Abstract
Staphylococcus aureus is an important pathogen that causes mastitis in cattle, and the emergence of methicillin-resistant S. aureus (MRSA) poses a threat to veterinary and human medicine. The aims of the study were to investigate the prevalence of MRSA and methicillin-resistant coagulase-negative staphylococci (MR-CoNS) isolated from clinical mastitis, their ability to form biofilms, and the antimicrobial susceptibility of S. aureus strains. In addition, the Staphylococcal Cassette Chromosome mec (SCCmec) type, spa type and the presence of Panton-Valentine Leucocidin in MRSA were evaluated. A total of 326 staphylococcal strains were screened by multiplex-PCR for S. aureus and Staphylococcus intermedius group (SIG) identification. The S. aureus strains (n = 163) were subjected to phenotypic testing for antimicrobial susceptibility and biofilm formation. Molecular analysis was performed on MRSA mecA-positive strains. Of 163 S. aureus isolates, 142 strains (87.1%) were resistant to at least one antibiotic, and all 19 MRSA strains were resistant to at least four out of five antibiotics tested. All S. aureus strains harboured the icaA gene and were biofilm producers. Nineteen MR-CoNS strains were also isolated. The most prevalent spa types among MRSA were t001 (57.9%) and t037 (31.6%), while one MRSA was type t008 and one was type t041. Most MRSA were SCCmec type I (63.2%) and III (31.6%) and only one strain was type IV. None of the MRSA isolates had the PVL gene. The prevalence of multidrug-resistant S. aureus in bovine mastitis is a serious concern. The finding of MRSA with spa types predominant in humans and infrequent in Italian cows and with SCCmec infrequently found in bovine milk or cheese suggest a human origin of these strains. The ability of MRSA and MR-CoNS involved in bovine mastitis to be transferred to humans and vice versa poses a public health concern.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bovine mastitis is an inflammation of the mammary gland, which is the most predominant disease in the dairy industry and causes major economic losses (Zaatout et al. 2020). Staphylococcus aureus has been reported to be one of the most relevant causative agents of bovine mastitis (Côté-Gravel and Malouin 2019). Its main reservoir seems to be the infected quarter, and its transmission between cows usually occurs during milking procedure (Monistero et al. 2018).
Staphylococcus spp. are common commensals of humans and animals, and important causes of opportunistic infections. In humans, S. aureus is the most important pathogen among the various Staphylococcus species and is the cause of widespread opportunistic and foodborne infections. Similarly, it is an important cause of infection of various animal species, including cattle. Other staphylococci are of varying clinical relevance, ranging from essentially harmless commensal to potentially important cause of disease. Moreover, S. pseudintermedius and other staphylococci of the Staphylococcus intermedius group (SIG) are important staphylococcal pathogens in animals and their phenotypic/genotypic resistance characterization is of great importance for dairy farming. Coagulase-negative staphylococci (CoNS), while typically considered to have limited pathogenicity, can cause disease under certain circumstances.
A common problem with all staphylococci is the tendency to acquire resistance to antimicrobials. Most of the attention has been paid to S. aureus, particularly with the emergence of methicillin-resistant S. aureus (MRSA), first in humans then in various animal species (Lee 2006). Methicillin-resistance in all staphylococci is due to the presence of the chromosomal mecA gene encoding a penicillin-binding protein with low affinity for β-lactams, resulting in resistance to virtually all beta-lactam antimicrobials. mecA resides on a Staphylococcal Cassette Chromosome mec (SCCmec), of which 8 types (I-VIII) are currently described (Rajeswari Anburaj and Raja 2020). The main route of transmission of MRSA is person-to-person, but pathogen cycling between humans and animals has been repeatedly reported (Weese et al. 2005; Rossi et al. 2016). The possibility of an interspecies transmission is a public health concern, which is clearly manifested by the increasing number of epidemiological studies of MRSA in animal populations reported in recent years (Kløve et al. 2022).
The occurrence and transmission of antimicrobial-resistant S. aureus itself or its genes has been suggested as one of the reasons for difficulty in antibiotic therapy. Therefore, the determination of susceptibility or resistance of strains to antibiotics is very important from a clinical and economic point of view. Moreover, this issue is of great public health importance because antibiotic treatment of infectious diseases in animals carries the risk of selection of resistant strains and introduction of these strains into the food chain and the environment, particularly the aquatic environment (White and McDermott 2001; Silva et al. 2023). MRSA detection and containment, associated with mastitis in dairy herds, is therefore essential. Epidemiological studies on MRSA associated with mastitis are documented (Lee 2006; Feltrin et al. 2016; Asadollahi et al. 2018). Likewise, reports on methicillin resistance in CoNS associated with mastitis have been demonstrated due to S. chromogenes, S. epidermidis and S. sciuri (Mello et al. 2020; Song et al. 2020; Silva et al. 2014; Devriese et al. 2002), and other CoNS, recently (Klibi et al. 2018; de Oliveira et al. 2022; Ibrahim et al. 2022).
Eradication of these microorganisms is also not always successful due to their ability to form biofilms. Extensive research concerning biofilm formation has been carried out using S. epidermidis as a model organism for staphylococci. There are several differences between S. epidermidis and S. aureus, with respect to biofilm formation. Most S. aureus strains reported so far contain the entire ica gene cluster (Cramton et al. 2001; Knobloch et al. 2002). The implication of biofilms in chronic infections has triggered an increasing interest in the characterization of genes involved in biofilm formation (Tormo et al. 2005).
The main objectives of this research work were (i) to evaluate the prevalence of methicillin-resistant staphylococci in milk sample of cows with clinical mastitis, (ii) to investigate the icaA gene in biofilm producer staphylococci strains, (iii) to determine the acquired multidrug resistance in S. aureus, and finally, (iv) to characterize the SCCmec of MRSA strains.
Materials and methods
Animals, samples, and bacterial strains isolation
According to National Mastitis Council (NMC) guidelines, a total of 813 quarter milk samples were aseptically collected from Holstein–Friesian dairy cows with clinical mastitis in 8 different farms of Marche and Umbria regions, Central Italy, during 2019 – 2020, including farm 1 (in the Macerata province; herd size ≥ 100; milking frequency, 2 times per day; n = 51 samples), farm 2 (in Macerata province; herd size ≥ 100; milking frequency, 2 times per day; n = 54 samples), farm 3 (in the Macerata province; herd size ≥ 200; milking frequency, 3 times per day; n = 47 samples), farm 4 (in the Ancona province; herd size ≥ 400; milking frequency, 3 times per day; n = 110 samples), farm 5 (in the Pesaro-Urbino province; herd size ≥ 700; milking frequency, 3 times per day; n = 201 samples, farm 6 (in the Perugia province; herd size ≥ 700; milking frequency, 3 times per day; n = 225 samples), farm 7 (in the Perugia province; herd size ≥ 100; milking frequency, 2 times per day; n = 43 samples), farm 8 (in the Terni province; herd size ≥ 200; milking frequency, 2 times per day; n 82 samples). All the randomly chosen lactating dairy cows were examined using clinical inspection of the udder as described by Quinn et al. (2002) and California Mastitis Test (CMT). For milk collection, the udders of cows with clinical mastitis were cleaned with water and dried. Cotton balls with 75% ethanol were used to disinfect the surface of the udder. The first few streams of milk were discarded. The collected milk, from each quarter, was kept in a sterile tube and transported refrigerated to the laboratory within 4 h.
Each specimen was cultured according to standard protocols plating 10 µl on Columbia Blood agar and Mannitol Salt agar (Liofilchem®, Italy), and incubated at 36 ± 1 °C in aerobic atmosphere for 24–48 h. All presumptive staphylococci were tested for coagulase (Staphylase Test kit, Oxoid) and identified by using the MALDI-TOF MS (SOP Direct Transfer Procedure Revision.4; Bruker Microflex Lt®, Bruker Daltonics, Germany).. Mass spectra were processed using Flex Analysis (version 3.4; Bruker Daltonics, Germany) and BioTyper software (version 3.1; Bruker Daltonics, Germany); The row spectra obtained, were compared with those in the Biotyper database and a log (score) of ≥ 2.0 indicated a secure genus identification and a highly probable species-level identification.
Genotypic identification
A molecular identification of strains was performed by amplifying the nuc gene of S. aureus and SIG using species-specific oligonucleotide primers (Baron et al. 2004). The mecA (Strommenger et al. 2003) and icaA (Zmantar et al. 2008) primers were used to detect the methicillin-resistant gene and a biofilm gene, respectively, by PCR. The oligonucleotide primers were used together with an internal positive control that targets a highly conserved region of 16S rDNA in a multiplex PCR (Baron et al. 2004). ATCC 33591 was used as MRSA control strain.
The oligonucleotide primer sequences, and PCR programs are summarized in Table 1. For DNA preparation, three colonies of freshly sub-cultured strains were suspended in 50 μl lysozyme 5 U/μl (Sigma, Germany). After incubation for 1 h at 37 °C, 0.75 μl proteinase K 20 μg/μl (Sigma, Germany) was added, and the suspension was re-incubated for 1 h at 56 °C. Finally, the proteinase K was inactivated through boiling of the mixture for 10 min followed by 5 min cooling on ice. After centrifugation at 8000 × g for 10 min the supernatant was used for PCR.
The PCR reaction mixture (30 µl) contained 1.4 µl, 1.7 µl, 1.6 µl, 1.5 µl and 0.3 µl of SInuc, SAnuc, mecA, icaA and 16S primers (10 pmol/µl) respectively (Table 1), 15 µl of Taq PCR mastermix 2x (Qiagen, Germany), 2 µl of MgCl2 25 mM (Sigma, Germany). Finally, 1 µl DNA preparation was added to each reaction tube. The tubes were then subjected to thermal cycling with the following programs: 1 × (95 °C 300 s), 35 × (94 °C 60 s, 55 °C 60 s, 72 °C 60 s), 1 × (72 °C 420 s). The presence of PCR products was determined by electrophoresis of 10 μl of reaction products in a 3% agarose gel (Qbiogene, Germany) with Tris–acetate EDTA buffer (TAE, 4.0 mmol/l Tris–HCl 1 mmol/l EDTA, pH 8.0) and visualized under UV light (Gel Logic 100, Imaging system). The primers used in this study are reported in Table 1.
Panton Valentine leukocidin (PVL) gene screening
Real time PCR for PVL with primers PVLSC-F, GCTCAGGAGATACAAG and PVLSC-R, GGATAGCAAAAGCAATG was performed on MRSA strains as described previously (Roberts et al. 2005). The Sybr Green PCR reaction was carried out by using the kit Premix Ex Taq DNA Polymerase (Takara Bio, Japan). According to the manufacturer's instructions, the 20 μl mixture contained 10 μl of Premix Ex Taq 2x, 1 μl of each 10 pmol/μl primer, 6 μl of PCR water and 2 μl of DNA. The cycling conditions were as follows: 98 °C for 5 min, 30 cycles of amplification (15 s of denaturation at 95 °C, 5 s of annealing at 55 °C and 10 s of extension at 72 °C). The melt curve analysis was carried out as follows: 5 s at 95 °C, 15 s at 65 °C, then ramp from 65 to 95 °C at 0.1 8C/s.
spa typing
The S. aureus isolates resulted MRSA, were used for spa typing. The SSR region of the spa gene was amplified and sequenced according to a method described by Shopsin et al. (1999). Sequences were analyzed by the eGenomics software (http://tools.egenomics.com) and were reported using a numerical system that corresponded to a spa type sequence. Ridom database equivalents were identified by the Ridom Spaserver website (http://www.spaserver.ridom.de) and were reported using a numerical coding system preceded by a ‘t’.
Characterization of SCCmec
The presence of SCCmec types I – IV was determined in MRSA by multiplex PCR as previously described (Zhang et al. 2005). In particular, the PCR reaction mix included two pairs of primers each for characterization of the mec gene (mecI-F, mecI-R, IS1272-F and mecR1-R) and of the ccr gene (ccrAB-2, ccrAB-2, ccrAB-3, and ccrAB-4) (Zhang et al. 2005). The PCR reaction mixture contained 25 µl of Taq PCR mastermix 2x (Qiagen, Germany), 0.08 µM of each primer, 2 µl of DNA and PCR water up to a final volume of 50 µl. The reaction condition were: 94 °C for 5 min followed by 10 cycles of 94 °C for 45 s, 65 °C for 45 s, and 72 °C for 90 s and another 25 cycles of 94 °C for 45 s, 55 °C for 45 s, and 72 °C for 1.5 min, ending with a final extension step at 72 °C for 10 min and followed by a hold at 4 °C. The PCR amplicons were visualized using a UV light box after electrophoresis on a 2% agarose gel.
Congo red agar method
The Congo Red Agar method (CRA) for phenotypic identification of biofilm production was done according to the Freeman protocol (Knobloch et al. 2002). Studies have demonstrated that this method has low accuracy, but it was chosen as support for PCR test (icaA gene detection) because is cheap and easy to perform and the evaluation criteria is based on visual analysis of the colour of the colonies that grow on the agar. The constituents of media were, Brain Heart Infusion broth (Oxoid, Milan, Italy) 37 g/l, sucrose (Carlo Erba Reagents, Milan, Italy) 50 g/l, Agar N. 1 (Oxoid, Milan, Italy) 10 g/l and Congo Red stain (Carlo Erba Reagents, Milan, Italy). A quantity of 0.8 g/l. Congo Red stain was prepared as a concentrated aqueous solution, autoclaved separately and added to the media when the agar had cooled to 55 °C. Plates of the medium were inoculated and incubated aerobically for 24 h at 37 °C. Biofilm-producing strains generated black colonies, which correspond to high biofilm production or brown colonies, which was considered evidence of low biofilm production, while biofilm-negative strains were red or pink.
Antimicrobial susceptibility testing
S. aureus strains were tested for susceptibility to a panel of five antimicrobials using the disc agar diffusion (Kirby-Bauer) method on Müeller Hinton agar (Liofilchem®, Italy) following the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines. The selected antibiotics, the most widely used for the treatment of clinical mastitis in the regions where the farms were located, were as follows: oxytetracycline (30 µg), gentamicin (10 µg), erythromycin (10 µg), clindamycin (10 µg) and cefoxitin (30 µg). The strains were classified as susceptible or resistant according to Clinical Breakpoint EUCAST Guidelines (EUCAST version 12.0, 2022).
Results
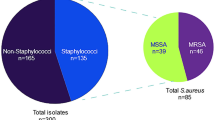
Totally, 326 Staphylococcus spp. were identified from the collected bovine milk samples; of these, 163 S. aureus (50%) and 19 SIG (5.8%) were identified. The remaining 144 staphylococci (44.2%), even if were speciated, were all considered coagulase-negative staphylococci (Table 2).
Nineteen (11.7%) out of 163 S. aureus isolates possessed mecA gene and were all phenotypically resistant to cefoxitin. All SIG isolates were mecA negative and 19 (13.2%) of the CoNS strains were mecA positive and considered methicillin-resistant (MR-CoNS). All S. aureus strains isolated from clinical mastitis harboured the icaA gene.
In CRA, 129 (79.1%) S. aureus produced black colonies, which correspond to high biofilm production and 34 (20.9%) produced brown colonies, which is considered evidence of weak biofilm production. The icaA gene was detected in 30 (20.8%) CoNS isolates. Twenty-five of them belong to the black colonies group and 5 to the brown colonies group. None of the SIG strains showed the icaA gene. All 19 MRSA isolates and 18 out 19 MR-CoNS produced black colonies on CRA.
S. aureus susceptibility data were presented in Table 3. Only 21 strains (12.9%) were susceptible to all the antibiotics. All 19 MRSA isolates were resistant to at least 4 of the tested antimicrobials and 9 (47%) were resistant to 5. Four isolates were resistant to clindamycin, erythromycin, gentamicin and oxytetracycline and one isolate to clindamycin, erythromycin, and gentamicin (Table 3).
Eleven MRSA isolates were spa type 385/t001 and SCCmecI, 6 were spa type 3/t037 and SCCmec III, 1 was spa type 388/t041 and SCCmec I and 1 strain was spa type 1/t008 and SCCmecIV (Table 4). All the MRSA strains were negative for the PVL toxin encoding gene.
Discussion
Staphylococcus spp. play an important role in causing clinical mastitis in cattle. Understanding prevalence and mechanisms of antimicrobial resistance and biofilm production is important to develop and assess measures to control or treat clinical mastitis and evaluate the potential for horizontal gene transfer of antimicrobial resistance genes.
MRSA is not commonly reported in bovine mastitis, but this has recently been identified, typically at a low prevalence. In this study, the prevalence of MRSA (11.7%) was remarkable compared to some previous reports (Bengtsson et al. 2009; Alves et al. 2009). Direct comparison of the prevalence of MRSA from clinical mastitis observed in this study with other studies should be performed with care because of potential differences in study population. Limited clinical data were available to determine whether the MRSA isolates were from infections that were chronic and/or had been previously treated with antimicrobials, but regardless, the data indicate that MRSA is common in bovine mastitis in these regions. These results are concordant with a survey carried out in European countries of prevalence of antimicrobial resistance in cattle, which reported that antimicrobial resistance was greater (> 10%) in Italy than European countries like Denmark and Sweden (Hendriksen et al. 2008). Resistance of MRSA to antimicrobials that are used in cattle, such as macrolides and lincosamides, is not particularly surprising. While the origin of MRSA in these cattle cannot be determined, the presence of strains commonly found in humans and resistance to a drug not used in cattle suggests that investigation of human-cattle transmission of MRSA is required. MRSA can be transmitted between animal species (Kaszanyitzky et al. 2007; Rossi et al. 2016) and given the close contact between human hands and the bovine udder during milking, human-animal transmission is certainly plausible.
Genes conferring resistance to one of the Macrolide-Lincosamide-Streptogramin B (MLSB) antibiotics may confer cross-resistance to others, because they have similar effects on bacterial protein synthesis inhibition. Bengtsson et al. (2009) reported only 1.9% MRSA were resistant to one or more antimicrobials, but in this study MRSA strains were all shown a multidrug resistance. All the S. aureus strains showed highly resistance to erythromycin, gentamicin, oxytetracycline and clindamycin and were probably related to the large use in veterinary medicine. Out of 163 isolates, 127 (77.9%) were resistant to one or more antimicrobials, and, like our results, Gentilini et al. (2000) and Wang et al. (2008) reported that high percentage (64% and 93.1%) of S. aureus isolates were resistant to one or more antimicrobials, respectively. Susceptibility to erythromycin (1.3%) in all tested strains was very low.
Kaszanyitzky et al. (2007) reported S. aureus isolated from subclinical mastitis was high susceptible to gentamicin, while our result showed high resistance to gentamicin. Overall, resistance rate against one of the antibiotics included in the group MLSB is high in our isolates and is like the resistance found in S. aureus isolates from clinical mastitis in China (Wang et al. 2008). The chance that the large use of some macrolide (e.g., erythromycin) and lincosamide (e.g., clindamycin) antibiotics in animals increases the rate of S. aureus resistant to other MLSB should be better evaluated to prevent the selection of multidrug resistant strains. There is the risk that livestock becomes a reservoir of S. aureus resistant to some streptogramin, that are used very carefully in humans as the last chance for the treatment of infections not responding to other antibiotics.
Information of biofilm production by coagulase positive S. aureus of intramammary origin is largely lacking. The ica locus has been detected in majority of the mastitis S. aureus isolates indicating its potential role as a virulence factor in the pathogenesis of mastitis in ruminants and all identified S. aureus in this study were harboured icaA gene. Similarly, Cramton et al. (1999) observed that icaA was present in all S. aureus strains studied. Similar percentages were observed by Pérez et al. (2020), Salina et al. (2020) and Felipe et al. (2017) reporting rates of 98.5%, 82%, and 100%, respectively. In contrast, Bouzidi et al. (2023) reported that the icaA gene was detected in 32.2% (n = 20) of S. aureus strains isolated from subclinical bovine mastitis in Algeria.
Spa typing is a cheaper and faster typing method than MLST that is currently used for typing S. aureus isolates. Although spa typing relies only on the assessment of the number of and sequence variation in repeats at the X region of the spa gene, this method exhibits excellent discriminatory power and is useful for epidemiological studies and exchanging data between laboratories. In this study, four different spa types were found in the MRSA strains: t001 (11 out of 19 strains), t037 (6 out of 19), t041 (1 out of 19) and t008 (1 out of 19). These spa types have been not so far described in MRSA strains isolated from bovine milk or cheeses in Italy (Tomao et al. 2020; Johler et al. 2018; Thiran et al. 2018; Basanisi et al. 2017; Luini et al. 2015). Spa types t0037, t008 and t001 are highly spread among human patients in different continents (Asadollahi et al. 2018). In particular, t0037 is the most prominent spa type found in Africa and is the second one found in Asia and in Australia, while it is not included in the group of the predominant spa types detected in Europe and America. The spa type t008 is the most predominant spa type in America and the second one detected in Europe. The spa type t001, which is the predominant type detected in our MRSA strains isolated from bovine, is not included among the most predominant spa types in Europe but is one of the most predominant types detected in Asia. All spa types found in the bovine MRSA isolates included in the present study have been already detected in human patients during a first nationwide survey about S. aureus epidemiology in 2012 in Italy (Campanile et al. 2015). These data shows that MRSA isolated from bovine milk with the same spa types of human strains are spread in Italy and attention should be paid about transmission of these strains from cattle to human and vice versa.
The association of the spa types found in our samples with SCCmec types is perfectly concordant with the data reported in literature (Asadollahi et al. 2018). Indeed, most strains with spa type t0037 isolated worldwide were associated with SCCmec type III, as in the six MRSA strains isolated in this study from bovine. The spa type t001 is most frequently associated with SCCmec type I in Asia, the spa type t041 is most frequently associated with SCCmec type I in Europe, and the spa type t008 is most frequently associated with SCCmec type IV worldwide. Although the most frequent SCCmec types found in MRSA strain isolated from bovine milk or cheese in Italy are types IV and V (Tomao et al. 2020; Basanisi et al. 2017; Luini et al. 2015), our MRSA strains showed mainly the SCCmec types I and III, suggesting that these strains are different than the strains previously found in dairy farms in Italy. None of the Staphylococcus isolates from bovine milk showed the PVL gene. The PVL gene is predominant in the community acquired MRSA and is frequently found in association with more severe diseases and a significant increased risk of sepsis (Ahmad et al. 2020). In the few studies carried out previously in Italy on MRSA in bovine milk, only one MRSA strain isolated from bulk tank milk from farms located in north-eastern Italy showed the PVL gene (Tomao et al. 2020), while 6 out of 16 MRSA isolated from milk samples and 14 out of 24 MRSA isolated from cheese in South Italy showed the PVL gene (Basanisi et al. 2017). Monitoring of PVL gene presence in S. aureus infecting food producing animals is required to prevent transmission of highly pathogenic strains from animals to humans.
Very little is known about SIG in bovine mastitis. A low prevalence (1–2%) has been identified in previous reports (Capurro et al. 1999). The 5.8% prevalence identified here is therefore somewhat higher than previous reports, although the reasons and relevance are unclear.
The literature often refers to CoNS as minor mastitis pathogens, and determination of the relevance of CoNS in clinical specimens can be challenging because of their commonness as commensals and the potential for contamination. MR-CoNS have been isolated from domestic animals although they are often found as commensals. The presence of methicillin-resistance does not increase the inherent virulence of CoNS, so determination of the clinical relevance of MR-CoNS remains problematic. The ability of some CoNS strains to produce biofilm raises concern about the potential for virulence, and the treatment of infections would be complicated by the presence of methicillin-resistance. Further study of the true role of MR-CoNS in mastitis is needed, as the investigation of the role of biofilm in the pathophysiology of disease.
To our knowledge this is the first report of icaA gene in CoNS isolated from bovine milk in Italy.
Conclusion
The present study revealed the high prevalence of Staphylococcus spp. from bovine mastitis samples, something that was expected. The high prevalence of biofilm production by S. aureus and the ability of some CoNS (including MR-CoNS) to produce biofilms raise questions about the potential role of these organisms in disease. While uncommon, SIG were isolated from clinical mastitis samples, and the potential role of these pathogenic staphylococci in mastitis requires study. While previous reports have typically indicated a low prevalence of methicillin-resistance in mastitis isolates, these data indicate that studies must be focused on evaluating the scope, epidemiology, and control of MRSA mastitis in cattle. The finding of MRSA with spa types more prevalent in humans than in cattle suggests that animal-to-human transmission and vice versa may occur and that biosecurity measures are required to prevent MRSA spread. Further studies should clarify the role of farm workers in spreading MRSA in dairy herds.
Data Availability
The data that support the findings of this study are available from the corresponding author, VC, upon reasonable request.
References
Ahmad NI, Yean Yean C, Foo PC, Mohamad Safiee AW, Hassan SA (2020) Prevalence and association of Panton-Valentine Leukocidin gene with the risk of sepsis in patients infected with Methicillin Resistant Staphylococcus aureus. J Infect Public Health 13(10):1508–1512. https://doi.org/10.1016/j.jiph.2020.06.018
Alves PD, McCulloch JA, Even S, Le Marechal C, Thierry A, Grosset N, Azevedo V, Rosa CA, Vautor E, Le Loir Y (2009) Molecular characterisation of Staphylococcus aureus strains isolated from small and large ruminants reveals a host rather than tissue specificity. Vet Microbiol 137(1–2):190–195. https://doi.org/10.1016/j.vetmic.2008.12.014
Asadollahi P, Farahani NN, Mirzaii M, Khoramrooz SS, van Belkum A, Asadollahi K, Dadashi M, Darban-Sarokhalil D (2018) Distribution of the Most Prevalent Spa Types among Clinical Isolates of Methicillin-Resistant and –Susceptible Staphylococcus aureus around the World: A Review. Front Microbiol 9:163. https://doi.org/10.3389/fmicb.2018.00163
Baron F, Cochet MF, Pellerin JL, Ben Zakour N, Lebon A, Navarro A, Proudy I, Le Loir Y, Gautier M (2004) Development of a PCR test to differentiate between Staphylococcus aureus and Staphylococcus intermedius. J Food Prot 67(10):2302–2305. https://doi.org/10.4315/0362-028x-67.10.2302
Basanisi MG, La Bella G, Nobili G, Franconieri I, La Salandra G (2017) Genotyping of methicillin-resistant Staphylococcus aureus (MRSA) isolated from milk and dairy products in South Italy. Food Microbiol 62:141–146. https://doi.org/10.1016/j.fm.2016.10.020
Bengtsson B, Unnerstad HE, Ekman T, Artursson K, Nilsson-Ost M, Waller KP (2009) Antimicrobial susceptibility of udder pathogens from cases of acute clinical mastitis in dairy cows. Vet Microbiol 136(1–2):142–149. https://doi.org/10.1016/j.vetmic.2008.10.024
Bouzidi S, Bourabah A, Cheriet S, Abbassi MS, Meliani S, Bouzidi H (2023) Occurrence of virulence genes and methicillin-resistance in Staphylococcus aureus isolates causing subclinical bovine mastitis in Tiaret area. Algeria. Letters in Applied Microbiology. 76(3):ovad003. https://doi.org/10.1093/lambio/ovad003
Campanile F, Bongiorno D, Perez M, Mongelli G, Sessa L, Benvenuto S, Gona F (2015) AMCLI – S. aureus Survey Participants; Varaldo PE, Stefani S. Epidemiology of Staphylococcus aureus in Italy: First nationwide survey, 2012. J Glob Antimicrob Resist 3(4):247–254. https://doi.org/10.1016/j.jgar.2015.06.006
Capurro A, Concha C, Nilsson L, Ostensson K (1999) Identification of coagulase-positive staphylococci isolated from bovine milk. Acta Vet Scand 40(4):315. https://doi.org/10.1186/BF03547011
Côté-Gravel J, Malouin F (2019) Symposium review: Features of Staphylococcus aureus mastitis pathogenesis that guide vaccine development strategies. J Dairy Sci 102(5):4727–4740. https://doi.org/10.3168/jds.2018-15272
Cramton SE, Gerke C, Schnell NF, Nichols WW, Gotz F (1999) The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun 67(10):5427–5433. https://doi.org/10.1128/iai.67.10.5427-5433.1999
Cramton SE, Gerke C, Gotz F (2001) In vitro methods to study staphylococcal biofilm formation. Methods Enzymol 336:239–255. https://doi.org/10.1016/S0076-6879(01)36593-X
de Oliveira RP, da Silva JG, Aragão BB et al (2022) Diversity and emergence of multi-resistant Staphylococcus spp. isolated from subclinical mastitis in cows in of the state of Piauí. Brazil Braz J Microbiol 53:2215–2222. https://doi.org/10.1007/s42770-022-00822-1
Devriese LA, Baele M, Vaneechoutte M, Martel A, Haesebrouck F (2002) Identification and antimicrobial susceptibility of Staphylococcus chromogenes isolates from intramammary infections of dairy cows. Vet Microbiol 87(2):175–182. https://doi.org/10.1016/s0378-1135(02)00047-0
Felipe V, Morgante CA, Somale PS, Varroni F, Zingaretti ML, Bachetti RA, Correa SG, Porporatto C (2017) Evaluation of the biofilm forming ability and its associated genes in Staphylococcus species isolates from bovine mastitis in Argentinean dairy farms. Microb Pathog 104:278–286. https://doi.org/10.1016/j.micpath.2017.01.047
Feltrin F, Alba P, Kraushaar B, Ianzano A, Argudín MA, Di Matteo P, Porrero MC, Aarestrup FM, Butaye P, Franco A, Battisti A (2016) A livestock associated, Multidrug-Resistant, Methicillin-Resistant Staphylococcus aureus Clonal Complex 97 lineage spreading in dairy cattle and pigs in Italy. Appl Environ Microbiol 82(3):816–821. https://doi.org/10.1128/AEM.02854-15
Gentilini E, Denamiel G, Llorente P, Godaly S, Rebuelto M, DeGregorio O (2000) Antimicrobial susceptibility of Staphylococcus aureus isolated from bovine mastitis in Argentina. J Dairy Sci 83(6):1224–1227. https://doi.org/10.3168/jds.S0022-0302(00)74988-5
Hendriksen RS, Mevius DJ, Schroeter A, Teale C, Meunier D, Butaye P, Franco A, Utinane A, Amado A, Moreno M, Greko C, Stark K, Berghold C, Myllyniemi AL, Wasyl D, Sunde M, Aarestrup FM (2008) Prevalence of antimicrobial resistance among bacterial pathogens isolated from cattle in different European countries: 2002–2004. Acta Vet Scand 50:1–10. https://doi.org/10.1186/1751-0147-50-28
Ibrahim ES, Dorgham SM, Mansour AS, Abdalhamed AM, Khalaf DD (2022) Genotypic characterization of mecA gene and antibiogram profile of coagulase-negative staphylococci in subclinical mastitic cows. Veterinary World 15:2186–2191. https://doi.org/10.14202/vetworld.2022.2186-2191
Johler S, Macori G, Bellio A, Acutis PL, Gallina S, Decastelli L (2018) Characterization of Staphylococcus aureus isolated along the raw milk cheese production process in artisan dairies in Italy. J Dairy Sci 101(4):2915–2920. https://doi.org/10.3168/jds.2017-13815
Kaszanyitzky EJ, Janosi S, Somogyi P, Dan A, van der Graaf-van Bloois L, van Duijkeren E, Wagenaar JA (2007) MRSA transmission between cows and humans. Emerg Infect Dis 13(4):630. https://doi.org/10.3201/eid1304.060833
Klibi A, Maaroufi A, Torres C, Jouini A (2018) Detection and characterization of methicillin-resistant and susceptible coagulase-negative staphylococci in milk from cows with clinical mastitis in Tunisia. Int J Antimicrob Agents 52(6):930–935. https://doi.org/10.1016/j.ijantimicag.2018.07.026
Kløve DC, Jensen VF, Astrup LB (2022) First Finding of a Methicillin-Resistant Staphylococcus aureus (MRSA) t304/ST6 from Bovine Clinical Mastitis. Antibiotics 11(10):1393. https://doi.org/10.3390/antibiotics11101393
Knobloch JK, Horstkotte MA, Rohde H, Mack D (2002) Evaluation of different detection methods of biofilm formation in Staphylococcus aureus. Med Microbiol Immunol 191:101–106. https://doi.org/10.1007/s00430-002-0124-3
Lee JH (2006) Occurrence of methicillin-resistant Staphylococcus aureus strains from cattle and chicken, and analyses of their mecA, mecR1 and mecI genes. Vet Microbiol 114(1–2):155–159. https://doi.org/10.1016/j.vetmic.2005.10.024
Luini M, Cremonesi P, Magro G, Bianchini V, Minozzi G, Castiglioni B, Piccinini R (2015) Methicillin-resistant Staphylococcus aureus (MRSA) is associated with low within-herd prevalence of intra-mammary infections in dairy cows: Genotyping of isolates. Vet Microbiol 178(3–4):270–274. https://doi.org/10.1016/j.vetmic.2015.05.010
Mello PL, Riboli DFM, Martins LA, Brito MAVP, Victória C, Calixto Romero L, Ribeiro de Souza da Cunha ML (2020) Staphylococcus spp. isolated from bovine subclinical mastitis in different regions of Brazil: Molecular typing and biofilm gene expression analysis by RT-qPCR. Antibiotics 9(12):888. https://doi.org/10.3390/antibiotics9120888
Monistero V, Graber HU, Pollera C, Cremonesi P, Castiglioni B, Bottini E, Ceballos-Marquez A, Lasso-Rojas L, Kroemker V, Wente N, Petzer IM, Santisteban C, Runyan J, Veiga Dos Santos M, Alves BG, Piccinini R, Bronzo V, Abbassi MS, Said MB, Moroni P (2018) Staphylococcus aureus Isolates from bovine mastitis in eight countries: genotypes, detection of genes encoding different Toxins and other virulence genes. Toxins 10(6):247. https://doi.org/10.3390/toxins10060247
Pérez VKC, Custódio DAC, Silva EMM, de Olivera J, Guimarães AS, Brito MAVP, Souza-Filho AF, Heinemann MB, Lage AP, Dorneles EMS (2020) Virulence factors and antimicrobial resistance in Staphylococcus aureus isolated from bovine mastitis in Brazil. Braz J Microbiol 51:2111–2122. https://doi.org/10.1007/s42770-020-00363-5
Quinn PJ, Markey BK, Carter ME, Donnelly WJ, Leonard FC (2002) Veterinary Microbiology and Microbial Disease. Blackwell Science Ltd, Blackwell Publishing Company pp 491–492.
Rajeswari Anburaj R, Raja A (2020) Colonization, epidemiology and genetic mechanism of methicillin resistant Staphylococcus aureus. J Pharm Sci Res 12(8):1018–1023
Roberts S, O’Shea K, Morris D, Robb A, Morrison D, Rankin S (2005) A real-time PCR assay to detect the Panton Valentine Leukocidin toxin in staphylococci: screening Staphylococcus schleiferi subspecies coagulans strains from companion animals. Vet Microbiol 107(1–2):139–144. https://doi.org/10.1016/j.vetmic.2005.01.002
Rossi G, Cerquetella M, Attili AR (2016) Amphixenosic aspects of Staphylococcus aureus infection in man and animals. In: Bagnoli F, Rappuoli R, Grandi G (eds) Staphylococcus aureus. Current Topics in Microbiology and Immunology 409:297–323. https://doi.org/10.1007/82_2016_2
Salina A, Guimarães FF, Richini Pereira VB, Menozzi BD, Rall VLM, Langoni H (2020) Detection of icaA, icaD, and bap genes and biofilm production in Staphylococcus aureus and non aureus staphylococci isolated from subclinical and clinical bovine mastitis. Arq Bras Med Vet Zootec 72:1034–1038. https://doi.org/10.1590/1678-4162-11284
Shopsin B, Gomez M, Montgomery SO, Smith DH, Waddington M, Dodge DE, Bost DA, Riehman M, Naidich S, Kreiswirth BN (1999) Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol 37(11):3556–3563. https://doi.org/10.1128/JCM.37.11.3556-3563.1999
Silva V, Araújo S, Monteiro A, Eira J, Pereira JE, Maltez L, Igrejas G, Lemsaddek TS, Poeta P (2023) Staphylococcus aureus and MRSA in Livestock: Antimicrobial Resistance and Genetic Lineages. Microorganisms 11(1):124. https://doi.org/10.3390/microorganisms11010124
Silva NCC, Guimarães FF, de P. Manzi M. et al. (2014) Characterization of methicillin-resistant coagulase-negative staphylococci in milk from cows with mastitis in Brazil. Antonie van Leeuwenhoek 106, 227–233. https://doi.org/10.1007/s10482-014-0185-5
Song X, Huang X, Xu H, Zhang C, Chen S, Liu F, Guan S, Zhang S, Zhu K, Wu C (2020) The prevalence of pathogens causing bovine mastitis and their associated risk factors in 15 large dairy farms in China: An observational study. Vet Microbiol 247:108757. https://doi.org/10.1016/j.vetmic.2020.108757
Strommenger B, Kettlitz C, Werner G, Witte W (2003) Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J Clin Microbiol 41(9):4089–4094. https://doi.org/10.1128/JCM.41.9.4089-4094.2003
Thiran E, Di Ciccio PA, Graber HU, Zanardi E, Ianieri A, Hummerjohann J (2018) Biofilm formation of Staphylococcus aureus dairy isolates representing different genotypes. J Dairy Sci 101(2):1000–1012. https://doi.org/10.3168/jds.2017-13696
Tomao P, Pirolo M, Agnoletti F, Pantosti A, Battisti A, Di Martino G, Visaggio D, Monaco M, Franco A, Pimentel de Araujo F, Palei M, Benini N, Motta C, Bovo C, Di Renzi S, Vonesch N, Visca P (2020) Molecular epidemiology of methicillin-resistant Staphylococcus aureus from dairy farms in North-eastern Italy. Int J Food Microbiol. 332:108817. https://doi.org/10.1016/j.ijfoodmicro.2020.108817
Tormo MA, Knecht E, Gotz F, Lasa I, Penades JR (2005) Bap-dependent biofilm formation by pathogenic species of Staphylococcus: evidence of horizontal gene transfer? Microbiology. 151(7):2465–2475. https://doi.org/10.1099/mic.0.27865-0
Wang Y, Wu CM, Lu LM, Ren GW, Cao XY, Shen JZ (2008) Macrolide-lincosamide-resistant phenotypes and genotypes of Staphylococcus aureus isolated from bovine clinical mastitis. Vet Microbiol 130(1–2):118–125. https://doi.org/10.1016/j.vetmic.2007.12.012
Weese JS, Archambault M, Willey BM, Hearn P, Kreiswirth BN, Said-Salim B, McGeer A, Likhoshvay Y, Prescot JF, Low DE (2005) Methicillin-Resistant Staphylococcus aureus in horses and horse personnel, 2000–2002. Emerg Infect Dis 11(3):430. https://doi.org/10.3201/eid1103.040481
White DG, McDermott PF (2001) Emergence and transfer of Antibacterial Resistance. J Dairy Sci 84:E151–E155. https://doi.org/10.3168/jds.S0022-0302(01)70209-3
Zaatout N, Ayachi A, Kecha M (2020) Staphylococcus aureus persistence properties associated with bovine mastitis and alternative therapeutic modalities. J Appl Microbiol 129(5):1102–1119. https://doi.org/10.1111/jam.14706
Zhang K, McClure JA, Elsayed S, Louie T, Conly JM (2005) Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol 43(10):5026–5033. https://doi.org/10.1128/JCM.43.10.5026-5033.2005
Zmantar T, Chaieb K, Makni H, Miladi H, Abdallah FB, Mahdouani K, Bakhrouf A (2008) Detection by PCR of adhesins genes and slime production in clinical Staphylococcus aureus. J Basic Microbiol 48(4):308–314. https://doi.org/10.1002/jobm.200700289
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
All authors have made substantial contributions to the following:
(1) ARA, SP and VC, the conception and design of the study, acquisition of data, analysis, and interpretation of data.
(2) ARA, SP and VC drafting the article and revising it critically for important intellectual content.
(3) ARA, SP and VC approved the final version to be submitted.
Corresponding author
Ethics declarations
Animal ethics
Since cattle captures and milk sampling were conducted as part of regular milk control measures in accordance with Annex IV of Regulation (EC) 854/04, no ethics committee approval was required.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Preziuso, S., Attili, AR. & Cuteri, V. Methicillin-resistant staphylococci in clinical bovine mastitis: occurrence, molecular analysis, and biofilm production. Vet Res Commun 48, 969–977 (2024). https://doi.org/10.1007/s11259-023-10268-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11259-023-10268-x