Abstract
Contigs with the highest sequence similarity (73%) to Apricot pseudo-chlorotic leaf spot virus (genus Trichovirus, family Betaflexiviridae) were identified by high-throughput sequencing from a symptomless sweet cherry accession. The complete genome sequence of this new virus is 7460 nucleotides, excluding the 3′ poly(A) tail. Its genome organization is very similar to several trichoviruses infecting fruit trees, with three open reading frames encoding putative replicase, movement protein and coat protein (CP). The virus shares amino acid sequence identities of 60–73% at replicase and 53–76% at CP with other trichoviruses. Phylogenetic analyses group it and other trichoviruses in a cluster. These results support that this virus, which is tentatively named cherry latent virus 1, should be considered a new member in the genus Trichovirus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sweet cherry (Prunus avium L.) is an economically important fruit crop worldwide. It is one of the most popular fresh fruit in many temperate countries since it has both desirable culinary attributes and high levels of bioactive compounds with antioxidant characteristics [1].
Many viruses infect sweet cherry [2]. Although most infections are latent, some, especially the mix infections, cause diseases that reduce yield and market value of the crop. Genus Trichovirus in the family Betaflexiviridae is a group of plant viruses consisting of a single stranded RNA genome of 5.9–9.5 kb encapsidated in flexuous filamentous particles of 640–890 × 10–12 nm [3]. Apple chlorotic leaf spot virus (ACLSV) and Cherry mottle leaf virus (CMLV) are two trichoviruses that have been reported to infect sweet cherry and cause diseases [4, 5]. Both viruses have filamentous morphology and are mechanically transmitted. However, the ACLSV genome contains only three open reading frames (ORF) and has no known insect vector [4], while CMLV encodes four putative proteins and is transmitted by eriophyid mites (Eriophyes inaequalis) [5].
In this study, a new member of the genus Trichovirus was identified from a sweet cherry accession. The complete genome of the virus was determined, and its relationship with other trichoviruses was analyzed.
Materials and methods
Virus source, bioassay and detection of known viruses
Two accessions of Prunus spp. that were imported to the U.S. from the Republic of Georgia were used in a high-throughput sequencing (HTS) evaluation study. One accession was cherry cultivar ‘Mskhvil Nakota’ and another was apricot cultivar ‘Farbel’. The two accessions were established by grafting scion buds onto peach rootstock. They were also grafted onto following three seedlings of each of eight indicator plants for symptom expression in the greenhouse: P. armeniaca ‘Tilton’, P. dulcis ‘Peerless’, P. persica GF305, P. salicina Shiro, P. serrulata ‘Kwanzan’ and P. avium ‘Bing’, ‘Sam’ and ‘Canindex’. The original plants were also tested by ELISA and RT-PCR/PCR using pathogen-specific or genus-specific primers for the presence of known viruses, viroids and phytoplasma (Supplementary Table 1).
RNA isolation, high-throughput sequencing and sequence analyses
Total RNAs were extracted by RNAeasy Plant Mini Kit (Qiagen) from leaves of both accessions and pooled into one sample, P3. The sample was subjected to HTS using Illumina NexSeq 500 system (SeqMatic). Analyses of total paired-end reads were performed using CLC Genomics Workbench 9.5.2 (Qiagen). The raw reads were filtered to remove failed reads, and qualified reads were assembled de novo into contigs with a cut-off of 200 nucleotides (nt). Contigs were annotated by Blastx comparisons to local Viruses_NR and viroid databases downloaded from NCBI Genbank and a phytoplasma database provided by Yan Zhao of USDA-ARS.
Virus genome amplification, cloning and sequencing
To obtain the genomic sequence of new virus identified by HTS, RT-PCR was conducted by SuperScript ™ III One-Step RT-PCR System (Thermo Fisher Scientific) using specific primers designed based on the contig sequences (Table 1). The 5′-end sequence was obtained using a 5′ RACE System Kit (Thermo Fisher Scientific), and the 3′-terminus was amplified by a virus-specific forward and oligo-dT primers. Amplicons were isolated and cloned into pGEM-T Easy Vector (Promega), and plasmid DNAs isolated from overnight cultures of selected colonies were sequenced by MCLAB.
Sequence and phylogenetic analyses
Sequence analyses were performed using CLC Genomics Workbench. The conserved domains were identified by Motif Scan at https://myhits.isb-sib.ch/cgi-bin/motif_scan. Phylogenetic analyses were performed by MEGA7.1 [6]. Recombination analysis was performed using RDP4.95 [7].
Survey of the virus in cherry germplasm
To detect for the new virus in additional cherry, total nucleic acids were extracted from 175 samples, 56 flowering cherries from the U.S. National Arboretum, Washington, D.C., 108 sour cherries from the USDA Plant Genetic Resources Unit in Geneva, NY and 11 sweet cherries from Maryland, were tested for CLV-1 by a CTAB method [8] and tested by RT-PCR using virus-specific primers designed according to the sequence obtained in this study (Table 1).
Results and discussion
Reactions of indicator plants and results of laboratory detection
Neither the original plant nor the indicator plants showed any symptoms in the screen house and green house conditions. Prune dwarf virus (PDV, genus Ilarvirus, family Bromoviridae) and hop stunt viroid (HSVd, genus Hostuviroid, family Pospiviroidae) were only detected by quarantine indexing in the sweet cherry accession.
Virus identification by high-throughput sequencing
A total of 36,327,573 RNA reads were obtained from the P3 sample. De novo assembly of the RNA reads generated 67,988 contigs (> 200 nt). A Blastx search of the contigs against the Virus_NR database revealed the following: three contigs (3265, 2506 and 1935 nt) with the highest amino acid (aa) sequence identities (95–97%) to the gene products of PDV; two contigs (2644 and 4799 nt) with the highest aa sequence identities (76–80%) to the gene products of Apricot pseudo-chlorotic leaf spot virus (APCLSV, genus Trichovirus, family Betaflexiviridae); a large contig (8349 nt) with high aa sequence identities (53–54%) to replicase of the four cherry robigoviruses (geuns Robigovirus, family Betaflexiviridae) [9]; and a short contig (292 nt) with the highest nt similarity (84%) to HSVd. The identities of the two betaflexiviruses identified in the P3 sample were less than the species threshold of 80% aa identity at replicase and coat protein (CP) when compared to other betaflexiviruses [3], suggesting that they might be new members of the family. No phytoplasma was identified in the sample.
RT-PCR detection of viruses and viroid
The presence of the viruses and viroid in each accession was determined by RT-PCR using pathogen-specific primers (Table 1). PDV, two novel viruses and HSVd identified by HTS in the P3 sample were detected only in the cherry accession (Supplementary Fig. 1). None of the viruses and HSVd was detected in apricot accession. In this study, the new trichovirus named cherry latent virus 1 (CLV-1) was characterized.
Analyses of genomic sequence
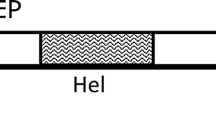
The complete genomic sequence of CLV-1 is 7460 nt (GenBank # MK770441), excluding the 3′ poly(A) tail. Its genome consists of three overlapping open reading frames (ORF) organized in the same arrangement as those of APCLSV, ACLSV, peach chlorotic leaf spot virus (PCLSV), grapevine berry inner necrosis virus and grapevine Pinot gris virus of the genus Trichovirus (Fig. 1a) [4, 5, 10,11,12,13]. ORF1 (nt 140-5783) encodes a putative replicase of 1872 aa (215 kDa) which contains viral methyltransferase (PF01660, aa 43–336), 2OG-Fe(II) oxygenase (PF13532, aa 723–826), peptidase_C34 (PF05413, aa 831–922), viral RNA helicase (PF01443, aa 1046–1294) and RNA-dependent RNA polymerase (PF009780, aa 1460–1774) domains. ORF2 (nt 5673–7043) codes for a putative movement protein of 456 aa (51 kDa), which might also act as a suppressor of RNA silencing [14]. ORF3 (nt 6730–7308) encodes a putative coat protein (CP) of 192 aa (21 kDa) that is crucial for infectivity [15].
Phylogenetic and recombination analyses
Comparisons of genomic and individual protein sequences among selected members of Betaflexiviridae confirm that CLV-1 is most closely related to APCLSV, ACLSV and PCLSV (Fig. 1b). The sequence identities between CLV-1 and other trichoviruses are 46.6–68.6% at the whole genome sequence level, 47.0–54.6% at the replicase aa sequence level and 32.8–75.5% at the CP aa sequence level. These values are below the species demarcation criteria (< 72% genome sequence identity or < 80% aa sequence identity of replicase or CP), indicating that CLV-1 is a novel virus [3]. Phylogenetic analyses showed that CLV-1 and five other trichoviruses infecting Prunus spp. form a clade distinct from two grapevine trichoviruses and members of the other genera in the Betaflexiviridae family (Fig. 2). Topologies of phylogenetic trees changed slightly when the aa sequences of the putative proteins were analyzed, but a close relationship of these trichoviruses was retained (data not shown).
Maximum likelihood tree based on replicase amino acid sequences of cherry latent virus 1, members of the genus Trichovirus and selected members of the family Betafexiviridae. Bootstrap analysis was applied using 1000 replicates. The diamond indicates the virus characterized in this study. Scale represents genetic distance. Indian citrus ringspot virus, a member of the family Alphaflexiviridae, was used as an outgroup
Recombination analysis conducted using the complete genomic sequences of CLV-1 and 107 other betaflexiviruses revealed that CLV-1 was a recombinant of two carlaviruses, Sambucus virus E (major parent) and Helleborus net necrosis virus (minor virus), at the region of nt 1438–1883. The recombination event was detected by GENEGONV (5.783 × 10–6), MaxChi (1.516 × 10–5) and Chimaera (5.776 × 10–3) of the RDP4 package. This event was also detected in the similar region of the APCLSV genome, supporting that the two viruses are most closely related.
Graft transmission and survey results
CLV-1 was detected by RT-PCR using primers P3CLV1DetF1 and P3CLV1DetR1 (Table 1) in three ‘Kwanzan’ cherry trees inoculated with infected cherry material (Fig. 3), indicating the virus is graft transmissible. The virus was not detected in any of the 175 samples, indicating its infection is not common. The genomic sequence obtained in this study contributes to the taxonomy of the genus Trichovirus, and the characterization and detection of CLV-1 alerts Prunus quarantine and clean stock programs about a new virus.
Detection of cherry latent virus 1 by RT-PCR in cherry trees grafted with an infected cherry accession from Republic of Georgia. Lanes M 1 kb plus DNA ladder, CO original cherry accession; K1–K3) cv. ‘Kwanzan’ trees grafted with the infected cherry. H uninoculated ‘Kwanzan’. Arrow indicates the size of the DNA marker
References
Blando F, Oomah BD (2019) Sweet and sour cherries: origin, distribution, nutritional composition and healthy benefits. Trends Food Sci Technol 86:517–529. https://doi.org/10.1016/j.tifs.2019.02.052
Rubio M, Martínez-Gómez P, Marais A, Sánchez-Navarro JA, Pallás V, Candresse T (2017) Recent advances and prospects in Prunus virology. Ann Appl Biol 171:125–138. https://doi.org/10.1111/aab.12371
Adams MJ, Candresse T, Hammond J, Kreuze JF, Martelli GP, Namba MN, Ryu KH, Saldarelli P, Yoshikawa N (2012) Family Betaflexiviridae. In: King AMQ, Adams MJ, Carstens EB, Lefknwitz EJ (eds) Virus taxonomy-ninth report of the international committee on taxonomy of viruses. Academic Press, San Diego, pp 920–941
Katsiani AT, Maliogka VI, Candresse T, Katis NI (2014) Host range studies, genetic diversity and evolutionary relationships of ACLSV isolates from ornamental, wild and cultivated rosaceous species. Plant Pathol 63:63–71. https://doi.org/10.1111/ppa.12058
James D, Jelkmann UC (2000) Nucleotide sequence and genome organization of cherry mottle leaf virus and its relationship to members of the Trichovirus genus. Arch Virol 145:995–1007. https://doi.org/10.1007/s007050050690
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003. https://doi.org/10.1093/ve/vev003
Li R, Mock R, Haung Q, Abad J, Hartung SJ, Kinard G (2008) A reliable and inexpensive method of nucleic acid extraction for the PCR-based detection of diverse plant pathogens. J Virol Methods 154:48–55. https://doi.org/10.1016/j.jviromet.2008.09.008
Wu LP, Liu H, Bateman M, Komorowska B, Li R (2019) First identification and molecular characterization of a novel cherry robigovirus. Arch Virol 164:3103–3106. https://doi.org/10.1007/s00705-019-04401-y
Liberti D, Marais A, Svanella-Dumas L, Dulucq MJ, Alioto D, Ragozzino A, Rodoni B, Candresse T (2005) Characterization of Apricot pseudo-chlorotic leaf spot virus, A Novel Trichovirus Isolated from Stone Fruit Trees. Phytopathol 95:420–426. https://doi.org/10.1094/PHYTO-95-0420
Zhou J, Zhang Z, Lu M, Xiao H, Habili N, Li S (2018) Complete nucleotide sequence of a new virus, peach chlorotic leaf spot virus, isolated from flat peach in China. Arch Virol 163:3459–3461. https://doi.org/10.1007/s00705-018-3984-6
Yoshikawa N, Iida H, Goto S, Magome H, Takahashi T, Terai Y (1997) Grapevine berry inner necrosis, a new trichovirus: comparative studies with several known trichoviruses. Arch Virol 142:1351–1363. https://doi.org/10.1007/s007050050165
Giampetruzzi A, Roumi V, Roberto R, Malossini U, Yoshikawa N, La Notte P, Terlizzi F, Credi R, Saldarelli P (2012) A new grapevine virus discovered by deep sequencing of virus- and viroid-derived small RNAs in Cv Pinot gris. Virus Res 163:262–268. https://doi.org/10.1016/j.virusres.2011.10.010
Yaegashi H, Takahashi T, Isogai M, Kobori T, Ohki S, Yoshikawa N (2007) Apple chlorotic leaf spot virus 50 kDa movement protein acts as a suppressor of systemic silencing without interfering with local silencing in Nicotiana benthamiana. J Gen Virol 88:316–324. https://doi.org/10.1099/vir.0.82377-0
Yaegashi H, Isogai M, Tajima H, Sano T, Yoshikawa N (2007) Combinations of two amino acids (Ala40 and Phe75 or Ser40 and Tyr75) in the coat protein of apple chlorotic leaf spot virus are crucial for infectivity. J Gen Virol 88:2611–2618. https://doi.org/10.1099/vir.0.82984-0
Acknowledgements
Authors thank Madeleine Chen, Sam Grinstead and Tom Kim for excellent technical assistance. This work was supported by the U.S. Department of Agriculture under ARS project 8042-22000-302-00D of National Plant Disease Recovery System to study newly emerging plant pathogens threatening National Agriculture.
Author information
Authors and Affiliations
Contributions
This work was designed by RL and MC; performed by EB, RL, MC, MB and BG; and written by RL and EB.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflicts of interest.
Ethical approval
This article does not contain any research involving human or animal participants.
Additional information
Edited by Karel Petrzik.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Brewer, E., Cao, M., Gutierrez, B. et al. Discovery and molecular characterization of a novel trichovirus infecting sweet cherry. Virus Genes 56, 380–385 (2020). https://doi.org/10.1007/s11262-020-01743-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-020-01743-7