Abstract
Emerging pseudorabies virus (PRV) variant has led to frequent outbreaks of PRV infection among Bartha-K61-vaccinated swine population in Chinese swine farms and caused high mortality in pigs of all age since late 2011. Here, we generated a gE/gI-deleted PRV (rPRVXJ-delgI/gE-EGFP) based on PRV variant strain (PRV-XJ) through homologous DNA recombination. Compared to parental strain, rPRVXJ-delgI/gE-EGFP showed similar growth kinetics in vitro. Its safety and immunogenicity were evaluated in weaned piglets. Our results showed that piglets immunized with rPRVXJ-delgI/gE-EGFP did not exhibit any clinical symptoms, and a high level of gB-specific antibody was detected. After lethal challenge with variant PRV (PRV-FJ strain), all vaccinated piglets survived without showing any clinical symptoms except slight fever within 7 days post-challenge. In unvaccinated piglets, typical clinical symptoms of pseudorabies were observed, and the piglets were all died at 5 days post-challenge. These results indicated that a live rPRVXJ-delgI/gE-EGFP vaccine could be a maker vaccine candidate to control the currently epidemic pseudorabies in China.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pseudorabies virus (PRV), also called Aujeszky’s disease virus, is a member of the family Herpesviridae, subfamily Alphaherpesvirinae, and genus Varicellovirus. It can infect many domestic and wild animals, and pigs are the natural host and reservoir. PRV infection causes high mortality in young piglets, growth retardation in growing pigs and reproductive failure in sows. The major clinical symptoms include fever, itchiness, respiratory symptoms, ataxia and tetany [1, 2].
Bartha-K61 is an attenuated vaccine strain of which the complete gE gene and part of gI gene have been deleted, playing an important role in controlling and eradicating PRV [3, 4]. With the wide use of Bartha-K61 vaccine and gE ELISA serologic tests, PRV has been deracinated in the United States, Canada, and some European countries in the past decades [5]. In China, PRV was first reported in 1947, and then widely spread in many provinces. In order to control PRV epidemic, the Bartha-K61 vaccine strain was imported from Hungary in 1979, and then PRV was under effective control from the 1990 to 2010 [6]. However, since October 2011, the PRV ourbreaks among Bartha-K61-vaccinated swine population spread rapidly to several provinces in China, which caused significant economic losses to the pig industry [7]. Virus genome analysis showed that the emerging PRV variants belonged to a relatively independent cluster in phylogenetic tree, which were different from the classical PRV strains on antigenicity and caused death in growing and finishing pigs, with a reported mortality rate of 10–30% [7,8,9]. This means that the Bartha-K61 vaccine cannot provide full protection against the emerging PRV variants.
In this study, we constructed a gI/gE-deleted mutant based on currently circulating variant strain (PRV-XJ), and its safety and protective effects were evaluated in weaned piglets for the purpose of developing a new candidate vaccine for PRV.
Materials and methods
All the animal experiments had been approved by the Laboratory Animal Management Committee of Sichuan Province (Approval Number SYXK(Chuan)2014-187).
Cells, viruses, and plasmid
PRV-XJ strain was isolated from the brain of a dead piglet which had been vaccinated from a PRV-infected pig farm in Sichuan province, China, and was preserved by the College of Veterinary Medicine, Sichuan Agricultural University (Chengdu, China). PRV-FJ strain were provided by Dr. Yuancheng Zhou at Livestock and Poultry Biological Products Key Laboratory of Sichuan Province (Chengdu, China), the LD50 is 107TCID50. BHK-21 cells were grown and maintained in RPMI1640 (Hyclone, USA), supplemented with 10% fetal calf serum (Gibco, USA) at 37 °C with 5% CO2 in a humidified incubator. The transfer plasmid pPI-2.EGFP was constructed and preserved by the College of Veterinary Medicine, Sichuan Agricultural University (Chengdu, China).
Generation of rPRVXJ-delgI/gE-EGFP
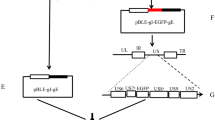
The genomic DNA of PRV-XJ strain was extracted by using saturated phenol–chloroform extraction method. BHK-21 cells were seeded in 12-well plates (1.0 × 105 cells/well). When the cells reached to 90% confluence, the transfer plasmid pPI-2.EGFP and PRV-XJ strain genomic DNA were co-transfected using Lipofectamine™ 3000 reagent (Invitrogen, USA) according to the manufacturer’s instructions. 1.5% low-melting agarose containing RPMI 1640 medium was preheated and added into the plates 6 h later. Based on the EGFP expression, plaque purification was carried out to obtain homogeneous viruses. The absence of the gE and gI genes were verified by PCR using gB-specific primers (gB-F/gB-R), deletion gene-specific primers (F/R), and right homologous-specific primers (F1/R1) (Table 1). The expected gE/gI-deleted virus was named as rPRVXJ-delgI/gE-EGFP (Fig. 1).
Flowchart to produce rPRVXJ-delgI/gE-EGFP. The genome of PRV-XJ (b) was co-transfected with the transfer vector pPI-2.EGFP (a) into BHK-21 cells, homologous recombination was conducted to insert Pcmv, EGFP, poly(A) and part of gI in lieu of gI and gE to generate the recombinant rPRVXJ-delgI/gE-EGFP strain
Indirect immunofluorescence assay
Detection of gE or gB expression in rPRVXJ-delgI/gE-EGFP and PRVXJ-infected cells was performed by indirect immunofluorescence assay. BHK-21 cells were infected with virus at an MOI of 1 for 24 h. Cells were fixed with formaldehyde and acetone (1:1) overnight at 4 °C, and then incubated with mouse anti-gE or anti-gB monoclonal antibody (Beijing TianTech Biotechnology, China) for 1 h at 37 °C. The cells were incubated with FITC-labeled goat anti-mouse IgG (Sigma-Aldrich, USA) for 1 h at 37 °C. Finally, the cells were observed under a fluorescence microscope (Nikon, Japan).
One-step growth kinetics
One-step growth kinetics was conducted to compare the growth kinetics of the rPRVXJ-delgI/gE-EGFP with the parental virus PRV-XJ. BHK-21 monolayers were infected with virus at a MOI of 1. Cell supernatants were harvested at successive intervals after infection and stored at −80 °C. Virus One-step growth curve was drawn based on 50% tissue culture infectious dose (TCID50).
Animals and experiment design
Twenty-five 4-week-old weaned piglets free of PRV, classical swine fever virus (CSFV), respiratory syndrome virus (PRRSV), and porcine circovirus 2 (PCV2) were randomly divided into five groups of 5 each. The piglets in group I were immunized intramuscularly (i.m.) with 105 TCID50 rPRVXJ-delgI/gE-EGFP suspended in 1 ml of RPMI 1640. The piglets in group II, III, and IV were immunized intranasally (i.n) with 105, 106, and 108 TCID50 rPRVXJ-delgI/gE-EGFP suspended in 1 ml of RPMI 1640, respectively. The piglets in group V were immunized intramuscularly with 1 ml PBS. All piglets were challenged intranasally with 5LD50 lethal variant (PRV-FJ strain) at 28 days post-vaccination (dpv).
Enzyme-linked immunosorbent assay (ELISA)
Blood samples were collected at 0, 7, 14, 21, 28, 35, and 42 dpv. The serum was separated by centrifugation after coagulation, and PRV-specific gE and gB antibodies in serum were detected using ELISA kits (IDEXX, USA) according to the manufacturer’s directions. Twofold dilution of standard sera (100 µl/well) were added. Serum samples (100 µl/well) were added, and then the plate was incubated for 1 h at 37 °C. After washing five times with washing solution, horseradish peroxidase (HRP)-labeled goat anti-mice IgG was added, and the plates was incubated for 30 min at 37 °C. After washing, TMB substrate solution were added 100 µl/well and then incubated for 15 min in the dark. The reaction was terminated by the addition of 50 µl stop solution. The optical density (OD) at 650 nm was measured by a microplate reader (Bio-Rad, USA).
Virus shedding
To detect virus shedding after vaccination and challenge, nasal and anal swabs from all piglets were collected at 7, 14, and 21 dpv and daily post-challenge (dpc). The swabs were serially diluted tenfold with PBS, and the dilutions were passed through a 0.45-μm filter and added into BHK-21 cells. After cytopathic effect (CPE) appeared, the virus DNA was extracted using saturated phenol–chloroform extraction method, and identified by PCR using gE F/R and gB F/R primers (Table 1).
Statistical analysis
The experimental data were analyzed by GraphPad Prism5.0 software, and error bars represent standard deviations. The results are expressed as mean ± standard deviation (SD), and p < 0.05 and p < 0.01 were considered as statistically high and extremely significantly, respectively.
Results
Generation of the recombinant virus rPRVXJ-delgI/gE-EGFP
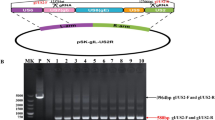
After co-transfecting transfer plasmid pPI-2.EGFP and genomic DNA of PRV-XJ strain into BHK-21 cells, the recombinant virus was distinguished from parental PRV-XJ strain based on the expression of EGFP (Fig. 2a). After plaque purification for several times, when all the cells expressed EGFP, virus was harvested, as rPRVXJ-delgI/gE-EGFP F1. The specific PCR products were observed from PRV-XJ F1, F5, F10 (2682 bp), while no PCR product was observed from rPRVXJ-delgI/gE-EGFP F1, F5, F10 with F/R primers. The specific PCR products were observed from rPRVXJ-delgI/gE-EGFP F1, F5, F10 and PRV-XJ F1, F5, F10 with gB F/R primers (264 bp), rPRVXJ-delgI/gE-EGFP F1, F5, F10 (1600 bp), while no PCR product was observed from PRV-XJ F1, F5, F10 with F2/R2 primers (Fig. 2b). gB protein was detected by IFA in BHK-21 cells infected with PRV-XJ and rPRVXJ-delgI/gE-EGFP, whereas the gE protein could only be detected in BHK-21 cells infected with PRV-XJ (Fig. 2c), which indicated that the recombinant rPRVXJ-delgI/gE-EGFP virus was successfully rescued.
Identification of rPRVXJ-delgI/gE-EGFP. a Identification of rescued RP virus from the CPE cells with green fluorescence under the fluorescent microscope. b Verification of gE/gI genes deletion in the genome of rPRVXJ-delgI/gE-EGFP by PCR. M DL2000 DNA Marker; 1–3 rPRVXJ-delgI/gE-EGFP F1, F5, F10 detection by F/R; 4–6 PRVXJ F1, F5, F10 detection by F/R; 7–9 rPRVXJ-delgI/gE-EGFP F1, F5, F10 detection by gB F/R; 10–12 PRVXJ F1, F5, F10 detection by gB F/R; 13–15 rPRVXJ-delgI/gE-EGFP F1, F5, F10 detection by F2/R2; 16–18 PRVXJ F1, F5, F10 detection by F2/R2; ‘−’ negative control. c The confirmation rPRVXJ-delgI/gE-EGFP of by detection of gE and gB protein using indirect fluorescence assay
The determination of growth kinetics
The growth features of reconstituted rPRVXJ-delgI/gE-EGFP were analogous to that of parental PRV-XJ in BHK-21cells (Fig. 3). The peak titer of rPRVXJ-delgI/gE-EGFP and PRV-XJ were 107.93TCID50/ml and 108.29TCID50/ml, respectively.
One-step growth curves of gene-deleted viruses and parent virus on BHK-21. BHK-21 cells grown in a 24-well plate were inoculated with PRV-XJ strain or rPRVXJ-delgI/gE-EGFP at an MOI of 1. The culture supernatant was collected at the indicated time points and used to determine the viral titers. Data were presented as mean ± SD
Protection of vaccinated piglets from virulent challenge
To investigate the protective efficacy of rPRVXJ-delgI/gE-EGFP as a candidate vaccine, 25 piglets were randomized in five groups and vaccinated with the recombinant virus. There were no clinical symptoms in all piglets after vaccination (data no shown). After 28 days, all piglets were intranasally challenged with 5LD50 virulent PRV (PRV-FJ strain). All piglets in unvaccinated group showed typical clinical symptoms, such as prolonged high fever, spirits atrophy and neurologic symptoms, and died within 5 dpc. On the contrary, no clinical symptoms were observed in vaccinated groups except fever. The piglets in group I showed fervescence between 2 and 5 dpc (up to 40.2 °C), and returned to normal body temperature at 7 dpc. All piglets in group II showed slight fever (no more than 40 °C) between 2 and 4 dpc, and then returned to normal body temperature. The temperature of all piglets in group III and group IV had no obvious change.
The gB-specific antibodies of vaccinated piglets were detected at 7 dpv, and kept steadily increasing until 21 dpv and then decreased slightly at 28 dpv. On the contrary, gB-specific antibodies failed to be detected in the unvaccinated piglets until death. The gB-specific antibodies in vaccination groups were significantly higher than that in the control group (p < 0.01), and there were no significant differences among vaccination groups (Fig. 4a). The gE-specific antibodies of vaccinated piglets and unvaccinated piglets were failed to be detected before PRV-FJ challenge. After PRV-FJ challenge, the gE-specific antibodies of vaccinated piglets increased, and all vaccinated piglets showed positive gE-specific antibody responses (Fig. 4b).
Protective efficacy of rPRVXJ-delgI/gE-EGFP immunization. a Development of PRV gB-specific Ab (a) and gE-specific Ab (b) in piglets vaccinated with rPRVXJ-delgI/gE-EGFP and challenged with PRV-FJ strain were detected with pseudorabies virus gB (gE) test kit (IDEXX, USA). If S/N ≤ 0.6, the sample is positive for gB antibodies or gE antibodies. S/N sample to negative control ratio, Data in a, b were presented as mean ± SD
Virus shedding
The isolated PRV from nasal and anal swabs was detected with gE-F/R and gB-F/R primers. The results showed that piglets inoculated with rPRVXJ-delgI/gE-EGFP did not shed virus. After challenge, the PRV shedding in all experimental groups were detected from 1 to 3 dpc. The gE gene could not be detected at 4 dpc. These results demonstrated that rPRVXJ-delgI/gE-EGFP may have certain protective effect of piglets from infection with virulent PRV (Table 2). The titers of virus shedding were shown in Fig. 5.
Vaccination reduced the amount and duration of virus shedding. Titers of shed virus in control group were 103.232TCID50/ml at 1 dpc and reach the maximum (106.518TCID50) at 3 dpc. In vaccination groups, the titers were ranged from 102.406 to 102.666TCID50 at 1 dpc and reached the maximum (102.315–102.893TCID50) at 3 dpc. The amount of excreted virus in the vaccination groups were significantly lower than that in the control group (p < 0.01) from 1 to 3 dpc, and there were no significant differences among vaccination groups. The piglets stopped to shed virus on 4 dpc, but in unvaccinated group virus shedding were still detected.
Discussion
PRV is prevalent on pig farms since it was first reported in China, but it had been well controlled after the use of Bartha-K61 vaccine. However, since late 2011, The outbreaks of PR among Bartha-K61-vaccinated pig population repeatedly happened, and the pigs showed typical clinical symptom of classical PR with 50% mortality among newborn piglets and 10–30% mortality among growing and finishing pigs. gE-antibody positive rates increased up to over 50% [10]. It has already demonstrated that Bartha-K61 vaccine could not provide protection against PRV variant strains, such as PRV HeN1 and TJ strains. The genome analysis of the TJ and HeN1 strains revealed extensive variations when compared with previous isolates worldwide, including substitutions, insertions, and/or deletions [7, 9, 11]. But its variant mechanism is not clear. Therefore, it is urgent to develop more efficient vaccines based on variant strain to control current PR in China.
So far, vaccination is still the most effective measure to control and eradicate PRV, especially gE-deleted vaccine, because of its capacity of infecting second- and third-order neurons of the olfactory and trigeminal routes in the central nervous system decrease significantly [12]. In order to solve the current problem, several vaccines have been developed based on the current PRV variants, including the gE-deleted PRV based on PRV HN1201 strain and PRV TJ strain. These vaccines were reported to protect piglets from variant PRV challenge [13, 14]. The gI gene is nonessential glycoprotein and plays an important role in spreading in non-neuronal cells and anterograde spreading in neurons [12]. Therefore, in this study, we deleted gE/gI gene based on a variant strain PRV-XJ, and the safety and efficacy were evaluated on weaned piglets.
The PRV variant strain (PRV-XJ strain) was isolated from the brain of a dead piglet which had been vaccinated Bartha-K61. The transfer vector pPI-2.EGFP was constructed by our laboratory, and in our previous study, recombinant PRV SA215/VP2 virus and PRV SA215 (gE-/gI-/TK-) have been constructed successfully using pPI-2.EGFP [15, 16]. In this study, rPRVXJ-delgI/gE-EGFP was obtained by homologous recombination between PRV-XJ and transfer vector pPI-2.EGFP. Compared to the parental PRV-XJ, rPRVXJ-delgI/gE-EGFP showed similar growth properties on BHK-21 cells.
Variant PRV infections usually cause high mortality rate of piglets [7]. In our study, the piglets inoculated with different doses of rPRVXJ-delgI/gE-EGFP showed no clinical PR symptoms. It has been demonstrated that PRV-deleted gE/gI genes could protect piglets from challenge with parental PRV strain [17, 18]. So we evaluated the protection and safety of rPRVXJ-delgI/gE-EGFP with a lethal variant, rather than parental strain. After PRV-FJ challenge, piglets in experimental groups showed slight fever without any other PR clinical symptoms, however, all piglets in unvaccinated group died within 5 dpc. This result indicated that rPRVXJ-delgI/gE-EGFP was safe and could provide protection against virulent PRV variant.
The glycoprotein gB of PRV are related to immunogenicity and viral protection [19]. Serological discrimination based on gE-ELISA is used for distinguishing infection with wild-type virus from vaccinated [20]. In this study, piglets vaccinated with rPRVXJ-delgI/gE-EGFP generated gB-specific antibodies at 7 dpv, and did not generate gE-specific antibodies. Posterior to virulent challenge, the level of the gB-specific antibody increased, and gE-specific antibody was tested positive within a week. Therefore, rPRVXJ-delgI/gE-EGFP is a maker vaccine candidate, making it possible to control and eradicate PRV in combination with the use of commercial gE-ELISA kit.
Virus shedding is one of the important parameters to evaluate the efficacy of vaccine [21]. In this study, all piglets did not excrete PRV post immunization. However, piglets in vaccinated groups excreted PRV at 1 dpc and lasted for 3 days. On the contrary, unvaccinated piglets excreted virus at 1 dpc and last until death. In previous study, a gE gene-deleted PRV based on PRV HN1201, a representative PRV variant, was generated and the protective efficacy was tested on 3-week-old pigs [13]. In this study, we deleted gE and gI genes based on PRV-XJ, a representative PRV variant. Compared with the PRV HN1201ΔgE, the shorter period of virus excretion in vaccinated pigs revealed that it is necessary to delete gI gene and induce better immune effect.
In conclusion, a gE/gI-deleted PRV strain (rPRVXJ-delgI/gE-EGFP) based on a circulating field isolate (PRV-XJ) was constructed. In vivo study proved that rPRVXJ-delgI/gE-EGFP is safe to piglets, and could provide protection against emerging PRV variant.
References
T. Müller, E.C. Hahn, F. Tottewitz, M. Kramer, B.G. Klupp, T.C. Mettenleiter, C. Freuling, Adv. Virol. 156(10), 1690–1705 (2011)
L.E. Pomeranz, C. Reynolds, J. Microbiol. Mol. Biol. Rev. 69(3), 462–500 (2005)
E.A. Petrovskis, J.G. Timmins, T.M. Gierman, L.E. Post, J. Virol. 60(3), 1166–1169 (1986)
B. Lomniczi, M.L. Blankenship, T. Ben-Porat, J. Virol. 49(3), 970–979 (1984)
T.C. Mettenleiter, Acta Vet. Hung. 42(2–3), 153–177 (1994)
G.Z. Tong, H.C. Chen, Chin. J. Vet. Sci. 19, 1–2 (1999). [in Chinese]
T.Q. An, J.M. Peng, Z.J. Tian, H.Y. Zhao, N. Li, Y.M. Liu, J.Z. Chen, C.L. Leng, Y. Sun, D. Chang, G.Z. Tong, Emerg. Infect. Dis. 19(11), 1749–1755 (2013)
Z. Gu, J. Dong, J. Wang, C. Hou, H. Sun, W. Yang, J. Bai, P. Jiang, Virus Res. 195, 57–63 (2015)
W. Rui, C. Bai, J. Sun, S. Chang, X. Zhang, J. Vet. Med. Sci. 14(3), 363 (2013)
Y. Wang, S. Qiao, X. Li, W. Xie, J. Guo, Q. Li, X. Liu, J. Hou, Y. Xu, L. Wang, C. Guo, G. Zhang, Virus Genes 50(3), 401–409 (2015)
Y. Luo, N. Li, X. Cong, C.H. Wang, M. Du, L. Li, B. Zhao, J. Yuan, D.D. Liu, S. Li, Y. Li, Y. Sun, H.J. Qiu, Vet. Microbiol. 174(1–2), 107–115 (2014)
R. Kratchmarov, T. Kramer, T.M. Greco, M.P. Taylor, T.H. Chng, I.M. Cristra, L.W. Enquist, J. Virol. 87(17), 9431–9440 (2013)
T. Wang, Y. Xiao, Q. Yang, Y. Wang, Z. Sun, C. Zhang, S. Yan, J. Wang, L. Guo, H. Yan, Z. Gao, L. Wang, X. Li, F. Tan, K. Tian, BioMed Res. Int. (2015). doi:10.1155/2015/684945
C.H. Wang, J. Yuan, H.Y. Qin, Y. Luo, X. Cong, Y. Li, J. Chen, S. Li, Y. Sun, H.J. Qiu, Vaccine. 32(27), 3379–3385 (2014)
Y. Chen, W. Guo, Z. Xu, Q. Yan, Y. Luo, Q. Shi, D. Chen, L. Zhu, X. Wang, Virol. J. 8(1), 1–8 (2011)
L. Zhu, Y. Yi, Z. Xu, L. Cheng, S. Tang, W. Guo, Virol. J. 8(1), 153–163 (2011)
W. Tong, G. Li, C. Liang, F. Liu, Q. Tian, Y. Cao, L. Li, X. Zheng, H. Zheng, G. Tong, Antivir. Res. 130, 110–117 (2016)
Z. Gu, J. Dong, J. Wang, C. Hou, H. Sun, W. Yang, J. Bai, P. Jiang, Virus Res. 195, 57–63 (2015)
T.C. Mettenleiter, Vet. Immunol. Immunopathol. 54(1–4), 221–229 (1996)
J.T. van Oirschot, M.J. Kaashoek, F.A. Rijsewijk, J.A. Stegeman, J. Biotechnology 44(1–3), 75–81 (1996)
A. Lipowski, Pol. J. Vet. Sci. 9(1), 75–79 (2006)
Acknowledgements
This work were supported by the Science and Technology program of Sichuan (Project Nos. 2014NZ0043 and 2017NZ0038), Key Projects in the National Science & Technology Pillar Program during the Twelfth Five-year Plan Period (2015BAD12B04-2.3) and Sichuan Crops and Animals Breeding Special Project during the Thirteenth Five-year Plan Period (2016NYZ0052).
Author contributions
Conceived and designed the experiments: Xiaowan Liu, Yue Yin, Zhiwen Xu. Performed the experiments: Xiaowan Liu, Yue Yin. Analyzed the data: Ping Li, Fan Yang. Contributed reagents/materials/analysis tools: Ping Li, Fan Yang, Yi Fan, Xiangang Sun. Wrote the paper: Yue Yin. All the authors had read and approved the article.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors in this paper declare they have no conflict of interest.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Additional information
Edited by Keizo Tomonaga.
Rights and permissions
About this article
Cite this article
Yin, Y., Xu, Z., Liu, X. et al. A live gI/gE-deleted pseudorabies virus (PRV) protects weaned piglets against lethal variant PRV challenge. Virus Genes 53, 565–572 (2017). https://doi.org/10.1007/s11262-017-1454-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-017-1454-y