 256:213–224. https://doi.org/10.1006/excr.2000.4825
Julıarena MA, Gutıerrez SE, Cerıanı C (2007) Determination of proviral load in bovine leukemia virus-infected cattle with and without lymphocytosis. Am J Vet Res 68:1220–1225. https://doi.org/10.2460/ajvr.68.11.1220
Dube S, Abbott L, Dube DK, Dolcını G, Gutıerrez S et al (2009) The complete genomic sequence of an in vivo low replicating BLV strain. Virol J 6:1–11. https://doi.org/10.1186/1743-422X-6-120
Murakamı H, Uchıyama J, Nıkaıdo S, Sato R, Sakaguchı M et al (2016) Inefficient viral replication of bovine leukemia virus induced by spontaneous deletion mutation in the G4 gene. J Gen Virol 97:2753–2762. https://doi.org/10.1099/jgv.0.000583
Montero Machuca N, Tórtora Pérez JL, González Méndez AS, García-Camacho AL, Marín Flamand E et al (2022) Genetic analysis of the pX region of bovine leukemia virus genotype 1 in holstein friesian cattle with different stages of infection. Arch Virol 167:45–46. https://doi.org/10.1007/s00705-021-05252-2
Acknowledgements
This manuscript represents part of a thesis submitted by SDY to the Department of Virology, Graduate School of Health Sciences, Ankara University, in fulfillment of the requirements for a PhD. degree.
Funding
This project was funded by the Ankara University Scientifc Research Projects Coordination Unit (Project no: 19L0239007).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study's conception and design. SDY: conducted the experiments and performed the molecular biology and bioinformatic analyses (alignments, phylogeny). SDY: drafted the manuscript. FA: was responsible for the supervision of the study. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
All authors declare that there are no financial or other relationships that might lead to a conflict of interest. All authors have seen and approved the manuscript and have contributed significantly to the work.
Ethical approval
This study was approved by the ethics committee of Ankara University with document number 2019–5-51.
Additional information
Edited by Nicola Decaro.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Duran-Yelken, S., Alkan, F. Molecular analysis of the env, LTR, and pX regions of bovine leukemia virus in dairy cattle of Türkiye. Virus Genes 60, 173–185 (2024). https://doi.org/10.1007/s11262-024-02058-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-024-02058-7