Abstract
Sapoviruses (SaVs) are enteric viruses and have been detected in various mammals. They are divided into multiple genogroups and genotypes based on the entire major capsid protein (VP1) encoding region sequences. In this study, we determined the first complete genome sequences of two genogroup V, genotype 3 (GV.3) SaV strains detected from swine fecal samples, in combination with Illumina MiSeq sequencing of the libraries prepared from viral RNA and PCR products. The lengths of the viral genome (7494 nucleotides [nt] excluding polyA tail) and short 5′-untranslated region (14 nt) as well as two predicted open reading frames are similar to those of other SaVs. The amino acid differences between the two porcine SaVs are most frequent in the central region of the VP1-encoding region. A stem-loop structure which was predicted in the first 41 nt of the 5′-terminal region of GV.3 SaVs and the other available complete genome sequences of SaVs may have a critical role in viral genome replication. Our study provides complete genome sequences of rarely reported GV.3 SaV strains and highlights the common 5′-terminal genomic feature of SaVs detected from different mammalian species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sapoviruses (SaVs), a member of Caliciviridae, have been detected in many mammals (i.e., humans, pigs, minks, dogs, sea lions, chimpanzees, bats, rats, hyena, lions, and foxes) [1, 2]. The SaV genome is a positive-sense, single-stranded RNA molecule of 7.1–7.7 kb in length, excluding the 3′-end poly (A) tail [1]. SaV genome commonly encodes two open reading frames (ORFs). The ORF1 encodes 7 non-structural proteins (NSs 1-7) and major capsid protein (VP1), and ORF2 encodes minor capsid protein (VP2) [3]. SaVs can be classified into multiple genogroups based on available VP1 sequences, and five genogroups (GI to GV) are well accepted [4, 5]. Furthermore, we proposed ten additional genogroups (GVI–GXV) recently [1, 6]. Currently, complete genome sequences for GI–GVIII, GXIII, and GXIV are available in the database [1]. GI, GII, and GV are further subdivided into multiple genotypes based on the VP1 sequence [4, 7].
We have previously reported the detection and partial genomic characterization of two porcine GV SaV strains, TYMPo31, and TYMPo239, which are genetically highly similar to each other in the capsid region sequences [8]. These two strains are genetically clustered independently with other GV strains [8]. GV SaVs are detected from humans, pigs, and sea lions, and currently subdivided into four genotypes, GV.1-4, based on the complete capsid sequence diversity, and TYMPo31 and TYMPo239 strains are classified as GV.3 [4]. We also have detected novel genetically unique GV WG194D-1 strain in swine; however, the genotype number is not assigned yet [1]. Six complete genome sequences of GV SaVs (three from human [DQ366344-GV.1-Ehime475, AY646856-GV.1-NK24, AB775659-GV.2-NGY-1], one from pig [KX000383-GV. not assigned genotype (NA) WG194D-1], one from sea lion [JN420370-GV.4-CSL9775]) are available on the DNA database [1, 4], but that of GV.3 is not available yet.
This study therefore was undertaken to determine the full genome sequences of two GV.3 porcine SaVs, TYMPo31, and TYMPo239, by using multiple techniques including Illumina MiSeq sequencing of RNA samples or PCR products of RT-PCR as well as 5′ RACE. We also analyzed the secondary structure in the 5′-end genome sequences of two newly determined GV.3 strains as well as that of representative strains for GV.1, GV.2, GV.3, GV-NA, and other genogroups of SaVs.
Materials and methods
Virus positive specimens
Swine fecal samples from the previous study were stored at −80 °C and were used in this study for further sequence analyses. SaV GV.3 strains, TYMPo31, and TYMPo239 were originally identified from a molecular survey of porcine caliciviruses in Toyama Prefecture, Japan, in May and December 2008, respectively, and 2.1 kb fragment covering from the putative entire NS7 and partial VP1 has been determined previously [8]. Then the partial genome sequences of approximately 3.9 kb fragment, covering from the putative entire NS7 encoding region to the 3′ end of the genome of these two porcine strains, were determined by using RT-PCR with gene-specific forward primer and oligo dT reverse primers (Nakamura and Takizawa, unpublished results), and were deposited in the DNA database (GV TYMPo31 [AB521772.1]; GV TYMPo239 [AB521771.1]).
Viral RNA extraction
Viral RNA was extracted from 140 µl of approximately 20% intestinal content suspensions in Dulbecco’s Phosphate Buffered Saline without Mg2+ and Ca2+ (Sigma) using RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions, except that the yeast RNA (Ambion) was added to AVL buffer instead of the carrier RNA which was supplied with the kit, and finally eluted in 60 µl of DNase-and RNase-free water (Invitrogen). The extracted RNAs were stored at −80 °C.
Library preparation from purified RNA for Illumina MiSeq
A 300-bp fragment library was constructed from the 13.5 µl of extracted RNA and using NEBNext Ultra RNA Library Prep Kit for Illumina v2.0 (New England Biolabs), purified using Agencourt AMPure XP magnetic beads (Beckman Coulter) and SPRIPlate 96R Ring Super Magnet Plate (Beckman coulter). The quality of the purified library was assessed on a MultiNA MCE-202 bioanalyzer (Shimadzu Corporation) and the concentration determined on a Qubit 2.0 Fluorometer using the Qubit HS DNA Assay (Invitrogen).
Samples were bar-coded for multiplexing using NEBNext Multiplex Oligos for Illumina Dual Index Primer Sets 1 (New England Biolabs).
Reverse transcription PCR
To amplify the fragment from putative NS2 encoding region to 3′ end of the viral genome, TYMPo31 and TYMPo239 cDNA were independently synthesized as follows: 9.5 µl of viral RNA solution was mixed with 2.5 µl of 20 pmol/µl reverse primer (5′-GACTAGTTCTAGATCGCGAGCGGCCGC CCT30-3′) (Fig. 1) complementary to the 3′-end polyA tail or the TYMPo31- and TYMPo239-specific reverse primer targeting the VP1 encoding region (nt position 5631-5655 of the viral genome, Fig. 1), and 1 µl of 10 mM dNTP. The mixtures were incubated at 65 °C for 5 min, cooled on ice, and then mixed with 4 µl of 5× SSIV buffer (Invitrogen), 1 µl of 100 mM DTT (Invitrogen), 1 µl of RNase OUT ribonuclease inhibitor (40 U/µl) (Invitrogen), and 1 µl of SuperScript IV reverse transcriptase (200 U/µl) (Invitrogen). This mixture was incubated first at 55 °C for 10 min and then inactivated at 80 °C for 10 min. Next, 1 µl of RNase H (2 U/µl) (Invitrogen) was added and incubated at 37 °C for 20 min, and then inactivated at 95 °C for 10 min.
Diagram for complete genome sequence determination of SaV TYMPo31 and TYMPo239 strains. Complete genome sequences of SaV, SaV TYMPo31 and TYMPo239 strains were determined by de novo assembly of Illumina MiSeq sequence data, 5′ RACE, and 3′-end genome region-targeting RT-PCR as described in the text. SaV genomic organization, the predicted non-structural (NS) and structural proteins VP1 and VP2, and the conserved amino acid motifs are shown. Asterisks indicate the different amino acid residue between TYMPo31 and TYMPo239 strains
The SaV genomic sequences spanning the putative NS2 to VP1 encoding region or the putative NS2 or NS7 to the genome end (assigned as middle to 3′ end in Fig. 1) were amplified by RT-PCR with the forward primer based on the partial sequence obtained by Illumina MiSeq sequencing (nt position 489–513 or 516–541, Fig. 1) or the forward primer designed previously that determined partial genome sequence (nt position 4774–4796, Fig. 1), and reverse primer complementary to the 3′-end polyA tail or targeting the VP1 encoding region (nt position 5594–5616 or 5564–5586, Fig. 1), using high-fidelity PCR enzyme, PrimeSTAR GXL DNA Polymerase (Takara). A final volume of 50 µ l of the PCR mixture contained 2 µl of the cDNA or the first PCR mixture, 10 µ l of 5× PrimeSTAR GXL DNA polymerase buffer, 4 µ l of 2.5 mM dNTPs, 2 µl of forward primer (10 pmol/µl), 2 µl of reverse primers (10 pmol/µl), and 1 µl of PrimeSTAR GXL DNA polymerase (1.25 U/µl). PCR cycling conditions were as follows: 98 °C for 10 s followed by 40 cycles of 98 °C for 10 s, 60 °C for 15 s, and 68 °C for 1 min 30 s, and a final extension at 67 °C for 7 min.
The 5′ part region-targeting RT-PCR was performed as follows: cDNA was synthesized using SuperScript IV reverse transcriptase with a primer corresponding to 779–803 nt (Fig. 1) of TYMPo31 or TYMPo239 genome cDNA as described above. After treatment with RNase H, semi-nested PCR was performed with KOD-Plus DNA polymerase (Toyobo) using the newly designed forward primer (5′-GTGATCACYTTGRGATGGCTTCMAAGCCA-3′) based on the alignment of GV SaVs 5′-terminal sequences of human and porcine origin (GV.1 NK24 [AY646856], GV.1 Ehime475 [DQ366344], GV.2 NGY1 [AB775659], and GV. not assigned WG194D-1 [KX000383]) and reverse primers (nt position 752–776 for the 1st PCR, and nt position 727–750 nt for the 2nd PCR) as summarized in Fig. 1. A final volume of 100 µl of the PCR reaction mixture contained 2 µl of the cDNA or the first PCR products or 1st PCR reaction mixture, 10 µl of 10 x PCR buffer for KOD plus, 10 µl of 2 mM dNTPs, 4 µl of 25 mM Mg2SO4, 2 µl of forward primer (10 pmol/µl), 2 µl of reverse primer (10 pmol/µl), and 2 µl of KOD-plus DNA polymerase (1.0 U/µl). PCR cycling conditions were as follows: 94 °C for 10 min followed by 45 cycles of 94 °C for 30 s, 55 °C for 30 s, and 68 °C for 45 s, and a final extension at 68 °C for 7 min.
The 5′-terminal nucleotide sequences of the TYMPo31 and TYMPo239 genomes (indicated as 5′ end in the Fig. 1) were finally confirmed by DNA–DNA ligation-based 5′ RACE as described [9]. cDNA was synthesized using SuperScript IV reverse transcriptase with a primer corresponding to nt position 551–581 of TYMPo31 or TYMPo239 genome cDNA as described above. After treatment with RNase H, the synthesized cDNA was purified and ligated to the 5′-phosphorylated and 3′-biotinylated anchor DNA primer (5′-TATAGTGAGTCGTATTAGGTACCGTCGAC-3′) by T4 RNA Ligase. The biotinylated anchor primer-ligated cDNA was captured by Dynabeads M-270 Streptavidin (Invitrogen) as described [9]. The 5′ end region of the TYMPo31 or TYMPo239 genome was amplified by semi-nested PCR: forward primer 5′ RACE-F (5′-GTCGACGGTACCTAATACGACTCACTATA-3′) complementary to the anchor, and a reverse primer corresponding to nt position 426–455 of TYMPo31 or TYMPo239 genome, for the first PCR; and forward primer (5′ RACE-F) and reverse primer corresponding to nt position 389–425 of TYMPo31 or TYMPo239 genome were used for the second PCR with KOD-Plus DNA polymerase. PCR conditions were identical as described above.
PCR product purification
The PCR products were separated by agarose gel electrophoresis, visualized under UV, purified using a QIAquick Gel Extraction Kit (Qiagen), and then eluted in 35 µl water.
Library preparation from purified PCR product for Illumina MiSeq
For PCR product longer than 700 bp, a 300-bp fragment library was constructed from 1 ng of the purified PCR products using Nextera XT kit (Illumina) according to the manufacturer’s instructions. Normalized library was purified as described above. Samples were bar-coded for multiplexing using Nextera XT index kit V2 (Illumina).
For PCR products shorter than 500 bp, a 300-bp fragment library were prepared from “Perform End Prep of cDNA Library” section of manufactures instruction of NEBNext Ultra RNA Library Prep Kit for Illumina v2.0 and purified the library. Samples were bar-coded for multiplexing using NEBNext Multiplex Oligos for Illumina Dual Index Primer Sets 1.
Illumina MiSeq sequencing
The DNA libraries were adjusted to 2 nM using the DNase-and RNase-free water. Then, 10 μl of the resulting 2 nM DNA library was denatured with 10 μl of 0.1 M freshly prepared NaOH solution. The denatured solution was diluted with pre-chilled HT1 (Illumina) to a final concentration of 6 pmol for sequencing. Nucleotide sequencing was performed on an Illumina MiSeq sequencer (Illumina) with a MiSeq Reagent Kit v2 (Illumina) to generate 151 paired-end reads.
The FASTQ files from MiSeq were analyzed and de novo assembled using CLC genomic workbench 8.0 (QIAGEN-CLC bio). For short PCR product corresponding to the 5′ end region of the genome, the reads including the adaptor sequence were retrieved from the sequence dataset.
Sequence editing and analysis
Sequence editing and assembly were performed using the Sequencher program v4.10.1 (GeneCodes), and nucleotide and amino acid sequences were analyzed by Genetyx-Mac software v16.0.4 (Genetyx Corporation). The secondary structure of the nucleotide was predicted using Genetyx-Mac software v16.0.4 (Genetyx Corporation) by “RNA 2ndary structure prediction” [10] with default settings. Nucleotide sequence alignment was determined using ClustalW version 2.1 and the genetic distance was calculated using Kimura’s two-parameter model using the online analysis tool (http://clustalw.ddbj.nig.ac.jp/top-j.html) with default settings. The neighbor-joining phylogenetic tree with 1000 bootstrap replications was illustrated using Njplot software.
Results
Firstly, only approximately 300-nt-assembled SaV-like sequence (corresponding to the nt position 487-796) was obtained from TYMPo31-positive specimen by Illumina MiSeq sequencing of the library prepared from the extracted RNA (Fig. 1). No SaV-like sequence was obtained from TYMPo239-positive specimen. However, secondly, a long PCR product (approximately 6.9 kb) was amplified from TYMPo31 cDNA (Fig. 1). The corresponding regions of TYMPo31 and TYMPo239 were also amplified separately as overlapping two PCR fragments (Fig. 1). Thirdly, the 5′ part regions of TYMPo31 and TYMPo239 were amplified in combination with the newly designed forward primer based on the available GV SaVs 5′-terminal sequence (29 nt in length) and gene-specific reverse primer, and finally,the actual 5′-terminal sequences were determined by 5′ RACE. The obtained sequence fragment was assembled to build up the complete genome sequences of SaV TYMPo31 and TYMPo239.
The complete genome sequences of SaV TYMPo31 and TYMPo239 have been updated in DNA database under the accession number GV TYMPo31 [AB521772.2]; GV TYMPo239 [AB521771.2]. These strains’ genomes both consist of 7494 nucleotides (nt), excluding the 3′-end poly (A) tail, and encodes two open reading frames (ORFs): nt 15-6905 (ORF1) and 6902-7417 (ORF2). The 5′- and 3′-end untranslated regions (UTRs) were 14 nt and 77 nt long, respectively (Fig. 1). The number of nucleotide and amino acid differences between TYMPo31 and TYMPo239 strains were found at residues 52 and 9, respectively. The amino acid differences between TYMPo31 and TYMPo239 strains were found only in the ORF1 encoding region (1 for NS1, 1 for NS7 [RdRp], and 7 for VP1, especially after “GWS” motif; Fig. 1). Based on the phylogenetic tree analysis, SaV TYMPo31 and TYMPo239 strains formed a branch distinct from other GV strains in both VP1 and complete genome sequences (Fig. 2). The 5′-terminal sequences were not strictly conserved even between porcine GV strains (Fig. 3). Instead, a single stem-loop structure was commonly predicted in the first 41 nt of the 5′-end nucleotide sequences among GV strains from multiple mammalian species (i.e., pigs, humans, and sea lion) (Fig. 3). The lengths of 3′ UTR were variable (77–155 nt) among GV SaVs, and those of TYMPo31 and TYMPo239 were shortest among GV SaVs.
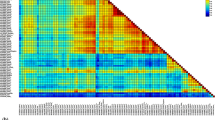
Phylogenetic tree of SaV based on complete VP1 nucleotide sequences or complete genome sequences. The newly determined SaV TYMPo31 and TYMPo239 strains (accession number AB521772 and AB521771) are shown in bold letters. Representative 32 SaV strains for 15 genogroups (GI, GII, GIII, GIV, GV, GVI, GVII, GVIII, GIX, GX, GXI, GXII, GXIII, GXIV, and GXV) classified based on complete VP1 sequences and representative 22 SaV strains for GI–GVIII, GXIII and GXIV for which complete genome sequences are available are included. Each SaV strain is shown in the following format: Genbank accession number-strain name (species) for VP1 nucleotide sequence tree, and VP1 sequence based on genogroup number-strain name for complete genome sequence tree. The nucleotide alignment was generated by ClustalW 2.1 using default settings (http://clustalw.ddbj.nig.ac.jp/) and a neighbor-joining method-based phylogenetic tree with 1000 bootstrap replications constructed was illustrated using NJPlot software (http://pbil.univ-lyon1.fr/software/njplot.html). The number on each branch indicates the bootstrap value, where values higher than 950 are indicated. The scale represents nucleotide substitutions per site
Predicted secondary structure in the 5′-end genome nucleotide sequences among GV SaVs. The secondary structure of the 5′-end nucleotide sequences of five GV, and representative strain for GI, GII, GIII, GIV, GVI, GVII, GVIII, GXIII, and GXIV SaVs are shown. Each SaV strain is shown in the following format: genogroup number-strain name (species). The predicted start codon for each GV SaV strain is boxed. The corresponding region sequences for GIX, GX, GXI, GXII, and GXV are not available yet
Discussion
The primer-independent next-generation sequencing method used in this study has already succeeded in determining nearly full genome sequence of genetically novel SaV from human fecal specimens by de novo assembly [11, 12]. However, porcine SaV TYMPo31 and TYMPo239 genome sequences were not determined well by this approach because we could detect partial genome sequences only for SaV TYMPo31. This result may be due to the low copy number of RNA for TYMPo31 and TYMPo239. However, following RT-PCR in combination with the forward primer designed based on the partial sequence of TYMPo31 strain that obtained from Illumina MiSeq, reverse primer targeting the 3′-end polyA tail the viral genome, and optional protocol of PrimeSTAR GXL provided from the manufacture (i.e., use twofold enzyme to allow DNA extension for 10 s/kb) allowed us to amplify a 6.9-kb PCR product covering the putative NS2 to 3′-genome end region of the TYMPo31. Then we could determine the complete genome sequences for TYMPo31 and TYMPo239 by further RT-PCR and 5′ RACE as summarized in Fig. 1. The most difficult part was to amplify the 5′ part region (Fig. 1). We could obtain the RT-PCR product when the used degenerated forward primer—described in Materials and methods—which was newly designed based on the available human and porcine GV SaV 5′-end genome sequence was introduced at the beginning of this study. However, we found a single nucleotide mismatch (the nucleotide M [A or C] was T in the TYMPo31 and 239) in the forward primer after we determined the 5′ termini of viral genome sequence by RACE.
We also used PrimeSTAR GXL or PrimeSTAR HS DNA polymerase in addition to KOD plus DNA polymerase to amplify the 5′ part region and 5′ termini of the viral genome sequence, but only KOD plus DNA polymerase could produce the PCR product (data not shown). Thus, we used different PCR enzymes in different regions of the TYMPo31 and TYMPo239 as indicated in Fig. 1.
Although partial genome sequences (3949 nt in length) of TYMPo31 and TYMPo239 (AB521771.1 and AB521772.1) have been determined previously (Fig. 1) ([8] and unpublished data), we finally determined complete genome sequence in this study. The lengths of the genome and UTR regions as well as two predicted ORFs of TYMPo31 and TYMPo239 strains were identical. The amino acid differences between TYMPo31 and TYMPo239 strains were mostly found after “GWS” motif in the VP1 protein. The after “GWS” motif was considered as putative variable protruding region of the VP1 (Fig. 1) [4, 13, 14]. TYMPo31 and TYMPo239 strains were detected in the same year (i.e., 2008 May for TYMpo31, and 2008 Dec for TYMPo 239) from different pigs [8]. Thus, the differences in their sequences suggest rapid diversity during circulation among the pigs in the same farm although further investigation is necessary for the future. GV.3 SaV distribution is also interesting because no highly similar sequences to TYMPo31 and TYMPo239 are reported on the DNA database.
The 5′ UTR of the GV SaVs including TYMPo31 and TYMPo239 strains are moderately conserved (8 out of 14 nt) and started with the consensus nucleotide sequence “GUGAU” (Fig. 3). The first three nucleotide sequence “GUG” is conserved among the available complete genome sequence of SaVs (Fig. 3) [1]. Recently, the predicted secondary structures at the 170-nt of the 5′ end of the GIII Cowden strain as well as representative GI, GII, GIV, and GV SaVs genome have been reported [15]. We also predicted single stem-loop structure in the TYMPo31 and TYMPo239 strains in the first 41 nt of the 5′ end (Fig. 3). A similar single stem-loop structure was also predicted in the same region of other GV strains (Fig. 3) as well as GVI, GVII, GVIII, GXIII, and GXIV in addition to GI, GII, GIII, and GIV SaVs (Fig. 3). Because the primary nucleotide sequence was not conserved and this common single stem loop was located very close to the 5′ end of the SaV genome (~41 nt), they may have a critical role in the viral genome replication, as well as in the possible interaction with cellular proteins for translation as discussed recently [15]. It is interesting to investigate the functional role of conserved 5′-end genomic feature of SaVs in the future.
Recently, SaVs nucleotide sequences were determined from multiple mammalian species (e.g., sea lions, dogs, chimpanzees, rodents, hyena, lion and fox, and humans) by NGS techniques [2, 11, 12, 16,17,18,19]. The method described in this study can be used to determine the complete or sufficient length of the SaV genome sequences for samples containing low amounts of viral RNA. Common SaV 5′-end genome sequence as well as the secondary structure has been highlighted across various genogroups detected from multiple mammalian species and may also worth estimating the sequence reliability of the determined SaV sequence.
References
T. Oka, Z. Lu, T. Phan, E.L. Delwart, L.J. Saif, Q. Wang, PLoS ONE 11, e0156373 (2016)
X.A. Olarte-Castillo, H. Hofer, K.V. Goller, V. Martella, P.D. Moehlman, M.L. East, PLoS ONE 11, e0163548 (2016)
K.Y. Green, Caliciviridae: The noroviruses: specific virus families, in Fields virology, 6th edn., ed. by D.M. Knipe, P.M. Howley (Lippincott Williams & Wilkins, Philadelphia, 2013), pp. 582–608
T. Oka, Q. Wang, K. Katayama, L.J. Saif, Clin. Microbiol. Rev. 28, 32–53 (2015)
T. Farkas, W.M. Zhong, Y. Jing, P.W. Huang, S.M. Espinosa, N. Martinez, A.L. Morrow, G.M. Ruiz-Palacios, L.K. Pickering, X. Jiang, Arch. Virol. 149, 1309–1323 (2004)
K.A. Scheuer, T. Oka, A.E. Hoet, W.A. Gebreyes, B.Z. Molla, L.J. Saif, Q. Wang, J. Clin. Microbiol. 51, 2344–2353 (2013)
X. Liu, H. Jahuira, R.H. Gilman, A. Alva, L. Cabrera, M. Okamoto, H. Xu, H.J. Windle, D. Kelleher, M. Varela, M. Verastegui, M. Calderon, G. Sanchez, V. Sarabia, S.B. Ballard, C. Bern, H. Mayta, J.E. Crabtree, V. Cama, M. Saito, H. Oshitani, J. Clin. Microbiol. 54, 1598–1604 (2016)
K. Nakamura, Y. Saga, M. Iwai, M. Obara, E. Horimoto, S. Hasegawa, T. Kurata, H. Okumura, M. Nagoshi, T. Takizawa, J. Clin. Microbiol. 48, 1215–1222 (2010)
T. Oka, H. Takagi, Y. Tohya, J. Virol. Methods 207, 178–181 (2014)
M. Zuker, Science 244, 48–52 (1989)
S. Shibata, T. Sekizuka, A. Kodaira, M. Kuroda, K. Haga, Y.H. Doan, R. Takai-Todaka, K. Katayama, T. Wakita, T. Oka, H. Hirata, Genome Announc. (2015). doi:10.1128/genomeA.01553-14
T. Oka, Y.H. Doan, K. Haga, K. Mori, T. Ogawa, A. Yamazaki, Jpn. J. Infect. Dis. 70, 223–224 (2017)
R. Chen, J.D. Neill, J.S. Noel, A.M. Hutson, R.I. Glass, M.K. Estes, B.V. Prasad, J. Virol. 78, 6469–6479 (2004)
N. Miyazaki, D.W. Taylor, G.S. Hansman, K. Murata, J. Virol. 90, 2664–2675 (2016)
M. Hosmillo, T.R. Sweeney, Y. Chaudhry, E. Leen, S. Curry, I. Goodfellow, K.O. Cho, J. Virol. 90, 5200–5204 (2016)
C. Firth, M. Bhat, M.A. Firth, S.H. Williams, M.J. Frye, P. Simmonds, J.M. Conte, J. Ng, J. Garcia, N.P. Bhuva, B. Lee, X. Che, P.L. Quan, W.I. Lipkin, MBio 5, e01914–e01933 (2014)
L. Li, P.A. Pesavento, T. Shan, C.M. Leutenegger, C. Wang, E. Delwart, J. Gen. Virol. 92, 2534–2541 (2011)
L. Li, T. Shan, C. Wang, C. Cote, J. Kolman, D. Onions, F.M. Gulland, E. Delwart, J. Virol. 85, 9909–9917 (2011)
I.M. Mombo, N. Berthet, C. Bouchier, J.N. Fair, B.S. Schneider, F. Renaud, E.M. Leroy, V. Rougeron, Genome Announc. (2014). doi:10.1128/genomeA.00680-14
Acknowledgements
This work was partly supported by Grants-in-Aid for Scientific Research (C) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and the Research Program on Emerging and Reemerging Infectious Diseases program from the Japan Agency for Medical Research and Development (AMED).
Author information
Authors and Affiliations
Contributions
Statement of author contributions
Oka T conceived and designed the project, performed PCR and RACE experiment, genetic analysis and wrote the paper. Doan YH and Haga K performed library preparation, Illumina MiSeq sequencing and analyzed the data. Shimoike T performed RNA 2ndary structure prediction and illustration. Takizawa T conceived this study.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Edited by Keizo Tomonaga.
Rights and permissions
About this article
Cite this article
Oka, T., Doan, Y.H., Shimoike, T. et al. First complete genome sequences of genogroup V, genotype 3 porcine sapoviruses: common 5′-terminal genomic feature of sapoviruses. Virus Genes 53, 848–855 (2017). https://doi.org/10.1007/s11262-017-1481-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-017-1481-8