Abstract
Among all the human cytomegalovirus (HCMV) gene families, US12 family is relatively undefined in their transcriptional profile and biological functions. In this study, the transcription pattern and characteristics of HCMV US12–US17 gene region were studied extensively. Twenty-three clones harboring US12 cDNA sequence were screened out from a late cDNA library of an HCMV clinical isolate, Han. Using a set of US12–US17 gene-specific probes, six transcripts from US12–US17 locus were detected by northern blot at late kinetics of the clinical isolate. One additional transcript was found in late RNA of HCMV strain AD169. No evidence showing these transcripts contain introns by reverse transcription PCR. 3′ and 5′ termini of these transcripts were confirmed by Rapid Amplification of cDNA Ends. A novel protein-coding region was predicted in the shorter US14 transcript with an alternative in-frame 5′ translation initiation site compared to that of the previously predicted US14 ORF. Our findings demonstrate the existence of a cluster of 3′ coterminal unspliced transcripts with distinct 5′ transcriptional initiation sites originated from US12–US17 gene region in the late infection phase of an HCMV clinical strain.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human cytomegalovirus (HCMV) primary infections usually cause mild and non-specific illness in the immunologically healthy populations but can cause life-threatening diseases in immunocompromised individuals. HCMV is also a leading cause of birth defects in newborns following congenital infection. Until now, mechanisms of HCMV pathogenesis remain to be uncovered, largely due to our unawareness of biological functions of HCMV gene products on modulating important viral and cellular processes.
HCMV has the largest and most complex genome among all characterized human herpesviruses [1]. HCMV genome consists of around 235 kb of double stranded DNA and is predicted to contain at least 165 protein-coding genes [2–5]. Several sets of HCMV genes share some sequence similarity and are thought to have originated from duplication and divergence events, resulting in the emergence of a number of distinct multigene families.
As one of the multigene families, US12 family comprised 10 tandem but non-overlapping open reading frames (ORFs), US12–US21. This family accounts for approximately 5 % of HCMV genomic content. Members in the US12 family are highly conserved in different clinical isolates and laboratory strains [4, 6], suggesting their biological importance. Except for some information obtained from computer analysis of viral DNA sequences, experimental data are limited and scattered on expression and functions of individual US12 family members. The products of these genes are noted by seven transmembrane segments and anti-apoptotic protein Bax inhibitor 1 domain but have limited similarity to classical cellular G protein-coupled receptors (GPCRs) superfamily and anti-apoptotic protein Bax inhibitor 1 family [7]. Although members in US12 family are non-essential for HCMV replication in human embryonic lung fibroblast cells [8, 9], inactivation of some US12 family members affects virus replication in other cell types. US16-deficient viruses failed to replicate in endothelial and epithelial cells [10]. A major defective growth phenotype was observed for US18-null virus in cultured human gingival tissues [11]. US20-deficient mutants of HCMV exhibited major growth defects in different types of endothelial cells [12]. Recent studies [13, 14] showed that US14, US17, and US18 encoded proteins may be related to virus assembly and release because of their localization in various zones within cytoplasmic virion assembly compartment. Inactivation of the US17 gene in producer fibroblasts results in increased production of non-infectious viral particles, suggesting the role of US17 in regulating adequate virion composition during HCMV maturation [15]. US12 family members are also found to be involved in immunomodulation. ΔUS17 mutant markedly blunted the host cell antiviral response at a very early time point after infection by downmodulating many interferon-stimulated transcripts and transcripts encoding proinflammatory chemokines and cytokines [15]. US18 and US20 encoded proteins were identified to have NK cell evasion functions capable of promoting independently MICA degradation by lysosomal degradation [16].
To verify the functions of US12 family members, it is important to make clear the transcriptional patterns in the US12–US21 regions. However, except for expression analysis of US18–US20 genes [17] and microarray data of all US12 family [18], detail information about transcriptional characteristics of US12–US17 gene locus is still unavailable so far. Our previous data have showed the existence of US12 cDNA in a late HCMV cDNA library [19]. In current study, HCMV transcripts from US12–US17 gene locus were analyzed during lytic infection in human embryonic lung fibroblasts. The findings may offer experimental basis for further research on biological functions of US12–US17 gene products in HCMV infection.
Materials and methods
Cell and virus
Human embryonic lung fibroblasts (HELF, passages 7–20) were maintained in Minimal Essential Medium (MEM) supplemented with 10 % fetal bovine serum (FBS), 100 IU/ml penicillin, and 100 μg/ml streptomycin. One HCMV clinical strain, Han (GenBank accession number: KJ426589.1), was isolated from urine sample of an HCMV-infected 5-month-old infant hospitalized in Shengjing Hospital of China Medical University. HCMV strain AD169 (GenBank Accession Number: FJ527563.1) was kindly provided by Professor Minhua Luo (Wuhan Institute of Virology, Chinese Academy of Sciences). Strains Han and AD169 were propagated in HELF cells and titrated by typical soft agar plaque assay.
RNA preparation
Following the protocols of Ma et al. [20], HCMV immediate-early (IE), early (E), and late (L) RNAs were prepared. HELFs were inoculated with HCMV Han and AD169 at a multiplicity of infection (MOI) of approximately 2 plaque forming unit (PFU) per cell. To prepare IE RNA, HELFs were treated with 100 μg/ml of cycloheximide (CHX) (Sigma, USA), an inhibitor of de novo protein synthesis, 1 h prior to HCMV inoculation and harvested at 24 h post infection (hpi). For E RNA preparation, DNA replication inhibitor phosphonoacetic acid (PAA) (Sigma, USA) was added immediately after virus inoculation at a final concentration of 100 μg/ml, and the cells were harvested at 48 hpi. L RNA was extracted from untreated HELFs that were harvested at 96 hpi. Uninfected cellular RNA was used as a mock infection control.
All RNAs were extracted from the cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The isolated total RNAs were treated with DNA-free reagent (TURBO DNA-free kit, Ambion, Austin, TX, USA) to remove possible contaminating genomic DNA. Quantity and quality of the RNA preparations were detected by Nanodrop 1000 spectrophotometer (Life Technology, USA) and electrophoresis in 1 % formaldehyde denatured agarose gel. All RNA preparations were stored at −80 °C for subsequent assays.
Screening of US12 cDNA clones in a HCMV Han cDNA library
A full-length cDNA library of L RNA from HCMV strain Han had been constructed previously in pBluescript II SK vector by the SMART technique (Clontech, USA) [19]. To screen out US12 specific cDNA clones, PCR identification was performed as previously described [21, 22]. US12 gene specific primers US12-S and US12-AS (Table 1; Fig. 1b) were used in the library screening. The PCR was performed as follows: initial denaturation at 98 °C for 5 min, 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min, followed by a final elongation at 72 °C for 10 min. The inserts of the identified clones were sequenced using the standard T7 primer of the pBluescript II SK vector with an ABI PRISM 3730 DNA analyzer (Applied Biosystems, Carlsbad, CA, USA).
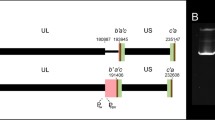
Graphic representation of HCMV Han genome, US12–US17 gene region, primers, and probes used. a The genome structure of HCMV strain Han (GenBank Accession Number: KJ426589.1) and schematic diagram of US12–US17 gene region. US12–US17 ORFs are shown by hollow arrows, and their positions are indicated. b The relative positions of the primers (5′ ends) and probes used in this study
Identification of transcripts from Han and AD169 US12–US17 locus by Northern Blot
To verify transcription of Han and AD169 US12–US17 gene locus, northern blot hybridization was performed using Digoxigenin (DIG) Northern Starter Kit (Roche, Indianapolis, IN, USA). The probes for detection of US12–US17 transcripts were generated according to the recommended protocol of the kit. Primers for generating the US12–US17 gene specific RNA probes were designed respectively within the predicted corresponding ORFs (Table 1; Fig. 1b) and used for amplification of gene specific fragments from Han and AD169 genomic DNA templates. The PCR products were purified using Wizard SV gel and PCR clean-up system (Promega Corporation, USA). The probes were synthesized in vitro using T7 RNA polymerase and labeled by DIG-11-UTP.
HCMV stage-specific RNAs and mock infected RNA were separated by electrophoresis (10 μg per lane) on a 1 % formaldehyde denatured agarose gel and transferred onto a positively charged nylon membrane by capillary blotting method. The nylon membrane was prehybridized in preheated hybridization solution at 65 °C for 30 min. Then, the membrane was incubated in hybridization solution at 65 °C overnight with gentle agitation (UVP HB-1000 hybridizer, hybridization Oven). After incubating with blocking solution and alkaline phosphatase conjugated anti-DIG antibody, the blots were developed gradually using the substrate of CDPStar for 20 min. The imaging was recorded by Bio-Rad molecular imager chemiDoc XRS with ImageLab software.
Identification of possible introns in US12–US17 gene region by RT-PCR
RT-PCR was performed using TaKaRa RNA PCR kit (AMV) ver 3.0 (TaKaRa, China). First-strand cDNA of HCMV L RNA was synthesized using random 9-mer primer and avian myoblastosis virus (AMV) reverse transcriptase at 30 °C for 10 min, 42 °C for 30 min, 99 °C for 5 min, and 5 °C for 5 min. The cDNA sequences were amplified using three pairs of primers of US12-R and US14-LF, US14-LR, and US15-F, as well as US15-R and US17-F, which will produce overlapping sequences of the US12–US17 gene region, respectively (Table 1; Fig. 1b). Reactions containing no AMV reverse transcriptase were performed to eliminate interference of possible DNA contamination in the RNA preparations. Meanwhile, viral DNA of the Han infected cells was isolated and amplified using the same primers as an unspliced transcript control.
Determination of ends of US12–US17 transcripts by Rapid Amplification of cDNA Ends
Rapid Amplification of cDNA Ends (RACE) was performed to precisely determine 5′ and 3′ boundaries of US12–US17 transcripts using 5′ Full Race Kit and 3′ Full RACE Core Set Ver.2.0 kits (TaKaRa, China). All primers for RACE were designed using Primer Premier 5.0 based on the sequence data of the HCMV Han (Table 1; Fig. 1b). L RNA of HCMV Han was used as templates.
All the experiments were carried out according to the manufacturer’s instructions. Briefly, for 3′ RACE, first-strand cDNA was synthesized with PrimeScript reverse transcriptase and 3′ RACE adaptor, which contains oligo-dT and adaptor primer binding sequence, under condition of 42 °C for 60 min and 70 °C for 15 min. Specific cDNA sequences were amplified using 3′ RACE adaptor outer primer (5′-TACCGTCGTTCCACTAGTGATTT-3′) and 3′ RACE gene specific primers of US12-3′, US13-F, or US14-LF (Table 1; Fig. 1b) as the following condition: 94 °C for 3 min, 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min, followed by final elongation at 72 °C for 10 min.
For 5′ RACE, L RNA of Han and AD169 was dephosphorylated using alkaline phosphatase (AP), decapped using tobacco acid pyrophosphatase (TAP), and ligated to 5′ RACE adaptor using T4 RNA ligase. After reverse transcription with M-MLV, nested PCR was done using 5′ RACE gene-specific primers (Table 1; Fig. 1b) and 5′ RACE adaptor primers provided by the kit (outer primer: 5′-CATGGCTACATGCTGACAGCCTA-3′ and inner primer: 5′-CGCGGATCCACAGCCTACTGATGATCAGTCGATG-3′). PCR reactions were performed using following parameters: 94 °C for 3 min, 25 cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 1 min, followed by final elongation at 72 °C for 10 min.
All RACE products were gel purified using Wizard SV gel and PCR clean-up system (Promega, USA) and cloned into pCR 2.1 vector (Invitrogen, USA). The plasmids were transformed into competent Escherichia coli DH5α. Clones containing the inserts in the transformed vectors were briefly identified by PCR. The inserted sequences of 10–25 randomly selected clones were sequenced using primers M13F (5′-TGTAAAACGACGGCCAGT-3′) and M13R (5′-CAGGAAACAGCTATGACC-3′), respectively.
Prediction of potential transcriptional regulation motifs and coding potential of the transcripts online
Sequences from RACE and cDNA library screening were aligned by comparing with Han and AD169 genomic sequence. All nucleotide positions in this study were in reference to the sequence of HCMV strain Han and AD169. Prediction of transcription regulation motifs in Han genome was performed using online transcription factor search tool (TFSEARCH) at http://www.cbrc.jp/research/db/TFSEARCH.html and transcription factor DNA-binding preferences (Jaspar Database) at http://jaspar.genereg.net/. NCBI ORF finder at http://www.ncbi.nlm.nih.gov/projects/gorf/ was used to search potential ORF in the identified transcripts.
Results
Identification of US12–US17 transcripts by cDNA library screening and sequencing
Twenty-three cDNA clones were identified to contain US12 sequence by PCR screening from the late cDNA library of Han strain. One 1762 bp cDNA sequence derived from a cDNA clone contained US12 and US13 sequences from nucleotide (nt) 208816–207055. Another 890 bp cDNA sequence derived from 22 clones was US12 specific, representing a monocistronic transcript. The 5′ end of the 890nt transcript was located at nt 207945–207948. The 3′ ends of the transcripts were located at nt 207055–207062 downstream of a polyA signal at nt 207082–207077.
Detection of Han and AD169 US12–US17 transcripts by Northern Blot analysis
Northern Blot was employed to identify the US12–US17 transcripts in IE, E, and L RNA preparations from HCMV Han and AD169 infected HELFs and RNA from mock infected cells. A set of digoxigenin-labeled gene specific RNA probes of US12-P, US13-P, US14-LP, US14-UP, US15-P, US16-P, and US17-P were used.
Using US12-P, 6 distinct transcripts with approximate lengths of 4600, 3600, 2800, 2100, 1800, and 900 nt, respectively, were detected in L RNA but not in IE, E RNAs, and mock RNA (Fig. 2a). However, one additional transcript with approximate 5600 nt in length was found in L RNA of strain AD169, and the 2100 nt transcript found in Han was obscured in L RNA of AD169 (Fig. 2b). Among these transcripts, the 900 nt transcript was the most abundant one, followed by the 1800 nt transcript.
Transcription analysis of US12–US17 gene region by Northern blot. Northern blots were performed using probe US12-P to detect specific transcripts in IE, E, and L RNAs of Han (a) and AD169 (b), respectively. Six transcripts with approximate 4600, 3600, 2800, 2100, 1800, and 900 nt in length, respectively, were observed in L RNA of Han. Additional transcript with approximate 5600 nt in length was found in L RNA of AD169 strain. The 2100 nt transcript observed in Han was obscured in L RNA of AD169. To identify the transcripts in detail, L RNA of Han was further detected by Northern blot using a set of gene specific probes as indicated in the c. Among the 6 transcripts detected by probe US12-P, 5 relatively longer transcripts, 4 longer transcripts and 3 longer transcripts were detected by probes of US13-P, US14-LP, and US14-UP, respectively. Two transcripts with approximate 3600 and 4600 nt in length were detected by US15-P, and the longest transcript with approximate 4600 nt in length was detected by US16-P. No transcript was detected by probe US17-P. The identified transcripts were indicated by black arrows. M indicates digoxigenin-labeled RNA Molecular Weight Marker I
To further validate the characteristics of transcripts from US12–US17 region, gene-specific probes of US13-P, US14-LP, US14-UP, US15-P, US16-P, and US17-P, complementary to the areas close to the possible 5′ ends of the 1800, 2100, 2800, 3600, 4600, and 5600 nt transcripts (Figs. 1b, 7), were used for detecting transcripts from the region in HCMV Han L RNA preparation. To detect the 2800 and 2100 nt transcripts which may contain the US14–US12 sequences and start upstream of the US14 ORF and within the US14 coding region respectively, two US14 probes, which were from nt 209304 to 209637 (US14-UP) and from nt 208929 to 209172 (US14-LP), were designed. As anticipated, 5 bands other than that of the 900 nt transcript were detected by probe US13-P; 4 bands of the 4, 600, 3600, 2800 and 2100 nt transcripts by probe US14-LP; 3 bands of the 4600, 3600, and 2800 nt transcripts by probe US14-UP; 2 bands of the 4600 and 3600 nt transcripts by probe US15-P; and 1 band of the 4600 transcript by probe US16-P. However, no band was detected by probe US17-P in L RNA of strain Han (Fig. 2c). The results were consistent in 3 different batches of RNA preparations.
These results suggest that a cluster of 3′ coterminal transcripts with distinct 5′ transcription initiation sites may be transcribed from the US12–US17 locus.
Identification of possible introns in US12–US17 gene region by RT-PCR
To detect possible introns in US12–US17 transcription unit, L RNA from Han infected HELFs was subjected to RT-PCR using three pairs of primers of US12-R and US14-LF, US14-LR and US15-F, as well as US15-R and US17-F, which will produce overlapping sequences of the US12–US17 gene region, respectively (Table 1; Fig. 1b). Results showed that the length of RT-PCR products from HCMV cDNA was the same as those of PCR products from viral genomic DNA for each set of primer pairs. The expected sizes of these PCR products were 1974, 1700, and 2317 bp, respectively. No amplicon was obtained from RNA treated without AMV reverse transcriptase (Fig. 3). The band with the length of approximate 500 bp, which was obtained from both the cDNA and genomic DNA for primers US15R/US17F, is likely to be a non-specific product.
RT-PCR result of transcripts from Han US12–US17 gene region. cDNAs reversely transcribed from Han L RNA with AMV RTase (cDNA), L RNA treated without AMV RTase (RT-) and HCMV Han genomic DNA (DNA) were amplified by PCR using three pairs of primers of US12-R and US14-LF (US12-R/US14-LF), US14-LR and US15-F (US14-LR/US15-F), as well as US15-R and US17-F (US15-R/US17-F), which will produce overlapping sequences of the US12–US17 gene region, respectively. The products from cDNAs were the same in length as those from HCMV genomic DNAs, which are expected to be 1974, 1700, and 2317 bp as indicated by the arrows, respectively. This result implies that there are no introns in the region. M indicates 15,000 and 2000 bp DNA markers
The results indicate that splicing events probably do not occur during late phase transcription of US12–US17 gene region in HCMV-infected HELFs.
Determination of ends of US12–US17 transcripts by rapid amplification of cDNA ends
Using 3′ adaptor outer primer and gene specific 3′ RACE primers of US12-3′ (US12), US13-F (US13), and US14-LF (US14), respectively, one predominant 600 bp band for US12 cDNA, 1400 bp band for US13 cDNA, and 2100 bp band for US14 cDNA were obtained, respectively (Fig. 4). Sequencing results of the recovered fragments showed that 3′ end of these cDNAs were located 20 bp downstream of the US12 ORF at nt 207055–207060 (Fig. 7; Table 2). The 3′ terminal location of the US13 and US14 transcripts was at nt 207055 in all sequenced clones. The results above were in agreement with that of library screening of US12 and US13 transcripts.
3′ RACE results of Han transcripts from US12–US17 gene region. 3′ ends of US12, US13, and US14 transcripts were amplified by 3′ RACE from L RNA of HCMV Han using 3′ adapter RACE outer primer and US12-3′, US13-F, or US14-LF primer respectively. M indicates 2000 and 15,000 bp DNA marker. The predominant bands are of 600, 1400, and 2100 bp in length for US12, US13, and US14 transcripts, respectively. Several weak bands in US13 and US14 were also obtained. Products in all the indicated bands (shown by arrows) were recovered, and sequences of the recovered DNA were cloned and sequenced
Except for the predominant band, several weaker bands were obtained from US13 and US14 cDNAs. Sequencing results showed that these weaker bands are all non-specific products.
The results of HCMV Han US12–US17 5′ RACE amplification using different 5′ RACE primers are shown in Fig. 5. Almost all the predominant bands obtained (Fig. 5) were recovered and the recovered DNA fragments were TA cloned and sequenced. Sequencing results showed that the US12 monocistronic transcript mainly initiated at nt 207948 of HCMV Han genome, and 5′ ends of the 1800 nt bicistronic transcript were located at nt 208810-208818, and those of the 2100 and 2800 nt transcripts located at nt 209132–209156 and nt 209856–210024, and those of the 3600 nt transcript and 4600 nt transcript were at nt 210330–210654 and nt 211601–211641, respectively (Table 2; Fig. 7). Although the 5600 nt transcript was detected in AD169 but not in Han by northern blot, transcription of Han US17 gene was proved by 5′ RACE. The 5′ termini of the US17 transcripts were located within a wide range at nt 212370–212650, but the most predominant one was from nt 212568 (Table 2).
5′ RACE results of the transcripts from US12–US17 gene region in HCMV Han. 5′ ends of US12–US17 transcripts were analyzed by 5′ RACE using L RNA of Han treated with both TAP and M-MLV. Nested PCR were performed using the 5′ RACE adaptor primers together with gene-specific primers of US12-5′O/US12-5′I, US13-5′O/US13-5′I, US14-5′O/US14-5′I, US15-5′O/US15-5′I, US16-5′O/US16-5′I, and US17-5′O/US17-5′I, respectively (a–f). L RNA without treatment of TAP (TAP-) and M-MLV (MLV-) were used as negative controls. M indicates 2000 bp DNA marker. Products of all the predominant bands indicated by white arrows were recovered, and sequences of the recovered DNA were cloned and sequenced
5′ RACE analysis for AD169 US17 transcripts was also performed. Four products, 1500, 400, 250, and 120 bp, were amplified using gene-specific primers of US17-5′O/US17-R (Fig. 6). As shown in Table 3, the 5′ terminal locations at nt 208472, 208356, and 208270 in AD169 were identical to those at nt 202568, 202449, and 212370 in Han. Those initiation sites are more believable and may reflect the dominant initiation sites of US17 mRNAs in this region. The transcript starting at nt 209524 is originated from a site within US18 ORF.
5′ RACE results of the transcripts from US17 gene in HCMV AD169. 5′ ends of US17 transcripts were analyzed by 5′ RACE using L RNA of AD169 treated with both TAP and M-MLV. Nested PCR was performed using the 5′ RACE adaptor primers together with gene specific primers of US17-5′O/US17-R. L RNA without treatment of TAP (TAP-) and M-MLV (MLV-) were used as negative controls. M indicates 2000 bp DNA marker. Products of all the predominant bands indicated by white arrows were recovered, and sequences of the recovered DNA were cloned and sequenced
The results of Northern Blot, RT-PCR, RACE, and cDNA library screening show that a cluster of unspliced transcripts with lengths of 889–894 nt, 1751–1764 nt, 2073–2102 nt, 2797–2970 nt, 3271–3600 nt, 4542–4587 nt, and 5311–5596 nt prior to polyadenylation are transcribed during late infection (Fig. 7).
Identified US12–US17 transcripts in HCMV Han in our study. The predicted ORFs are marked in the transcripts by hollow arrows. The 5′ and 3′ ends of the transcripts are labeled on the right and left side of the transcripts, respectively. The approximate length of the transcripts achieved by Northern Blot and RACE are labeled on the right side in the brackets. Positions of TATA elements (shown by hollow triangles) and polyA signals (shown by black triangles) found in the US12–US17 gene region are marked
Prediction of potential transcriptional regulation motifs and potential ORF
Two consensus polyA signals (AATAAA) were predicted by TFSEARCH downstream of US18 ORF at nt 212766–212761 and of US12 ORF at nt 207082–207077, respectively, in US12–US21 locus (Fig. 7). This result indicates that the US12–US17 gene region is probably an independent transcription locus and all transcripts from the region share the polyA signal at nt 207082–207077. But 3′ end position of the AD169 transcript originated from nt 209524 remains to be identified.
Two canonical TATA boxes (TATAAA) are present at immediate upstream of the US12 gene at nt 207977–207972 and US17 gene at nt 212681–212676. Another 2 non-canonical TATA elements, TATAAG at nt 208844–208839 and TCTAAA at 211672–211667, were identified just upstream of the US13 and US16 genes, respectively (Fig. 7). No TATA motifs were found upstream of or within the predicted US14 and US15 genes.
In the 2100 nt transcript, a novel US14 ORF was predicted by means of NCBI ORF finder at http://www.ncbi.nlm.nih.gov/projects/gorf/ to possibly initiate from an alternative putative in-frame AUG at nt 209087–209085 and end at nt 208857. The internal US14 ORF shares the termination site with the canonical US14 ORF.
Discussion
Defining HCMV gene products and their functions is essential to reveal its pathogenic processes. Because HCMV has complex transcriptomics, annotations of HCMV predicted ORFs based on in silico analyses [2, 3] may not represent the full set of translational products. In a recent study, 751 ORFs were identified in HCMV by ribosome profiling [5]. Therefore, precise experimental analyses of HCMV transcripts in infected cells can contribute to genome annotations and further proteomic investigations.
Until now, transcriptional patterns of nearly half of the HCMV predicted ORFs have not been investigated extensively [23]. US12–US17 region is one of the genomic loci of which transcriptional characteristics have not been acquired as yet. Two transcripts from US18–US20 gene region, an adjacent transcriptional unit in US12 family, were found to share 3′ end at US18 polyA signal and have different 5′ initiation sites [17]. 3′ coterminal polycistronic transcripts with alternative transcriptional start sites are a major feature frequently encountered among HCMV transcripts [24–26]. Our evidence suggests that transcripts from the US12–US17 locus conform with this paradigm.
In our study, six transcripts from HCMV US12–US17 region were detected in a clinical strain by northern blot using US12 specific probe. Among them, the US12 and US13 transcripts (Named according to their starting gene) were more abundant than the other four transcripts, which was in line with HCMV deep sequencing transcription profile [27]. Results of cDNA library screening and RACE defined precise termini of these transcripts. RT-PCR results almost ruled out the possibility of existence of introns in the US12–US17 gene region. Although direct evidences for 3′ ends of US15, US16, and US17 transcripts were absent because of their poor 3′ RACE amplification results (data not shown), it is still reasonable to speculate that these transcripts utilize the same poly A signal which is exclusively located downstream of US12 ORF. The 5311–5596 nt transcripts of Han confirmed by 5′ RACE were not detected by northern blot. US17 transcription in cells infected with Han appears to be at low level and initiated at multiple start sites. Therefore, at least seven mRNAs are transcribed in US12–US17 regions in HCMV Han. Northern blot results of AD169 RNA preparations showed a minor difference from that to Han. A 5600 nt US17 specific band was detected, but the 2100 nt band found in Han was weak in AD169. The fact that there is no strong and classical promoter motif upstream of the alternative US14 ORF for the 2100 nt transcript might explain the inconsistency.
Based on RT-PCR data, the identical lengths of cDNA and template DNA amplicons imply a probability that there are no introns in this region. These results are also supported by the previous evidence [5, 27].
Combining the results of RT-PCR and RACE, lengths of the US12–US17 transcripts were predicted. The results were consistent with the transcripts identified by cDNA library screening and matched exactly the size of the mRNA detected by northern blot. These data confirmed the previously annotated US12–US17 ORFs with one exception in Han strain. An unrecognized US14 ORF was predicted in the 2100 nt transcript of Han strain with an alternative putative in-frame AUG at nt 209087–209085. But some follow-up studies should be done to prove the coding potential of the novel ORF at protein level. Six alternative ORFs within the US12–US17 locus have been identified by ribosome profiling [5].
Three of these ORFs (ORFS340C.iORF1, ORFS342C.iORF1, ORFS343C.iORF1) can be found in the transcripts identified in our current study. The predicted novel ORF in the truncated US14 transcript was not reported in the Stern-Ginossar study [5]. Whether this ORF encodes protein or not needs to be confirmed by future experimental evidence.
In the present study, all the US12–US17 transcripts were detected at the late stage of HCMV infection. The expression kinetics across the US12–US17 gene locus was different from that identified by high-throughput chip microarray analysis in Towne strain [18]. In that study, early transcripts were detected across US12–US17 region except for US15 (Early-Late). It has been reported that chip results showed approximately 10 % disagreement overall with published studies (e.g., US18 [17] and UL102 [28]. Further experiments were required to confirm the kinetics of these transcripts as far as HCMV strains and host cell context are concerned. Late and synchronized expression kinetics of US12–US17 ORFs indicate that they might be under identical regulation mechanisms and be associated with assembly and morphogenesis of the virions, which has been proven by previous evidence [13, 14].
Result of DNA sequence blast between Han and AD169 showed 99 % sequence identity in the US12–US17 region. Northern blot analyses showed that the overall transcription pattern of these structurally polycistronic genes in AD169 was similar to that in Han, except for the US17 transcript. These results demonstrate that the expression products of US12–US17 gene locus are relatively conserved among clinical and laboratory strains.
In cDNA library screening, no clones with cDNAs from the US14, US15, US16, and US17 transcripts were obtained from the late cDNA library. Besides their lower abundance than the US12 and US13 transcripts, limitation of the cDNA library failed to construct long mRNA clones might be the main explanation. The US17 transcript, which was found in AD169 by northern blot and was proved to exist in strain Han by 5′ RACE, was not detected in Han by northern blot. The repeatable northern blot results are surely contradictory to the existence of classical TATA sequence upstream of the predicted US17 transcription initiation site. This may be attributed to its low transcriptional level and unstable expression in strain Han. Before any conclusion can be made, further research about regulation of US17 gene transcription should be done.
Conclusions
A cluster of 3′ coterminal unspliced unidirectional transcripts with distinct 5′ transcriptional initiation sites were identified to be transcribed from US12–US17 gene region during late stage infection in an HCMV clinical isolate. The data provide basic information about transcripts from US12–US17 gene locus and will be helpful for further analysis of HCMV transcriptomics and proteomics.
Abbreviations
- HCMV:
-
Human cytomegalovirus
- ORF:
-
Open reading frame
- GPCR:
-
G protein-coupled receptor
- HELF:
-
Human embryonic lung fibroblast
- FBS:
-
Fetal bovine serum
- MEM:
-
Minimal essential medium
- MOI:
-
Multiplicity of infection
- IE:
-
Immediate-early
- PFU:
-
Plaque forming unit
- CHX:
-
Cycloheximide
- PAA:
-
Phosphonoacetic acid
- DIG:
-
Digoxigenin
- RACE:
-
Rapid amplification of cDNA ends
- AP:
-
Alkaline phosphatase
- TAP:
-
Tobacco acid pyrophosphatase
- AMV:
-
Avian myoblastosis virus
References
M.S. Chee, A.T. Bankier, S. Beck, R. Bohni, C.M. Brown, R. Cerny, T. Horsnell, C.A. Hutchison 3rd, T. Kouzarides, J.A. Martignetti, Curr. Top. Microbiol. Immunol. 154, 125–169 (1990)
A.J. Davison, A. Dolan, P. Akter, C. Addison, D.J. Dargan, D.J. Alcendor, D.J. Mcgeoch, G.S. Hayward, J. Gen. Virol. 84, 17–28 (2003)
E. Murphy, I. Rigoutsos, T. Shibuya, T.E. Shenk, Proc. Natl. Acad. Sci. USA 100, 13585–13590 (2003)
A. Dolan, C. Cunningham, R.D. Hector, A.F. Hassan-Walker, L. Lee, C. Addison, D.J. Dargan, D.J. Mcgeoch, D. Gatherer, V.C. Emery, P.D. Griffiths, C. Sinzger, B.P. Mcsharry, G.W. Wilkinson, A.J. Davison, J. Gen. Virol. 85, 1301–1312 (2004)
N. Stern-Ginossar, B. Weisburd, A. Michalski, V.T. Le, M.Y. Hein, S.X. Huang, M. Ma, B. Shen, S.B. Qian, H. Hengel, M. Mann, N.T. Ingolia, J.S. Weissman, Science 338, 1088–1093 (2012)
E. Murphy, D. Yu, J. Grimwood, J. Schmutz, M. Dickson, M.A. Jarvis, G. Hahn, J.A. Nelson, R.M. Myers, T.E. Shenk, Proc. Natl. Acad. Sci. USA 100, 14976–14981 (2003)
M. Lesniewski, S. Das, Y. Skomorovska-Prokvolit, F.Z. Wang, P.E. Pellett, Virology 354, 286–298 (2006)
W. Dunn, C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, F. Liu, Proc. Natl. Acad. Sci. USA 100, 14223–14228 (2003)
D. Yu, M.C. Silva, T. Shenk, Proc. Natl. Acad. Sci USA 100, 12396–12401 (2003)
M. Bronzini, A. Luganini, V. Dell’oste, M. De Andrea, S. Landolfo, G. Gribaudo, J. Virol. 86, 6875–6888 (2012)
R. Hai, A. Chu, H. Li, S. Umamoto, P. Rider, F. Liu, Virol. J. 3, 84 (2006)
N. Cavaletto, A. Luganini, G. Gribaudo, J. Virol. 89, 11092–11106 (2015)
S. Das, P.E. Pellett, Virology 361, 263–273 (2007)
S. Das, Y. Skomorovska-Prokvolit, F.Z. Wang, P.E. Pellett, J. Virol. 80, 1191–1203 (2006)
S.J. Gurczynski, S. Das, P.E. Pellett, J. Virol. 88, 2168–2182 (2014)
C.A. Fielding, R. Aicheler, R.J. Stanton, E.C. Wang, S. Han, S. Seirafian, J. Davies, B.P. Mcsharry, M.P. Weekes, P.R. Antrobus, V. Prod’homme, F.P. Blanchet, D. Sugrue, S. Cuff, D. Roberts, A.J. Davison, P.J. Lehner, G.W. Wilkinson, P. Tomasec, PLoS Pathog. 10, e1004058 (2014)
Y.W. Guo, E.S. Huang, J. Virol. 67, 2043–2054 (1993)
J. Chambers, A. Angulo, D. Amaratunga, H. Guo, Y. Jiang, J.S. Wan, A. Bittner, K. Frueh, M.R. Jackson, P.A. Peterson, M.G. Erlander, P. Ghazal, J. Virol. 73, 5757–5766 (1999)
Y.P. Ma, Q. Ruan, Y.H. Ji, N. Wang, M.L. Li, Y. Qi, R. He, Z.R. Sun, G.W. Ren, Genet. Mol. Res. 10, 566–575 (2011)
Y.P. Ma, S. Gao, L. Wang, N. Wang, M.L. Li, B. Zheng, Y. Qi, Z.R. Sun, W. Liu, Q. Ruan, Virol. J. 10, 65 (2013)
Y. Qi, Y.P. Ma, R. He, N. Wang, Q. Ruan, Y.H. Ji, M.L. Li, Z.R. Sun, G.W. Ren, Microbiol. Immunol. 55, 95–99 (2011)
R. He, Y.P. Ma, Y. Qi, N. Wang, M.L. Li, Y.H. Ji, Z.R. Sun, S.J. Jiang, Q. Ruan, Virol. J. 8, 299 (2011)
Y.P. Ma, N. Wang, M.L. Li, S. Gao, L. Wang, B. Zheng, Y. Qi, Q. Ruan, Future Microbiol. 7, 577–593 (2012)
C. Smuda, E. Bogner, R.K. Adsak, J Gen. Virol. 78, 1981–1992 (1997)
B.A. Wing, E.S. Huang, J. Virol. 69, 1521–1531 (1995)
P. Akter, C. Cunningham, B.P. Mcsharry, A. Dolan, C. Addison, D.J. Dargan, A.F. Hassan-Walker, V.C. Emery, P.D. Griffiths, G.W. Wilkinson, A.J. Davison, J. Gen. Virol. 84, 1117–1122 (2003)
D. Gatherer, S. Seirafian, C. Cunningham, M. Holton, D.J. Dargan, K. Baluchova, R.D. Hector, J. Galbraith, P. Herzyk, G.W. Wilkinson, A.J. Davison, Proc. Natl. Acad. Sci. USA 108, 19755–19760 (2011)
J.A. Smith, G.S. Pari, J. Virol. 69, 1734–1740 (1995)
Acknowledgments
This work was supported by the National Natural Science Foundation of China (30672248, 81171580, 81171581 81201274 and 81371788), the Specialized Research Fund for the Doctoral Program of Higher Education (20112104110012), and the Outstanding Scientific Fund of Shengjing Hospital and the Natural Science Foundation of Liaoning Province, China (2013021008).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Edited by Hartmut Hengel.
Qiang Ruan is the first corresponding author for this paper.
Rights and permissions
About this article
Cite this article
Lu, Y., Ma, Y., Liu, Z. et al. A cluster of 3′ coterminal transcripts from US12–US17 locus of human cytomegalovirus. Virus Genes 52, 334–345 (2016). https://doi.org/10.1007/s11262-016-1308-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-016-1308-z