Abstract
The molluscum contagiosum virus (MCV) uses a variety of immune evasion strategies to antagonize host immune responses. Two MCV proteins, MC159 and MC160, contain tandem death effector domains (DEDs). They are reported to inhibit innate immune signaling events such as NF-κB and IRF3 activation, and apoptosis. The RxDL motif of MC159 is required for inhibition of both apoptosis and NF-κB activation. However, the role of the conserved RxDL motif in the MC160 DEDs remained unknown. To answer this question, we performed alanine mutations to neutralize the arginine and aspartate residues present in the MC160 RxDL in both DED1 and DED2. These mutations were further modeled against the structure of the MC159 protein. Surprisingly, the RxDL motif was not required for MC160′s ability to inhibit MAVS-induced IFNβ activation. Further, unlike previous results with the MC159 protein, mutations within the RxDL motif of MC160 had no effect on the ability of MC160 to dampen TNF-α-induced NF-κB activation. Molecular modeling predictions revealed no overall changes to the structure in the MC160 protein when the amino acids of both RxDL motifs were mutated to alanine (DED1 = R67A D69A; DED2 = R160A D162A). Taken together, our results demonstrate that the RxDL motifs present in the MC160 DEDs are not required for known functions of the viral protein.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Molluscum Contagiosum Virus (MCV), a common, dermatotropic poxvirus, exclusively infects human keratinocytes. In otherwise healthy individuals, MCV lesions persist for up to 12 months [1]. Immune-compromised patients are susceptible to uncontrolled lesion formation which results in excessive scarring of the skin [1, 2]. The persistence of MCV has been attributed to the production of MCV immune evasion molecules, which enable MCV to limit host immune responses. However, only a few of the predicted MCV immune evasion molecules have been studied [3].
Two MCV proteins, MC159 and MC160, contain two tandem death effector domains (DEDs). Both MC159 and MC160 belong to a group of proteins collectively known as viral FLICE-like Inhibiting Proteins (vFLIPs) [3–5]. These MCV vFLIPs antagonize a variety of host-mediated immune responses including apoptosis and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) activation [6–12]. Expression of MC159 prevents both Fas- and tumor necrosis factor-α- (TNF-α)-induced apoptosis. MC159 associates with fas-associated protein with death domain (FADD) and procaspase-8, preventing the oligomerization of death-inducing signaling complexes (DISC) [12, 13]. MC159 also inhibits the activation of the host pro-inflammatory transcription factor NF-κB by associating with host signaling proteins IκB kinase-γ (IKKγ) and TNF receptor-associated factor 2 (TRAF2) [11]. MC160 expression blocks activation of the IKK complex presumably by associating with both procaspase-8 and heat shock protein 90 (Hsp90), thereby inhibiting TNF-α induced NF-κB activation [7, 8]. However, despite the MC160 protein’s ability to bind both FADD and procaspase-8 through its DEDs, MC160 does not have a detectable effect on apoptosis [8, 12].
Recently, MC159 and MC160 were each reported to inhibit Mitochondria Antiviral Signaling (MAVS) adapter protein-mediated activation of the interferon (IFNβ) promoter [10, 14]. MAVS is present in the mitochondria and forms a complex with viral dsRNA sensing proteins retinoic acid inducible gene 1 (RIG-I) and melanoma differentiation-associated factor gene (MDA5) [15–20]. The complex leads to the activation of the kinases TANK binding kinase 1 (TBK1) and IKKε, which phosphorylate and activate the interferon regulatory factor 3 (IRF3) resulting in activation of IFNβ promoters [21–23]. The MC159 protein binds to both TBK1 and IKKε to inhibit IRF3 activation [10]. MC160 expression inhibits MAVS-mediated IRF3 activation, but the molecular mechanisms of how the MC160 functions in this pathway is not well understood [10].
Both the MC159 and the MC160 DEDs contain conserved RxDL motifs. The RxDL motif is found in several DED-containing proteins including procaspase-8, FADD, and the ORF-K13 protein from Human Herpes Virus 8 [13, 24–27]. Structural analysis of the MC159 DEDs revealed that the arginine and aspartate residues of the RxDL motif interact with either an upstream aspartate or an upstream glutamate. This interaction is collectively referred to as the charged triad. Charged triads are a key element in DED protein folding and provide a network of hydrogen bonding that is necessary for protein–protein interactions [24, 25]. In the MC159 protein, mutating the RxDL motif of the charged triad in the first DED results in mutant proteins that no longer inhibit either apoptosis or NF-κB activation [11, 24, 28]. However, the MC160 RxDL motifs have not yet been characterized. In the current study, we investigated whether RxDL motifs of MC160 are required for MC160 function. We utilized alanine mutations to substitute the arginine and aspartate residues present in the MC160 RxDL in both DED1 and DED2 to neutralize the charged triad. Based on MC159 studies, we hypothesized that mutation of residues within the charged triad would disrupt local MC160 structure and diminish its function. MC160 mutant constructs were assessed for their effect on NF-κB activation, IFNβ activation, as well as the ability to associate with procaspase-8, a known target of MC160. Finally, these mutations were modeled against the structure of the MC159 protein.
Materials and methods
Cells, plasmids, and reagents
HEK 293T cells were purchased from the ATCC and maintained in DMEM supplemented with 10% FCS and 1% Pen/Strep. Recombinant human TNF-α was purchased from Sigma Aldrich. pHA-MC160, pHA-MC160 N, and pHA-MC160D2 have been described previously [8]. pMAVS, encoding a Flag tagged MAVS protein and pTBK1 encoding a FLAG-TBK1 protein, were gifts from Fanxiu Zhu (Florida State University, Tallahassee, FL) and Siddharth Balachandran (Fox Chase Cancer Center, Philadelphia), respectively.
MC160 mutagenesis
To query the importance of conserved RxDL motifs in each DED of MC160, site-directed mutagenesis was used with HA-tagged-MC160/pCI and HA-tagged-MC160 N-terminal/pCI plasmid. MC160 DED1 arginine and aspartic acid at residues 67 and 69, respectively, were both altered to alanine using the MC160 R67A D69A forward primer (5′-GAGAACAGCAAAAGCCCGGAGC-3′) with either NREV (5′-CCGTCCGCTCTAGATTACCCCGCGGA-3′) for pHA-MC160 N/pCI or CREV (5′-CGTCTAGACGCTCGCTAGTAGG-3′) for the full-length pHA-MC160/pCI; and MC160 R67A D69A reverse primer (5′-GAGAACAGCAAAAGCCCGGAGC-3′) with NFOR (5′-CGAGAATTCGCCACCATGTATCCA-3′) for both pHA-MC160/pCI and pHA-MC160 N/pCI. MC160 DED2 arginine and aspartic acid at amino acid sites 160 and 162, respectively, were both altered to alanines using the MC160 R160A D162A forward primer (5′-GCCGTTTGCGCTTATGCTCTTTCC-3′) with either NREV for the pHA-MC160/pCI or CREV for pHA-MC160/pCI; and MC160 R160A D162A reverse primer (5′-GGAAAGAGCATAAGCGCAAACGGC-3′) and NFOR for both pHA-MC160/pCI and pHA-MC160 N/pCI. DNA alterations are underlined within the primer sequence to highlight alanine substitutions. Separate MC160 products were combined and amplified using PCR primers NFOR with either CREV or NREV to produce the MC160 and MC160 N-terminal RxDL mutants, respectively. To create mutations on both DEDs of MC160, site-directed mutagenesis through PCR was used with newly created DED1 mutant sequences with the MC160 R160A D162A forward and reverse primers. Purified products were inserted back into pCI vector through restriction enzyme digestion with EcoRI and XbaI and ligase reaction. DNA sequencing confirmed the presence of the mutations (Genewiz).
Luciferase assays
HEK 293T cells were transfected with 500 ng of pFLAG-MAVS, 450 ng of pIFNβ-Firefly luciferase, 50 ng of pRL-null, and 500 ng of either empty vector or the indicated pHA-MC160 wild-type or mutant construct. Transfections were performed using the Trans-IT 2020 reagent according to the manufacturer’s protocol. At 24 h post transfection, cells were lysed with the 1x passive lysis buffer (Promega). For TNF-α-induced luciferase experiments, cells were transfected with 450 ng of pNF-kB-Firefly luciferase and 50 ng of pRL-null and 500 ng of either pCI or the indicated pHA-MC160 construct. Following 24 h incubation, the cells were stimulated with 10 ng/mL of TNF-α. Following 4 h, the cells were lysed with 1x passive lysis buffer (Promega). Lysates were analyzed in triplicate using the Dual Luciferase assay (Promega) on the Spectromax 5 (Molecular Dynamics). The relative luciferase activity was calculated as the ratio of Firefly to Renilla luciferase values. Luciferase values were normalized to values obtained from untreated cells, transfected with vector alone.
Immunoblotting
HEK293T cells were transfected with the indicated amounts of either pCI, pHA-MC160, pHA-MC159, or the indicated mutant construct. Following 24-h incubation, the cells were lysed with DED lysis buffer [11], and the total protein concentration of the clarified lysates was determined using the bicinchoninic acid assay (Pierce). Lysates were mixed with 1× sample buffer and 5% β-Mercaptoethanol and boiled for 5 min. 10 µg total protein were resolved on 10%-SDS-PAGE and transferred to a PVDF membrane. Membranes were probed with antiserum using the following antibodies: anti-HA (1:1000, Sigma Aldrich), anti-caspase-3 (1:1000, Cell Signaling Technologies), anti-FLAG (1:1000, Sigma Aldrich), and anti-actin (1:500, Santa Cruz). Secondary antibodies of either goat anti-Rabbit HRP (Invitrogen) or goat anti-mouse HRP (Invitrogen) were used at a concentration of 1:5000. Bands corresponding to the indicated protein were visualized via chemiluminescence using the SuperSignal West Pico substrate (Pierce). All images were obtained using the Fluorchem E system (Protein Simple).
Immunoprecipitations
Immunoprecipitation reactions were performed as previously described [8]. Briefly, HEK 293T cells were co-transfected with one µg of procaspase-8 C360S/pCI and one µg of either pCI, pHA-MC160/pCI, pHA-MC160 DM/pCI, or pMC163/pcDNA3.1. Upon 24 h post transfection, cellular monolayers were collected and lysed in 150 µL of DED IP lysis buffer supplemented with protease inhibitors. Lysates were centrifuged for 10 min at 14,000×g at 4 °C, and 50 µL of the supernatants were removed and used to verify protein expression. The remaining lysates were removed, placed in a new tube, and incubated with 1 µg of rabbit anti-procaspase-8 monoclonal antibodies (Santa Cruz) overnight under constant rotation. Fifty microliters of protein G-Sepharose beads were added and incubated with the lysates for an additional 6 h. Beads were collected by centrifugation (14,000×g for 2 min) and washed three times with 500 µL of DED IP lysis buffer. Immunoprecipitated complexes were mixed in 30 µL of 2× Laemmli buffer containing 5% 2-mercaptoetanol. Complexes were boiled for 5 min and immunoblotted using anti-HA (Sigma Aldrich) and anti-procaspase-8 antibodies (Santa Cruz).
Molecular modeling
Computations were performed using a Mac Pro 6-core Intel Xenon X5 processor with Macintosh Operating System (El Capitan), and the software utilized for this study was Schrödinger (2015-1) Maestro. Protein structures were initially refined using Protein Preparation Wizard [29] using the default parameters. Homology modeling was performed using Prime, which is a component of Maestro [30]. MC160 sequence was scanned for homology using Blast Homology Search. PDB with the highest homology (MC159 – PDB ID: 2BBR) [24] was selected (Identities—43%, Positives—60%, and Gaps—2%) from the scan results. Secondary structure prediction was performed using Prime STA, and the model was built using knowledge-based approach, which was available as an option in the program. Thus, constructed homology model was refined using Protein Preparation Wizard by maintaining default parameters. The homology model of MC160 was then analyzed using Ramachandran plot (available as a tool in Maestro 2015-1) after loading the refined homology model in the workspace.
Protein structure alignment was performed using Maestro 2015-1, and proteins were aligned using default parameters. Mutations were performed using Maestro 2015-1, by selecting alanine as the mutant amino acid residue. Further, the mutated proteins were refined using Protein Preparation Wizard, with default parameters. These models were used to interpret the experimental data.
Results
Characterization of MC160 mutant proteins lacking a functional RxDL motif
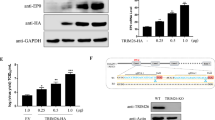
The RxDL motif in DEDs is highly conserved and is present in several DEDs including cellular procaspase-8 and FADD, as well as other vFLIPs such as the Human Herpes Virus 8 ORF-K13 protein (Fig. 1a). The MC159 protein RxDL motif in DED1 was previously shown to be required for MC159 mediated apoptosis as well as its ability to dampen TNF-α-induced NF-κB activation [11, 24, 25]. Therefore, we reasoned that this motif would be required for MC160 anti-inflammatory properties. To explore this possibility, site-directed mutagenesis was used to change the arginine and aspartate in the RxDL motifs in MC160 DED1 and DED2 to alanines (R67A, D69A in DED1 and R160A, D162A in DED2, Fig. 1b). Interestingly, the MC160 constructs containing either mutated RxDL motifs were not expressed as abundantly as compared to the full-length MC160 wild-type or truncated MC160 N, a construct that contains only the MC160 DEDs (a.a. 1-220) (Fig. 1c). A similar trend was observed when the RxDL motifs were mutated in the MC159 protein [28], suggesting that this RxDL motif is important for protein stability. This decrease in MC160 protein expression was not due to uneven protein loading because actin levels were similar for all samples (Fig. 1c).
Expression of HA-tagged mutant MC160 protein. a Alignment of MC159, MC160, ORK13, FADD, and procaspase-8 DED1 and DED2 (Clone Manager Professional). Conserved amino acids residues are indicated in gray. The RxDL motifs in DED1 and DED2 are indicated by a solid black line. An asterisk denotes the location of mutations made in MC160: R67A;D69A (DED1), R160A;D162A (DED2). b Representation of the full-length (FL) and truncated (N) MC160 constructs. Amino acid residues are indicated above. c 1 µg of plasmids expressing HA-tagged MC160 wild-type or mutated versions of the MC160 protein were transfected into HEK 293T cells. pCI denotes the empty vector control, WT wild-type MC160, RxD1 R67A;D69A, RxD2 R160A;D162A, double mutant (DM) = R67A;D69A/R160A;D162A. Mutations were made in either the full-length (pHA-MC160) or the DED-containing region lacking the C-terminus (pHA-MC160N). 24 h post transfection, 25 µg of total protein from each cellular lysates were resolved on 10% SDS-PAGE and proteins were transferred to a PVDF membrane. Bands corresponding to wild-type or mutant proteins were visualized with anti-HA antiserum and detected via chemiluminescence. Blots were re-probed with anti-actin antibodies as a loading control. Molecular weight (MW) markers are indicated to the left of the panel
The RxDL motif of MC160 DEDs is not required to inhibit TNF-α-induced NF-κB activation
We next analyzed the effect of these mutations on the MC160 protein’s ability to dampen TNF-α-induced NF-κB activation. NF-kB activation was quantified using a luciferase reporter assays. HEK 293T cells were transfected with the indicated vector as well as a Firefly reporter plasmid under the control of an NF-κB enhancer element. Upon 24 h post transfection, cells were stimulated with 10 ng/mL of TNF-α for 6 h. MC159 constructs possessing mutated RxDL motifs in DED1 were greatly compromised in their ability to inhibit both apoptosis and NF-kB activation [11, 24, 28]. In contrast, the mutation of the first (RxD1) or second (RxD2) RxDL motif of MC160 did not alter the NF-kB inhibitory properties of MC160 (Fig. 2a). Furthermore, MC160 retained its inhibitory function if both RxDL motifs were mutated simultaneously (construct MC160 DM; Fig. 2b). Neither MC160 WT nor DM induced NF-κB activity in the absence of TNF-α (Fig. 2b), suggesting that the mutations in the RxDL motif did not grossly alter MC160 function. As a control, we over-expressed a procaspase-8 mutant that lacks proteolytic activity. In contrast to results obtained using MC160, overexpression of procaspase-8 activated NF-κB activation and enhanced TNF-α-induced NF-κB activation (Fig. 2b). This observation is consistent with previous reports of procaspase-8 activating NF-κB [8, 29–33]. Therefore, inhibition of TNF-α-induced luciferase activity is specific to MC160 and its mutants and not a result of overexpressing a DED-containing protein.
The MC160 RxDL motif is not required for TNF-α-induced luciferase activity. HEK 293T cells were co-transfected with pNF-κB Firefly luciferase, pRL-null, and either pCI the full-length pHA-MC160 (FL) (a, b), procaspase-8 C360S (b), or truncated pHA-MC160 N/pCI (c) expression vectors. 24 h post transfection, cells were stimulated with 10 ng/mL TNF-α. Following 6 h post stimulation, cells were lysed, and the ratio of Firefly:Renilla luciferase values were calculated. Values were normalized to unstimulated pCI transfected cells. All assays were performed in triplicate, n = 3. Asterisks denote statistically significant inhibition as determined by the Student’s t test (p < 0.05)
Previously, we reported that the C-terminus of MC160 (residues 224–371) inhibits TNF-α-induced NF-κB activation when expressed independently from the MC160 DEDs [8]. Therefore, we reasoned that the presence of this C-terminal region in the above constructs might mask any loss of function in the N-terminal region of the MC160 protein. Thus, these assays were repeated using MC160 constructs in which the C-terminus was absent (Fig. 2c), and the RxDL motifs were mutated in one or both DEDs. Under these conditions, MC160 still retained its ability to inhibit TNF-α-induced NF-κB regulated luciferase activity (Fig. 2c). Therefore, we conclude that the MC160 RxDL motif is not required for inhibition of TNF-α-induced NF-κB activation.
Characterization of the MC160 RxDL motif on MAVS-induced IFNβ activation
Recently, MC160 expression was reported to antagonize the activation of the IFNβ promoter when cells over-express MAVS or TBK1 [10], as measured by the expression of a luciferase reporter gene controlled by the natural IFNβ enhancer [34]. Using this same reporter assay, we asked if our same mutant MC160 proteins still antagonized IFNβ activation. The mutated MC160 proteins dampened MAVS-induced IFNβ enhancer activation similar to wild-type MC160 (Fig. 3a). Both the C-terminal and N-terminal regions of the MC160 protein independently antagonize MAVS-induced IFNβ-activation when expressed independently [10]. Therefore, one concern was that the presence of the MC160 C-terminal region might mask any loss of function in the DED-region of MC160. However, as was the case with TNF-α-induced NF-κB activation, mutations in the RxDL motifs in the HA-MC160 N truncation mutant had no effect on MAVS-induced activation of the IFNβ enhancer element (Fig. 3a, pHA-MC160 N). A similar result was obtained when cells were stimulated instead by TBK1 overexpression (Fig. 3b). As a negative control, we transfected the cells with pHA-MC160D2/pCI. This mutant expresses only the second death effector domain and was previously reported to not inhibit TBK1-induced IFNβ activation. Consistent with results from Randall et al. [10], cells transfected with HA-MC160D2 did not inhibit TBK-1-induced IFNβ activation (Fig. 3b). Therefore, we conclude that the RxDL motifs are not required for MC160′s ability to inhibit activation of IFNβ.
RxDL motif in either DED of MC160 is not required to inhibit either MAVA or TBK-1-mediated IFNβ activation. HEK 293T cells were co-transfected with the IFNβ luciferase reporter plasmid, pRL-null, and either pCI, pHA-MC160/pCI, or the indicated MC160 mutant. IFNβ activation was achieved by overexpressing either pMAVS (a) or pTBK1 (b). The ratios of Firefly:Renilla were normalized to pCI transfected cells in the absence of either pMAVS or pTBK1. All experiments were performed in triplicate, n = 3. Asterisks * denote statistically significant inhibition (p < 0.05)
The RxDL motif of MC160 is dispensable for binding procaspase-8
The MC160 protein interacts with procaspase-8 through the MC160 DEDs [8]. While this interaction does not enable MC160 to inhibit apoptotic responses, it remains possible that the MC160-procaspase-8 interaction plays a role in MC160’s ability to antagonize additional pathways such as TNF-α-induced NF-κB activation and MAVS-mediated IFNβ-induction. Therefore, we asked if the RxDL motif in the MC160 DEDs was required for MC160 binding to target proteins. To this end, we co-transfected HEK 293T with a plasmid expressing an inactive procaspase-8 (C360S) and either empty vector, pHA-MC160, the MC160 Double Mutant (DM), or pMC163-HA. pMC163-HA expresses the MC163-HA protein and serves as the negative control as MC163 lacks any predicted region that would indicate procaspase-8 interaction. Procaspase-8, HA-MC160 wild-type, the double mutant, and MC163 were readily detected in the lysates (Fig. 4). Both the MC160 wild-type and double mutant immunoprecipitated with procaspase-8 indicating that the MC160-procaspase-8 interaction was not affected by mutating the RxDL motif. As predicted, MC163 did not immunoprecipitate with procaspase-8 ruling out the possibility of nonspecific interactions in our assay. Therefore, the MC160 RxDL motif is not required for MC160’s ability to interact with procaspase-8.
MC160 mutants lacking the RxDL motif interact with procaspase-8. HEK 293T cells were transfected with procaspase-8 C360S/pCI and either pHA-MC160, pHA-MC160DM, or pMC163C-HA. Following 24 h post transfection, cells were lysed and a portion of the cellular lysates was used to verify protein expression. The remaining lysates were immunoprecipitated using anti-procaspase-8 antibodies bound to protein G-Sepharose beads. Both immunoprecipitation reactions and lysates were resolved on 10% SDS-PAGE and proteins were transferred to a PVDF membrane. Bands corresponding to procaspase-8, HA-MC160, or MC163-HA were visualized by probing with anti-procaspase-8 and anti-HA antisera. Bands were visualized by chemiluminescence. Molecular weight markers (MW) are indicated to the left of the blots
Molecular modeling
Homology modeling of v-FLIP MC160 protein
Our results were surprising given that similar mutations in the MC159 charged triad significantly altered the function of MC159. Therefore, we next wanted to perform in silico homology modeling of the 3D structure of the MC160 DEDs to determine if there were significant structural differences between MC159 and MC160. The 3D structure of tandem DEDs in MC160 was derived by subjecting its amino acid sequence to homology modeling with the v-FLIP MC159 protein as a template (PDB ID: 2BBR) [24]. Subsequently, the refined homology model of MC160 (Fig. 5a) was analyzed for its quality using Ramachandran plot, which suggested that majority of amino acids were placed under the favored region indicating a good quality 3D structure of the homology model (Fig. 5b). Protein structure alignment was performed (Figs. 6a, 6b) thereafter with the homology model of MC160 aligned onto the template v-FLIP MC159 protein. Structures of the proteins MC160 and MC159 were aligned using the C-alpha atoms and an alignment score of 0.012 Å was obtained, which is indicative of a reasonably good alignment (a score of 0.6 and less indicates a good alignment) [35]. The DED domains of MC160 were studied for the key interactions, focusing mainly on the RxDL domains located in both DED1 and DED2 domains. Key hydrogen bonding interactions present in the RxDL motif of DED1 were conserved within the homology model of MC160 (Fig. 6c). Interestingly, a change in the orientation of aspartate 109 was observed within DED2 of MC160 (Fig. 6d). Overall, our modeling results suggest that the structure of MC160’s DEDs is predicted to be similar to that of the MC159 protein. However, the projected structure of MC160 and accuracy of molecular modeling data will need to be confirmed experimentally by X-ray crystallography or nuclear magnetic resonance (NMR).
Homology model of MC160 obtained by using v-FLIP MC159 as a template. a Representation of the homology model of MC160 in ribbons. b Ramachandran plot of the homology model. Orange represents favored region; Yellow represents allowed region and white represents disallowed region (Color figure online)
Results of protein structure alignment of MC160 onto MC159 by using MC159 as a template. a Representation of the structural alignment of proteins MC160 and MC159 with C-alpha atoms. (C α-Helix; E β-Sheet and C coil). b 3D representation of the superimposed proteins as ribbon models. MC159 is represented in Cyan and MC160 in green color. c, d Superimposition of RxDL motifs located within the DED1 and DED2 domains of proteins MC159 and MC160, respectively. The key amino acids are represented as ball and stick model. Carbon atoms are represented in green color in case of MC160 and gray color for MC159. Oxygen and nitrogen atoms are represented in red and blue colors, respectively, for both MC159 and MC160. Hydrogen bonds are shown as yellow broken lines (Color figure online)
Alanine mutation of RxDL charged triad in the DED domains of MC160
Since experimental mutational studies of DED domains in the v-FLIP MC160 demonstrated no variation in its normal function (Figs. 2, 3,, 4), we proceeded to carryout alanine mutations with residues present in the RxDL motifs of the two DED domains computationally with (R67A and D69A in DED1; R160A and D162A in DED2), one DED domain at a time and both the domains together (R67A, D69A, R160A and D162A). Subsequently, we performed protein structure alignment of the wild-type MC160 with the DED1 and DED2 single domain mutations (Fig. 7a, b) and double mutants (Fig. 8a, b) MC160 protein wherein we noticed negligible difference in the structural integrity of the overall protein (see Fig. S1a, S1b and S1c in supporting information) and the orientation of the additional residues located in the RxDL domain of the mutated (Figs. 7a, b, 8a, b) versus wild-type MC160 of the two domains in comparison to the wild-type MC160. To investigate the effect of mutation in MC159, we sought to mutate the RxDL motif located in DED1 domain of MC159, by replacing R69 and D71 with alanine residues and according to the experimental mutational studies done by Yang et al. [ [24].], mutating RxDL motif with alanine in DED1 led to a loss of apoptosis protection by the v-FLIP protein. Surprisingly, we did not notice a significant variation in the structural integrity (see Fig. S2 in supporting information) of the overall protein or the orientation of the associated amino acids in the RxDL motif of DED1 mutant MC159 (Fig. 8c). Predicted structures of the mutated MC160 RxDL motifs will need to be verified experimentally.
Single mutations of the RxDL motifs present within the DED domains of MC160. Mutated residues are represented in green color and the other amino acids follow the same representation pattern as described in Fig. 5. a Aligned structure of ARG 67 ALA and ASP 69 ALA mutant MC160 protein with wild-type MC160. b Aligned structure of ARG 160 ALA and ASP 162 ALA mutant MC160 protein with wild-type MC160
Representation of mutant DED domains of MC160 and DED1 mutant MC159. All the figures are depicted in the same fashion as shown in Fig. 6. a Double mutant DED1 RxDL motif of MC160 containing ARG 67 ALA and ASP 69 ALA superimposed onto the wild-type MC160. b Double mutant DED2 RxDL motif of MC160 containing ARG 160 ALA and ASP 162 ALA superimposed onto the wild-type MC160. c DED1 mutant RxDL motif of MC159 containing ARG 69 ALA and ASP 71 ALA aligned onto the wild-type MC159
Computational studies lead us to a conclusion that the role of MC159 in inhibiting apoptosis is mainly achieved probably because of the interactions supplemented by RxDL motif in the formation of ternary complex and that the structural integrity of the protein is maintained in the absence of key residues located within the RxDL motif. However, in MC160, the RxDL motif in DED1 might not be playing a significant role in known MC160 functions.
Discussion
While the conserved RxDL motifs present in MC159 DEDs are required for many MC159 functions, our data indicates that the RxDL motifs present in MC160 DEDs are dispensable for the MC160 protein’s ability to inhibit both TNF-α-induced NF-κB activation and MAVS-mediated activation of IFNβ. Further, an MC160 mutant with alanine substitutions in both RxDL motifs in DED1 and DED2 retained its ability to bind to procaspase-8, suggesting that these regions are not required for MC160 DEDs to associate with a known cellular target. Molecular modeling predicted no apparent loss of structural integrity in the MC160 protein. Taken together, we conclude that the charged triad is not required for any known function of the MC160 protein.
Unlike the MC160 protein, the structure of MC159 and its interactions with target proteins have been extensively studied. The charged triad in both MC159 DEDs provides a point of organization for surrounding amino acids that ensures proper folding of the vFLIP structures that orients additional surface amino acids. Alanine substitutions in the RxDL motif of either MC159 DEDs result in MC159 mutant proteins that could no longer inhibit apoptosis [28]. Interestingly, Yang et al. reported that the first DED1 RxDL motif was most critical in forming a ternary complex with FADD, whereas mutations in the second DED2 of MC159 were better tolerated in terms of binding [24]. Further, alterations in the RxDL motif in DED1 of MC159 resulted in a mutant protein that could no longer inhibit TNF-α-induced NF-κB activation suggesting that this motif is required for other functions of the MC159 protein [11].
While the requirement of the charged triad has not been determined for all functions of the MC159, the hydrogen bonding network formed by the charged triad properly orients amino acids in such a way that allows critical regions to contact target proteins [24]. However, in our predicted molecular models where alanine substitutions were introduced in MC160 DED1 and DED2, we did not observe a significant impact on the overall structural integrity of MC160 as well as amino acid residues within the RxDL domain of the mutant when compared to the wild-type MC160. This observation combined with the experimental data leads us to conclude that the RxDL motif is not required for the known functions of the MC160 protein.
It is of interest that MCV codes for two tandem DED-containing proteins. However, despite their sequence similarities, MC159 and MC160 have different biological properties when expressed in host cells. For example, MC159 binding to FADD and procaspase-8 and prevents FADD oligomerization and thus inhibits apoptosis [28]. Although MC160 binds to both FADD and procaspase-8, MC160 expression does not have a detectable effect on apoptosis. Both MC159 and MC160 inhibit TNF-α-induced NF-κB activation and MAVS-induced IRF3 activation, but the viral proteins target different areas in the pathway to elicit their effects. Of note, given that both FADD and procaspase-8 are involved in MAVS signaling complexes, it remains possible that MC159 and MC160’s ability to associate with these proteins is at least partially responsible for the ability of MCV vFLIPs to antagonize the pathway. Based on the observation that our MC160 mutants retain their ability to both bind procaspase-8 and inhibit MAVS and TBK1-mediated IFNβ activation, we cannot rule out the potential of the MC160-procaspase-8 interaction as a potential means by which MC160 elicits its effects in host cells.
Unlike MC159, the molecular mechanism of MC160 has been difficult to pin down. While we have previously shown MC160 interacts with several host proteins, none of these interactions has been validated by identifying loss of function MC160 mutants incapable of binding these proteins. Based on our molecular modeling projections, the predicted structural homology between MC159 and MC160 death DED domains is considerably high. Therefore, future studies targeting amino acid residues predicted to be present on the surface of the MC160 protein based on similarity with MC159 may lead us in the identification of the means by which MC160 inhibits activation of host innate immune pathways.
References
C.M. Randall, J.L. Shisler, Future Virology 8, 561–573 (2013)
X. Chen, A.V. Anstey, J.J. Bugert, Lancet. Infect. Dis 13, 877–888 (2013)
J.L. Shisler, Adv. Virus Res. 92, 201–252 (2015)
T.G. Senkevich, J.J. Bugert, J.R. Sisler, E.V. Koonin, G. Darai, B. Moss, Science 273, 813–816 (1996)
T.G. Senkevich, E.V. Koonin, J.J. Bugert, G. Darai, B. Moss, Virology 233, 19–42 (1997)
L.E. Murao, J.L. Shisler, Virology 340, 255–264 (2005)
D.B. Nichols, J.L. Shisler, J. Virol. 80, 578–586 (2006)
D.B. Nichols, J.L. Shisler, J. Virol. 83, 3162–3174 (2009)
C. Randall, J. Shisler, Future Virol. 8, 561–573 (2013)
C.M. Randall, S. Biswas, C.V. Selen, J.L. Shisler, Proc. Natl. Acad. Sci. U.S.A. 111, E265–272 (2014)
C.M. Randall, J.A. Jokela, J.L. Shisler, J. Immunol. 188, 2371–2379 (2012)
J.L. Shisler, B. Moss, Virology 282, 14–25 (2001)
J. Bertin, R.C. Armstrong, S. Ottilie, D.A. Martin, Y. Wang, S. Banks, G.H. Wang, T.G. Senkevich, E.S. Alnemri, B. Moss, M.J. Lenardo, K.J. Tomaselli, J.I. Cohen, Proc. Natl. Acad. Sci. U.S.A. 94, 1172–1176 (1997)
S. Balachandran, T. Venkataraman, P.B. Fisher, G.N. Barber, J. Immunol. 178, 2429–2439 (2007)
R.B. Seth, L. Sun, C.K. Ea, Z.J. Chen, Cell 122, 669–682 (2005)
T. Kawai, K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K.J. Ishii, O. Takeuchi, S. Akira, Nat. Immunol. 6, 981–988 (2005)
M. Yoneyama, M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, T. Fujita, Nat. Immunol. 5, 730–737 (2004)
V. Hornung, J. Ellegast, S. Kim, K. Brzozka, A. Jung, H. Kato, H. Poeck, S. Akira, K.K. Conzelmann, M. Schlee, S. Endres, G. Hartmann, Science 314, 994–997 (2006)
A. Pichlmair, O. Schulz, C.P. Tan, T.I. Naslund, P. Liljestrom, F. Weber, C. Reis e Sousa, Science 314, 997–1001 (2006)
H. Kato, O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K.J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C.S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, S. Akira, Nature 441, 101–105 (2006)
K.A. Fitzgerald, S.M. McWhirter, K.L. Faia, D.C. Rowe, E. Latz, D.T. Golenbock, A.J. Coyle, S.M. Liao, T. Maniatis, Nat. Immunol. 4, 491–496 (2003)
S. Sharma, B.R. tenOever, N. Grandvaux, G.P. Zhou, R. Lin, J. Hiscott, Science 300, 1148–1151 (2003)
G. Oganesyan, S.K. Saha, B. Guo, J.Q. He, A. Shahangian, B. Zarnegar, A. Perry, G. Cheng, Nature 439, 208–211 (2006)
J.K. Yang, L. Wang, L. Zheng, F. Wan, M. Ahmed, M.J. Lenardo, H. Wu, Mol. Cell 20, 939–949 (2005)
F.Y. Li, P.D. Jeffrey, J.W. Yu, Y. Shi, J. Biol. Chem. 281, 2960–2968 (2006)
P.E. Carrington, C. Sandu, Y. Wei, J.M. Hill, G. Morisawa, T. Huang, E. Gavathiotis, M.H. Werner, Mol. Cell 22, 599–610 (2006)
C. Shen, H. Yue, J. Pei, X. Guo, T. Wang, J.M. Quan, Biochem. Biophys. Res. Commun. 463, 297–302 (2015)
T.L. Garvey, J. Bertin, R.M. Siegel, G.H. Wang, M.J. Lenardo, J.I. Cohen, J. Virol. 76, 697–706 (2002)
2016-4 S.R. Epik; Impact; and Prime (Schrodinger LLC, New York, 2016)
2016-4 S.R. Prime (Schrodinger, LLC. New York, 2016)
S. Kreuz, D. Siegmund, J.J. Rumpf, D. Samel, M. Leverkus, O. Janssen, G. Hacker, O. Dittrich-Breiholz, M. Kracht, P. Scheurich, H. Wajant, J. Cell Biol. 166, 369–380 (2004)
H. Su, N. Bidere, L. Zheng, A. Cubre, K. Sakai, J. Dale, L. Salmena, R. Hakem, S. Straus, M. Lenardo, Science 307, 1465–1468 (2005)
A. Oberst, D.R. Green, Nat. Rev. Mol. Cell Biol. 12, 757–763 (2011)
M. Sato, H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, T. Taniguchi, Immunity 13, 539–548 (2000)
A.S. Yang, B. Honig, J. Mol. Biol. 301, 665–678 (2000)
Acknowledgements
We would like to thank Joanna Shisler. This work was funded by Seton Hall University and the Seton Hall University Research Council. Structure coordinates of the homology model of MC160 are available upon request.
Author information
Authors and Affiliations
Contributions
Author contributions
MB, UKV, SW, CS, TTT, and DBN have made substantial contributions to work described herein.
Corresponding author
Ethics declarations
Conflicts of interest
The authors have no conflicts of interest to declare.
Ethical approval
The research described in the paper does not use any human or animal subjects.
Additional information
Edited by Wolfram Gerlich.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Beaury, M., Velagapudi, U.K., Weber, S. et al. The molluscum contagiosum virus death effector domain containing protein MC160 RxDL motifs are not required for its known viral immune evasion functions. Virus Genes 53, 522–531 (2017). https://doi.org/10.1007/s11262-017-1456-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-017-1456-9