Abstract
To date, the genetic replication and translation mechanisms as well as the pathogenesis of duck hepatitis A virus type 1 (DHAV-1) have not been adequately characterized due to the lack of a reliable and efficient cell culture system. Although the full-length infections clone system is the best platform to manipulate the virus, it is relatively difficult to assemble this system due to the lack of a suitable cell line. It has been proven that the minigenome system an efficient reverse genetics system for the study of RNA viruses. In some cases, it can be used to displace the infectious clone of RNA viruses. Here, we generated a minigenome for DHAV-1 with two luciferase reporter genes, firefly luciferase (Fluc) and Renilla luciferase (Rluc). The Rluc gene was used as a reference gene for the normalization of the Fluc gene expression in transfected cells, which provided a platform for studying the regulatory mechanisms of DHAV-1. Furthermore, to investigate the role of DHAV-3′UTR in the regulation of viral protein translation, deletions in the 3′UTR were introduced into the DHAV-1 minigenome. Luciferase activity, an indicator of virus translation, was then determined. These results showed that a minigenome system for DHAV-1 was successfully constructed for the first time and that the complete or partial deletion of the DHAV-3′UTR did not affect the expression level of the reporter gene, indicating that DHAV-1 translation may not be modulated by the viral genomic 3′UTR sequence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Duck hepatitis virus serotype 1 (DHV-1) can cause an acute, contagious, and highly fatal disease of young ducklings known as duck viral hepatitis (DVH). Recently, DHV-1 has been renamed Avihepatovirus A, which has been classified as a member of the genus Avihepatovirus in the family Picornaviridae (http://www.picornaviridae.com/). The DHAVs have been divided into three serotypes or genotypes, including DHAV-1 (classical serotype 1), DHAV-2 (a serotype isolated in Taiwan) and DHAV-3 (a serotype isolated in South Korea and China) or DHAV genotypes A, B, and C [1, 2].
Of these three DHAV serotypes, the most virulent and widespread is DHAV-1, which can cause mortality of up to 95 % in young ducklings within 1 week of age.
Although DVH was first reported in Long Island in 1949, the complete genome of the causative agent was not determined until 2006 [3]. The DHAV-1 genome is 7691 nucleotides long and contains a single open reading frame (ORF) that encodes a large polyprotein precursor of 2249 amino acids (aa) that is further cleaved into 12 structural and nonstructural proteins. The ORF is flanked by two untranslated regions (UTR) at the 5′ and 3′ ends. The 626-nucleotide (nt)-long 5′UTR of the DHAV-1 genome contains an internal ribosome entry site (IRES) element, which is essential for the initiation of viral protein synthesis [4, 5]. In contrast to the 5′UTR, little is known about the role of the 3′UTR in viral replication and translation.
A better understanding of the genetic replication and translation mechanisms as well as the molecular pathogenesis of DHAV is a prerequisite for vaccine development and specific antiviral therapy. However, the lack of sufficient molecular tools has greatly impeded research on the molecular mechanisms of DHAV pathogenicity and propagation. Reverse genetics systems, including a full-length clone system and a number of life cycle modeling systems, are experimental tools that enable the production and subsequent replication and transcription of viral genomes from complementary DNA (canal). Although a full-length infectious clone derived from DHV-1 has been constructed by Yun et al. [6], the relative instability and heterogeneity of the RNA transcripts synthesized in vitro increases the difficulty of genetic manipulation in practice and reduces the transfection efficiencies. A minigenome (MG) system, a life cycle modeling systems, is considered to be a model system for exploring the processes of virus replication and transcription without biosafety concerns from the use of infectious virus [7].
In this study, a bicistronic DHAV-1 minigenome system containing two types of luciferase reporter genes, firefly luciferase (Fluc) and Renilla luciferase (Rluc), was developed as a platform for studying the regulatory mechanism of DHAV-1. The 3′UTR of the picornaviruses genome has been proven to be associated with viral genome translation and replication [8, 9]. However, whether these conclusions are applicable to DHAV-1 remains unclear. Here, a series of deletion mutants were constructed to evaluate the role of 3′UTR in translation of the DHAV-1 genome.
Materials and methods
Cells
BSR-T7/5 cells, baby hamster kidney (BHK) cells stably expressing T7 RNA polymerase, were seeded at a density of 1 × 106 cells/ml in Dulbecco’s modified Eagle medium (DMEM, Gibco) with 10 % fetal bovine serum (FBS, Gibco), 100 units/ml penicillin (Gibco) and 100 mg/ml streptomycin (Gibco) at 37° in a 5 % CO2 incubator. The parental DHAV strain ZJ-A was derived from chicken embryos from the adapted attenuated descendent of the DHAV strain SY05 [10].
Virus and plasmids
The pBluescript II SK (+) vector was maintained in our laboratory. We generated a recombinant plasmid pDHAV containing the full-length genome of DHAV strain ZJ-A with hammerhead ribozyme (HRz) immediately upstream of the 5′UTR and hepatitis delta virus ribozyme (HdvRz) immediately downstream of the 3′UTR in our laboratory. The plasmid pRluc-Fluc was generated from the modified pDHAV by replacing the ORF of DHAV with the Fluc gene and inserting the Rluc gene upstream of the 5′UTR in our previous study [11]. The Rluc gene was used as an internal reference gene for the normalization of Fluc gene expression in transiently transfected cells.
Plasmid construction
The pBluescript II SK (+) (pSK) plasmid has been modified to exhibit a T7 promoter that was cloned immediately upstream, and HdvRz was inserted downstream of the multi-cloning sites to precisely generate the target transcripts [12, 13]. This modified plasmid was renamed pSK-Rz and was initially used as a basic vector for the construction of the minigenome plasmid. The minigenome fragment Rluc-5′UTR-Fluc-3′UTR was amplified from pRluc-Fluc using a pair of specific primers (Rluc-BamHI-T7-F and NCR-NotI-R) (Table 1), which were then introduced into the multiple cloning site (MCS) of pSK-Rz using the restriction enzymes BamHI and NotI. The resulting plasmid was designated as pSK-RLuc/Fluc and contained two different reporter genes encoding Rluc and Fluc.
To examine the efficiency of the minigenome system for pSK-RLuc/Fluc and to analyze the role of the 3′UTR elements in the regulation of viral protein translation, a series of deletion mutants were constructed based on pSK-Rluc/Fluc. For example, a mutant with an entire 3′UTR deletion was created by PCR amplification from pSK-Rluc/Fluc with primers Δ3′UTR–XhoI-F and Δ3′UTR–NotI-R (Table 1), and the PCR product was then inserted into pSK-Rluc/Fluc, which had been digested using the same restriction enzymes. The resulting plasmid was designated pSK-Δ3′UTR. In addition to the poly (A) tail, 315 nucleotides of the 3′UTR were folded into two large stem-loops (SL) based on the predicted secondary structure using software RNA Draw (Fig. S1): SL1 (1–150 nt) and SL2 (151–315 nt). Using the same strategy as described above, three additional UTR deletion mutants, pSK-Δ3′UTR (1-150), pSK-Δ3′UTR (151-315), and pSK-Δ5′UTR, were constructed with the corresponding primers (Table 1). PSK-Δ5′UTR and pSK-Rz were considered to be the negative and empty control plasmids, respectively, while pSK-Rluc/Fluc was used as a positive control.
Transfection
BSR-T7/5 cells were plated in 35-mm diameter culture dishes the day before transfection to obtain 70–80 % confluent monolayers. Next, 2 μg of each of the above recombinant plasmids containing pSK-Rz were transfected into cells using Effectene Transfection Reagent (QIAGEN, Germany) according to the manufacturer’s protocol.
RT-PCR
At 24 h post-transfection, total RNA was extracted from the transfected cells using Trizol reagent (Invitrogen, USA) according to the manufacturer’s instructions and then resuspended in nuclease-free water (Takara, Japan). The first-strand canal of plus strand RNA from transfected and non-transfected cells were, respectively, synthesized using the specific primers Fluc-R and β-actin-R as well as the M-MLV reverse transcriptase (PR omega, Madison, WI) according to manufacturer’s instructions. The PCR reaction was performed using 2 all of canal with Ex Taw polymerase (Tamale, Japan) for 30 cycles. Next, the PCR products were analyzed on a 2 % agars gel. The primers used for PCR detection are listed in Table 1.
Western blotting analysis
Expression of the luciferase gene was confirmed using western blotting. Briefly, at 24-h post-transfection, the cells were collected and lysed in 50 all of reporter lists buffer (PR omega, USA). After incubation for 15 min on ice, the cell lysates were centrifuged at 12,000×g for 2 min at 4 °C. Protein (20 μg) extractions of the supernatants were analyzed using 10 % SDS-PAGE and transferred onto a nitrocellulose membrane (Merck Millipore, USA). After blocking with 5 % skimmed milk in PBST (0.05 % Tween-20 in PBS) overnight at 4 °C, the membrane was incubated with the following antibodies: goat anti-Fluc and mouse anti-β-actin monoclonal antibodies (PR omega, USA) for 1 h at 37 °C, followed by horseradish peroxidase (HRP)-conjugated rabbit anti-goat and anti-mouse antibodies (Dao Cytomation, Glostrup, Denmark). Finally, the results were visualized using chemiluminescence reagents (Pierce).
Indirect immunofluorescence assay (IFA)
Expression of the luciferase reporter gene was further determined using IFA. At 24-h post-transfection with pSK-Rluc/Fluc (3 μg) as described above, BSR-T7/5 cells were fixed with 4 % paraformaldehyde, followed by staining with primary antibodies and the goat polyclonal antibody against a firefly luciferase and mouse anti-Renilla reniformis luciferase antibody (PR omega, USA), respectively. After incubation with the primary antibodies, the cells were subjected to the corresponding secondary antibodies, fluorescein isothiocyanate (FITC)-conjugated rabbit anti-goat IgG and FITC- conjugated goat anti-mouse IgG (Santa Cruz, California, USA). Samples were counterstained with DAPI (Invitrogen) and observed using a fluorescence microscope (BX60; Olympus, Japan).
Translation assay in cell culture
BSR-T7/5 cells were grown overnight in 14-mm diameter culture dishes prior to transfection and repeated three times for each time point. After transfection with pSK-Rluc/Fluc (250 ng), the cells were incubated at 37 °C in a humidified atmosphere of 5 % CO2. The cells were harvested at 5, 16, 24, 30, 38, 46, 54, 66, 76, 88, 100, and 112 h post-transfection and stored at −80 °C. Luciferase and Renilla reniformis luciferase activities were monitored by a FB12 Luminometer (Berthold, Germany) using a dual-luciferase reporter assay system (PR omega) according to the manufacturer’s instructions.
Luciferase assay
Using 14-mm diameter culture dishes, the BSR-T7/5 cells were lysed by the addition of the reporter lists buffer (PR omega, USA) (500 all per well) after several washes with PBS 1 day after transfection with the constructed deletion mutants (250 ng). This was repeated three times for each sample. The cells were then incubated for 15 min at room temperature with shaking. Subsequently, the cell suspension was transferred to a microcentrifuge tube and the cell samples were clarified by centrifugation at 12,000×g for 2 min at 4 °C. Next, 20 all of each supernatant was assayed for luciferase and Renilla reniformis luciferase activity as previously described.
Statistical analysis
All statistical analyses was performed using GraphPad Prism Software Version 5.00 (GraphPad Software Inc., San Diego, CA, USA). Differences in luciferase activities produced by pSK-RLuc/Fluc and its mutant derivatives were compared using one-way analysis of variance (ANOVA). Differences were considered statistically significant at P < 0.05. All data were reported as the mean ± standard deviation (SD). The DHAV-1 3′UTR secondary structure was predicted using RNA Draw Software Version 0.2.0.1 (Department of Computer Science University Leipzig, Germany).
Results
Construction of the DHAV minigenome plasmid and deletion mutants
Construction of the DHAV minigenome system and the derived DHAV-1 deletion mutants are outlined in the Materials and Methods and shown schematically in Fig. 1. To generate the DHAV-1 minigenome plasmid pSK-RLuc/Fluc, the Rluc-5′UTR-Fluc-3′UTR was cloned into pSK-Rz using the restriction sites BamHI and NotI. On the basis of pSK-RLuc/Fluc and the secondary structure of the DHAV-1 3′UTR, four deletion mutants, pSK-Δ3′UTR, pSK-Δ3′UTR (1–150), pSK-Δ3′UTR (151–315), and pSK-Δ5′UTR, were constructed in this study. All of the resulting plasmids were confirmed to be correct by restriction mapping and DNA sequencing.
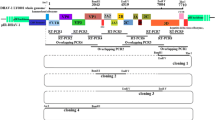
Schematic diagrams of the organization of the DHAV-1 minigenome and the derived DHAV-1 minigenome deletion mutants. The plasmid pBluescript II SK(+) (pSK) was modified to harbor a T7 promoter immediately upstream and HdvRz immediately downstream of the multi-cloning sites to precisely generate the target transcripts and was renamed pSK-Rz. The structure of the DHAV-1 genome is shown together with the minigenome plasmid constructs. The original fragment, Rluc-5′UTR-Fluc-3′UTR, was inserted into pSK-Rz, resulting in the plasmid pSK-Rluc/Fluc. This plasmid was then modified to generate a series of deletion mutants, pSK-Δ5′UTR, pSK-Δ3′UTR, pSK-Δ3′UTR (1–150), and pSK-Δ3′UTR (151–315), by deletion of the 5′UTR, 3′UTR, 3′UTR(1–150), and 3′UTR (151–315), respectively
Verification of the DHAV-1 minigenome system
Following the transfection of BSR-T7/5 cells with pSK-RLuc/Fluc, the availability of the DHAV-1 minigenome system was examined using RT-PCR, Western blotting analyses and IFA. However, total RNA was extracted from the cells and then subjected to standard RT-PCR to detect the accumulation of mRNA at 24 h post-transfection. The results in Fig. 2 showed that the specific band for the Fluc gene could be detected in transfected cells, while it was not observed in the non-transfected cells. Moreover, the band for the internal reference gene β-actin could be detected in both transfected and non-transfected cells.
Detection of mRNA using RT-PCR. The lane of pSK-Rluc/Fluc shows that the reporter gene Fluc and the internal reference gene β-actin is detectable in BSR-T7/5 cells transfected with pSK-Rluc/Fluc. The lane of untransfected/Psk-Rz shows that the internal reference gene β-actin can be detected from mock-transfected BSR-T7/5 cells, while the reporter gene Fluc cannot be detected
To further determine the efficiency of the minigenome system, expression of the reporter genes, Fluc and Rluc, was examined in transfected BSR-T7/5 cells with pSK-Rluc/Fluc using Western blotting and the IFA assay. Fluc and Rluc proteins expression was readily detectable using anti-Fluc and anti-Rluc antibodies (PR omega, USA); however, cells transfected with pSK-Rz failed to cross-react with the anti-Fluc and anti-Rluc antibodies (Figs. 3, 4).
Expression of the Fluc and Rluc reporter genes and the internal reference gene β-actin in BSR-T7/5 cells. Expression of Rluc and Fluc reporter genes and the internal reference gene β-actin is recognized by the corresponding antibody in BSR-T7/5 cells transfected with pSK-Rluc/Fluc. However, only expression of the internal reference gene β-actin could be observed in the mock-transfected BSR-T7/5 cells
Expression of the Fluc (a) and Rluc (b) genes was detected using an indirect immunofluorescence assay (IFA). BSR-T7/5 cells were transfected with pSK-Rluc/Fluc and pSK for 24 h, followed by staining with the following primary antibodies: mouse anti-Renilla reniformi polyclonal antibody and goat anti-Firefly luciferase polyclonal antibody, and the corresponding secondary antibodies, fluorescein isothiocyanate (FITC)-conjugated rabbit anti-goat IgG and FITC-conjugated goat anti-mouse IgG. pSK served as a negative control. Finally, the samples were counterstained with DAPI (blue color). Merged images are shown (×400)
Times course of Fluc activity
To monitor the expression dynamics of the Fluc reporter gene, BSR-T7/5 cells were transfected with pSK-Rluc/Fluc and then assayed for Fluc activity normalized against Rluc activity at 5, 16, 24, 30, 38, 46, 54, 66, 76, 88, 100, and 112 h post-transfection. These results showed that Fluc activity in cells reached a maximum value at 24-h post-transfection and then gradually declined (Fig. 5).
The time course of the DHAV-1 minigenome-derived luciferase activity. During the time course with the pSK-Rluc/Fluc experiments, cells were transfected with 250 ng of pSK-Rluc/Fluc, and each time point was repeated three times followed by lysis at different time points post-transfection. The Fluc activity was measured and normalized against Rluc activity. The Fluc activity reached a maximum value at 24 h post-transfection and then gradually decline
DHAV translation initiation is independent of the 3′UTR
To evaluate the effect of the 3′UTR on significant viral translation, BSR-T7/5 cells were transfected with equal amounts of pSK-RZ, pSK-Rluc/Fluc, and a series of deletion mutants were constructed in this study. These results showed that Fluc activity from BSR-T7/5 cells transfected with the complete or partial 3′UTR deletion mutants was consistent with the activity obtained from pSK-Rluc/Fluc-transfected cells, while the 5′UTR deletion resulted in a remarkable reduction in Fluc activity (Fig. 6).
Luciferase activity in BSR-T7/5 cells transfected with the DHAV-1 minigenome and the derived deletion mutants. The luciferase activities in cells transfected with several deletion mutants and the parental pSK-Rluc/Fluc were measured at 24-h post-transfection. pSK-Rz was used as a negative control. The Fluc activity in the cells transfected with the complete or partial 3′UTR deletion mutants was performed according to the protocol used for the pSK-Rluc/Fluc-transfected cells. The 5′UTR deletion resulted in a remarkable reduction of the Fluc activity. Fluc activity was measured and normalized against Rluc activity. Asterisks (***) represent P < 0.05 and indicate significant differences compared to parental pSK-Rluc/Fluc
Discussion
DHV serotype I exhibits the longest 3′UTR in the family Picornaviridae [14]. The importance of the 3′UTR on the in vivo replication of picornavirus has been confirmed in many other viruses [15–17]. However, little is understood about the structure and function of the DHAV 3′UTR. The lack of effective molecular tools has severely limited the exploration of the molecular mechanisms of DHAV replication and translation. The reverse genetics system (RGS) is considered to be a powerful tool used to study the molecular mechanisms of RNA viruses. Although the infectious clone is the best of the different types of the RGS, the characteristics of instability and low efficiency limit its wide application. The minigenome system is a good substitute of the infectious clone in studying the replication or translation mechanisms of DHAV-1. Here, we report a bicistronic reporter minigenome system containing two expression cassettes encoding Fluc and Rluc, which are separated by the 5′UTR of DHAV-1. This minigenome plasmid pSK-RLuc/Fluc contains T7 promoter and HdvRz located immediately upstream and downstream of the DHAV minigenome, respectively [12, 13]. Although the minigenome system for DHAV-1 has been constructed in our previous study [11], we generated two different minigenome systems with a different vector and promoter. However, the previous minigenome efficiency was low under the control of the CMV promoter, indicating that the CMV promoter is not capable of the stable expression of foreign genes in eukaryotic cells, which may be due to promoter silencing in cells by epigenetic conditions [7, 18]. However, the T7 promoter without the constraint of the cellular silencing mechanism is more efficient than the CMV promoter in expressing external proteins. Thus, we considered establishing a T7 promoter-driven minigenome system for DHAV-1 in this study.
The expression of reporter genes in BSR-T7/5 cells transfected with pSK-Rluc/Fluc was demonstrated using RT-PCR, Western blotting analyses and IFA. In addition, a translation assay in cell culture was performed at different time points post-translation. Taken together, these results indicated that we successfully constructed a minigenome system for DHAV-1. To the best of our knowledge, this is the first report of such a system developed for DHAV-1.
Using this system, we performed a functional analysis of the DHAV-1 3′ UTR. Previous studies have shown that the 5′UTR of the viral genome harbored the hepatitis C virus like (HCV-like) IRES element [4, 19]. The IRES element is responsible for the internal initiation of translation of viral RNA [20]. Thus, the plasmid pSK-Δ5′UTR was used as a negative control corresponding to a reduction in Fluc activity. Indeed, it has been shown that the HCV 3′UTR enhances IRES activity and that the encephalomyocarditis virus (EMCV) IRES is also enhanced by the HCV 3′-UTR or by a poly(A)-tail in different cell types [21]. Thus, the specific regulation of IRES activity by the 3′UTR is illustrated by specific nucleotide interactions, which is inconsistent with results reported by previous groups [22, 23]. An absence of regulation of the HCV IRES activity by the 3′UTR was demonstrated by Fang and Moyer [22] in vitro translation system [22]. In addition, various mutations were constructed to analyze the effect on translation efficiency by Friebe and Bartenschlager [24]. These results showed that a complete deletion of the variable domain, the poly (U/UC) tract or the 3′X domain did not modify the HCV IRES activity [24]. Imbert et al. [23] also showed that HCV translation is not modulated by the viral genomic 3′UTR sequence, even in the presence of HCV structural or non-structural proteins [23]. Translation is directed by the HCV IRES and HCV-like IRES of DHAV-1, which belongs to a distinct family of picornaviruses. The results in this study demonstrated that the complete or partial deletion of the 3′UTR elements could not reduce the expression level of Fluc.
In summary, we successfully constructed a bicistronic reporter minigenome system of DHAV, and using this platform, a series of UTR deletion mutants were constructed to analyze the role of the 3′UTR in DHAV IRES-mediated translation initiation. Our data demonstrated that neither the complete nor a partial deletion of DHAV-3′UTR had any apparent effect the expression level of the reporter gene.
References
Y. Fu, M. Pan, X. Wang, Y. Xu, H. Yang, D. Zhang, Vet. Microbiol. 131, 247–257 (2008)
L. Wang, M. Pan, Y. Fu, D. Zhang, Virus Genes 37, 52–59 (2008)
M.C. Kim, Y.K. Kwon, S.J. Joh, A.M. Lindberg, J.H. Kwon, J.H. Kim, S.J. Kim, J. Gen. Virol. 87, 3307–3316 (2006)
G. Liu, E. Yanguez, Z. Chen, C. Li, Virol. J. 8, 147 (2011)
M. Pan, X. Yang, L. Zhou, X. Ge, X. Guo, J. Liu, D. Zhang, H. Yang, J. Virol. 86, 1129–1144 (2012)
T. Yun, Z. Ni, G.Q. Liu, B. Yu, L. Chen, J.G. Huang, Y.M. Zhang, J.P. Chen, Virus Res. 147, 159–165 (2010)
A. Groseth, H. Feldmann, S. Theriault, G. Mehmetoglu, R. Flick, J. Virol. 79, 4425–4433 (2005)
J.B. Rohll, D.H. Moon, D.J. Evans, J.W. Almond, J. Virol. 69, 7835–7844 (1995)
S.L. de Quinto, M. Saiz, D. de la Morena, F. Sobrino, E. Martinez-Salas, Nucleic Acids Res. 30, 4398–4405 (2002)
Y. Wang, C. Li, Z. Chen, B. Xu, G. Li, G. Liu, Virus Genes 45, 398–401 (2012)
R.Y. Liang, W. Hu, N. Li, Q.H. Miao, Z.L. Bi, C.C. Meng, C.F. Li, Z.Y. Chen, G.Q. Liu, Construction of the Mini-genome of Duck Hepatitis A Virus. Acta Veterinaria et Zootechnica Sinica 45, 609–613 (2014)
C. Estevez, D. King, B. Seal, Q. Yu, Virus Res. 129, 182–190 (2007)
H. Feng, D. Wei, G. Nan, S.J. Cui, Z.N. Chen, H. Bian, Arch. Virol. 156, 611–616 (2011)
C.Y. Ding, D.B. Zhang, Bing du xue bao = Chinese journal of virology/[bian ji, Bing du xue bao bian ji wei yuan hui] 23, 312–319 (2007)
C.C. Kok, G.G. Au, Arch. Virol. 158, 765–773 (2013)
R.Y. Liang, C.F. Li, C.C. Meng, Z.Y. Chen, G.Q. Liu, Bing du xue bao = Chinese journal of virology/[bian ji, Bing du xue bao bian ji wei yuan hui] 30, 463–469 (2014)
J. Zoll, H.A. Heus, F.J. van Kuppeveld, W.J. Melchers, Virus Res. 139, 209–216 (2009)
C. Teschendorf, K.H. Warrington Jr, D.W. Siemann, N. Muzyczka, Anticancer Res. 22, 3325–3330 (2002)
C. Ding, D. Zhang, Virology 361, 9–17 (2007)
S. Garcia-Nunez, M.I. Gismondi, G. Konig, A. Berinstein, O. Taboga, E. Rieder, E. Martinez-Salas, E. Carrillo, Virology 448, 303–313 (2014)
C. Bung, Z. Bochkaeva, I. Terenin, R. Zinovkin, I.N. Shatsky, M. Niepmann, FEBS Lett. 584, 837–842 (2010)
J.W. Fang, R.W. Moyer, J. Hepatol. 33, 632–639 (2000)
I. Imbert, M. Dimitrova, F. Kien, M.P. Kieny, C. Schuster, J. Gen. Virol. 84, 1549–1557 (2003)
P. Friebe, R. Bartenschlager, J. Virol. 76, 5326–5338 (2002)
Acknowledgments
This study was supported by the Science and Technology Project in Shanghai (No. 13391901602), Special Fund for Agro-scientific Research in the Public Interest (No. 201303046), and the Chinese Natural Sciences Foundation (31270194).
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by Keizo Tomonaga.
Ruiying Liang and Chuanfeng Li have contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liang, R., Li, C., Jin, H. et al. Duck hepatitis A virus serotype 1 minigenome: a model for studying the viral 3′UTR effect on viral translation. Virus Genes 51, 367–374 (2015). https://doi.org/10.1007/s11262-015-1255-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-015-1255-0