Abstract
Insulin-like growth factor-binding protein acid-labile subunit (IGFALS) encodes a protein which binds to IGF1 and IGFBP-3 to regulate the growth, differentiation, and other physiological processes. The aim of this study was the identification of allelic polymorphisms of the IGFALS gene using the PCR-RFLP technique and evaluation of their association with growth traits. For this end, 120 blood samples were randomly collected from each breed. Following amplification of an 1113-bp fragment of exon 1 and a part of intron 1 of the IGFLAS gene, genotyping was conducted by three restriction enzymes including HinfI, MscI, and PvuII. The results showed that only one allele was observed in IGFALS-PvuII site, while in IGFLAS-MscI site, three AA, AB, and BB genotypes with the frequencies of 17.5%, 32%, and 50.5% and 11%, 37.5%, and 51.5% were observed in Makouei and Ghezel sheep breeds, respectively. Additionally, in the IGFLAS-HinfI site, two AB and BB genotypes with the frequency of 34.2% and 65.8% were observed in Makouei sheep and AA, AB, and BB genotypes with the frequency of 9%, 21%, and 70% were observed in Ghezel sheep. So that, Makouei sheep with AB genotype had more chest girth (CG) compared with other genotypes. Furthermore, a significant association was observed between the genotypes of IGFLAS-HinfI with birth weight (BW) in Ghezel and BW, weaning weight (BW3), and CG in Makouei sheep. Haplotype analysis revealed an association between paternal haplotypes and BW in both Ghezel and Makouei breeds. So that, AAB and ABB haplotypes showed more BW than others in Makouei and Ghezel sheep, respectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Given the important role of sheep in providing the main part of meat consumption in the country and a high diversity of breeds in Iran as well as the use of wool of this animal in the carpet industry, studying the genes and economic traits in sheep are particularly interested in animal breeding programs. In Iran, sheep meat is more consistent with Iranian people’s tastes and temperament than beef (Valipour-Koutanaee et al. 2019). Nowadays, sheep products have an eminent role in the agricultural economy worldwide (Thornton 2010). The most common sources of meat in Iran are domesticated animal species such as sheep. The importance of sheep is strengthened by existing of more than 50 million sheep from several breeds in Iran (FAO 2016). Therefore, sheep are one of the most important candidates to provide meat and protein products (FAO 2016).
Makouei sheep is a wool meat breed, and its main habitat is in northwestern Iran. This sheep is suitable for carpeting because of its white wool production. Also due to its proper weight gain, the Makouee breed is a good candidate for meat production. Resistance to nomadic and low nutrition conditions and cold environmental are other characteristics of this breed (Karimzadeh et al. 2016). The Ghezel sheep is also a dairy meat breed, and its habitat is northwestern Iran. The wool colors of these animals are white, and its neck, head, and legs are brown. In this breed, the head, legs, and abdomen are usually wool-free and covered with hair. The rate of growth and fecundity in this breed is desirable in Iran (Karimzadeh et al. 2016).
Identification of major genes affecting growth traits is one of the main purposes of the sheep breeding program (Liu et al. 2014) IGFLAS gene firstly was cloned in a mouse in 1996 (Boisclair et al. 1996). The gene covers spanning about 3.3 Kb of mouse genomic DNA and includes two exons and an 1126-bp intron, while this gene is located on chromosome 16 in human and chromosome 24 in sheep with 3 exons (NC_019481.2 Reference Oar_v4.0 Primary Assembly) that encodes ALS protein.
The ALS protein binds to IGF-I and IGFBP-3 and forms a ternary complex in the circulation and regulates the growth, development, and other physiological/pathophysiological processes. The structural features of ALS are almost conserved among all species, including 12–13 cysteine residues, 6–7 asparagine with glycosylated sites, and 18–20 leucine-rich domains with 24-amino acid length. These leucine-rich domains cover about 75% of the mature protein. ALS has belonged to a super-family with leucine-rich repeats that contribute to protein-protein interaction (Janosi et al. 1999). The ALS is synthesized by liver, and, like IGF-I and IGFBP-3, its production is induced by GH secretion. After birth, the presence of ALS simultaneously is associated with the activity of GH, due to increased secretion of GH and GH receptor. The concentration of ALS remains largely unaffected in adulthood, after an initial increase of ALS puberty. Although ALS has no affinity for binding to IGF-I and IGF-II and also a very low affinity for non-complex IGFBPs, it can easily bind to the binary complexes composed of IGF-I and IGFBP-3 (Twigg et al. 1998). It is notable that the ability of ALS to form a ternary complex under acidic conditions (pH < 4.5) is irreversibly destroyed (Holman and Baxter 1996). The protein, as a growth hormone mediator, plays a critical role in bone and muscle growth, as well as interacting with growth hormone, increases their half-life in circulation (Leong et al. 1992).
Analysis of IGFLAS-mRNA and the level of protein expression in the placenta demonstrated an increase in IGF-I, IGFBP-3, and IGFLAS mRNA expression. Additionally, protein content varied between normal infants and preterm or premature infants (gestational age < 37). These studies suggest that mutations in IGFLAS and, as a result, different levels of expression of the mRNA and its protein may have a functional effect on growth characteristics (Iniguez et al. 2011).
In sheep, a low level of ALS mRNA was detected after birth, but it suddenly increased on day 7 of postnatal age. This expression pattern in sheep was due to forming the primarily 50-kDa complex of IGFs before birth, and circulation of a 150-kDa complex 1 week after birth (Butler and Gluckman 1986). Courtland et al. (2010) showed the association between ALS and body/bone size at different ages and gender. The researchers using normal mice and knockout mice with ALS deficiency at the age of 4–16 weeks found that knockout mice had small size compared with normal groups so that female rats at the lower age (4 weeks) had diminished size in the body than the male rats (Courtland et al. 2010). Tseng et al. (2014) measured the serum levels of ALS, IGF-I, and IGFBP-3 proteins in 98 infants. They showed that there was a positive association between the plasma levels of ALS proteins with birth weight and the average circumference of the newborn’s head. They concluded that, in addition to IGF-I, the plasma ALS was one of the important factors in the IGF-I circulation system to predict anthropometry (a measurement of different parts of the body) during pregnancy, and IGF could promote fetally growth (Tseng et al. 2014). Studying the effect of IGFALS gene polymorphisms on growth traits using the PCR-RFLP method on 300 beef cattle revealed that IGFALS can be used as a genetic marker for the selection of growth characteristics in beef cattle (Liu et al. 2014). Poukoulidou et al. (2014) reported a case with an early-deficient IG-I, and the sequencing result of the IGFLAS gene showed a novel homozygous mutation, which was associated with a delay in growth and puberty. They concluded that the mutation in this gene altered the structure of the protein and preventing its binding to form a ternary complex (Poukoulidou et al. 2014).
Since the important role of ALS protein to determine the effects of endocrine IGF on target tissues have not been well documented in comparison with other members of the circulating IGF systems such as IGFBPs. In the present study, the allelic variation of IGFALS gene in three marker sites of HinfI, MscI, and PvuII in Makouei and Ghezel sheep was investigated using PCR-RFLP method and their associations with some growth and production traits.
Material and methods
Flock management
In the breeding stations of the studied breeds, rams are releasing in the herd every year in late September for three estrus periods. There are 20 to 25 ewes for each ram considering relativeness and kinship to avoid inbreeding depression. Estrus synchronization is done using Sider that contains progesterone. In the second and third estrus periods, the teaser was used to scanning the estrus and find the ewes that are in estrus will mate with selected rams. Over time of pregnancy, the necessary care has been taken of pregnant ewes and birth began in late February and continued until late April. After birth, the lambs have weighted immediately weight loss after drying and the number is listened to, and information such as gender, type of birth, parent’s number, date of birth, and birth weight are registered in computer programs. The lambs stay for a week with their mother in the boxes and are kept in solitary confinement and receive special care. After this period, they are kept in groups with their mother. From the age of 3 weeks, they are separated from their mothers and fed Alfalfa leaves and oatmeal start. During the feeding, the lambs are taken to their mothers twice a day for breastfeeding. This continues for 3 months. After 3 months, they are completely cut off from breast milk.
Sample collection and DNA extraction
In the present study, 240 blood samples from the jugular vein of Ghezel (120 samples, breeding stations of Maku city in West Azerbaijan Province) and Makouei sheep breeds (120 samples, breeding station of Miandoab city west Azerbaijan Province) were randomly collected in tubes containing ethylene diamine tetraacetic acid (EDTA) and transferred to the laboratory and stored at − 20 °C. Genomic DNA was extracted from peripheral blood by modified salting-out method (Miller et al. 1988). To assess the quantity and quality of extracted DNA, both spectrophotometry and agarose gel electrophoresis methods were applied.
Polymerase chain reaction
Firstly, a pair of specific primers (forward 5′-GTGAAAGCAAACAGAGCAG-3′ and reverse 5′-CATTGACCACTGGAGACTG-3) was designed using OLIGO7 software to amplify an 1113-bp fragment, including exon I and part of the intron I of IGFALS gene. PCR was performed in a 20 μl reaction mixture by T100 Thermocycler (Bio-Rad Company). The reaction mixture was comprised of 1.5 μl of DNA template (150 ng), 1 μl of each primer (10 μmol), 10 μl of 2× PCR Master Mix (Yekta Tajhiz Azuma Co. Tehran, Iran), 2.5 μl distilled water, and 4 μl of GC-rich buffer. Then, the PCR thermal cycles were performed as initial denaturation at 95 °C for 5 min and 35 cycles of denaturing at 95 °C for 45 s, annealing at 57 °C for 45 s, extension at 72 °C for 45 s, and a final extension at 72 °C for 7 min.
Genotyping by RFLP technique
To identify the polymorphisms in the IGFAL gene, three different restriction enzymes including MscI, PuvII, and HinfI were used. These enzymes have 2, 3, and 1 restriction sites in amplified sequence, respectively. The digestion reaction was prepared in 15 μl volume, containing 5 μl of PCR product, 1.5 μl of the digestive buffer, 0.3 μl of enzyme, and 8.2 μl of distilled water. The reaction mixture was incubated at 37 °C for 14 h. Then, it was loaded on a 2.5% agarose gel, and after staining with ethidium bromide, DNA bands were detected in the gel documentation device (Bio-Rad Company).
Statistical analysis
Population genetic indices including allelic and genotype frequency, effective allele number, mean heterozygosity, and Shannon index for each marker site were estimated using POPGENE software version 1.32. Then, using the Phase software, the haplotype and their frequencies were determined. Additionally, the comparison of allelic, genotype, and haplotype frequencies between studied populations in both marker sites was performed using chi-squared test. Also, association studies between genotypes and haplotypes with growth traits including birth weight (BW), weaning weight (BW3), weight at 6 months (BW6), weight at 9 months (BW9), and yearling weight (BW12) and body measurements including height of withers (HW), height of rump (HR), body length (BL), chest girth (CG), and thigh girth (TG) were analyzed using the GLM procedure in SAS (9.1) software using the following statistical model:
where Yijk is the value of the observed trait, μ is mean of studied trait, Si is an effect of sex, G(H)j is effect of genotypes (haplotypes), and eijk is a random error. Means of different genotypes and haplotypes were compared using the Tukey test. The five body dimensions measured at 1 year of age as follows: BL was considered as the distance between the point of the shoulder and pinbone, CG was measured behind the shoulder, HW was measured as the distance from the floor to between the shoulders, HR was taken on a flat surface and was the distance between the floor to the back of the animal, and TG was measured as the size of the circumference of right back leg. All measurements were measured by tape. The effects of classificatory factors provided by the breeding center included the year of birth, a season of birth, and sex of the lambs. The number of lambs that were born twins was very small and therefore not included in the analysis. Also, the studied traits were measured in lambs uniformly (at 1 year of their age). Initially, the effects of classificatory or fixed effects (year and season of birth, sex of lambs and herds) on studied traits were statistically analyzed and the only significant effect was related to lamb sex which was included in the final model. The significance level in this study was 0.05.
Results
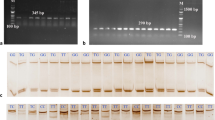
The 1113-bp fragment was amplified by specific primers from 240 extracted DNA samples. Then, the PCR products were digested by PvuII (Fig. 1), MscI, and HinfI (Fig. 2a, b) for genotyping.
The results of PCR-RFLP for PCR product of IGFALS gene on 2.5% agarose gel. a IGFALS-HinfI: BB = 869 bp + 264 bp; AB = 1113 bp + 869 bp + 264 bp; AA = 1113 bp; M, DNA ladder mi-M8200 (MetaBion). b IGFALS-MSCI: BB = 714 bp + 256 bp + 143 bp; AB = 1113 bp + 714 bp + 256 bp + 143 bp; AA = 1113 bp; M, 100 bp DNA Ladder: PR911653 (CinnaGene)
Comparison of allelic and genotypic frequencies among studied populations
The allelic and genotypic frequencies of each marker site were compared between two populations using Fisher exact test and chi-squared statistics. Our results showed a significant difference between genotype frequencies in IGFALS-HinfI marker site between Makouei and Ghezel sheep. However, no significant the difference was found between to breeds in IGFALS-MscI marker site (Table 1).
Haplotype frequency of IGFALS gene in studied marker sites
In the present study, haplotype frequencies of three marker sites including IGFALS-PvuII, IGFALS-HinfI, and IGFALS-MscI were determined in both Makouei and Ghezel sheep using Phase software. The findings have shown that the highest haplotype frequency belonged to the ABB haplotype in both Ghezel and Makouei sheep (Table 2).
Effect of gender on studied traits
The effect of gender on BW9, BW12, and biometric traits such as HR and RT in the Makouei sheep was significant. In this regard, the male had higher weight and body length compared with a female (Table 3).
Association analysis between observed genotypes of IGFALS gene and studied traits
IGFALS-MscI marker site
Association analysis for IGFALS-MscI marker site in Makouei population showed that IGFALS had a significant association with CG and WW (P < 0.05). So that the AB genotype showed the highest TG and BB genotype had the highest WW. The relationship between observed genotypes and other traits was not significant. This marker site in Ghezel population also had a significant relationship with BW as a result, the genotype BB and AA had the highest and the lowest birth weight, respectively (Table 4). It should be noted that in Ghezel sheep, the only available record was body weight. Therefore, other analyzes related to biometric traits in this breed have not been carried out.
IGFALS-HinfI marker site
In statistical analyses for IGFALS-HinfI marker site in Makouei sheep, a significant relationship with CG and BW was observed (P < 0.05). The BB genotype had the highest TG and BW. The relationship between observed genotypes and other traits was not significant. In this way, only two genotypes were observed in the Makouei sheep. This position was also significant in Ghezel population with BW (P < 0.0001), so that the genotype BB and AA showed the highest and the lowest BW, respectively (Table 4).
Statistical association between haplotypes and studied traits
In the statistical analysis of haplotypes, there was a significant association between paternal haplotypes with BW in Ghezel breed. The results showed that the ABB haplotype had the highest BW compared with other haplotypes (Table 5). This haplotype was also significantly associated with BW in Makouei population; as a result, AAB and ABA had the highest and the lowest birth weight, respectively (Table 5). None of the biometric traits had a significant relationship with observed haplotypes.
Discussion
Growth in humans and animals is controlled by a complex process. This process is controlled by the activity of growth factors and regulatory elements. Insulin-like growth factors I and II (IGFs) play a key role in cellular processing, proliferation, differentiation, and prevention of apoptosis (Zapf 1995). ALS is a vital compound that in free mode increases the half-life of IGFs for up to 10 min and in binary and triple complexes of 30–90 min and 12 h, respectively (Zapf 1997). Despite the fact that research on the genes associated with growth traits in sheep has been carried out, in the present study, the IGFALS gene polymorphisms and their effects on growth traits in sheep have evaluated for the first time. It is notable that the effects of this gene on growth and maturation in humans, mice, and cows have been already proven. According to the importance of IGFALS in the formation of a triple complex in mammals, the current study can be a rewarding introduction for the extensive research of this gene and its relation with growth traits in sheep breeding programs.
Our results showed that in the IGFALS-PvuII marker site, no polymorphism was observed in both populations of Makouei and Ghezel sheep. It seems that this marker site has no important role in determining the growth traits in the studied populations, while in the IGFALS-MscI marker site, three genotypes AA, AB, and BB were observed. After statistical analysis, a significant association was found between genotypes and weight gain and CG (P < 0.05) in Makouei sheep and birth weight trait in Ghezel sheep (P < 0.0001). Moreover, AB genotype had the greater CG and BB genotype had the highest BW and WW. Regarding the association of the AB genotype with the largest extent of the CG, it is likely that a mutation has occurred in this gene. Although observed mutation in our study did not change the amino acid sequences of the protein, as previously reported, it could lead to functional changes in the protein through processes such as mRNA stability (Duan et al. 2003) and splicing events (Pagani et al. 2005). Although there has not yet been a report on the significant relationship between this SNP and the wool weight, the remarkable association in the present study showed the role of SNP in controlling the wool weight in sheep beside to body growth. Also, using the HinfI enzyme, AB and BB genotypes were observed in the Makouei population with the genotype frequency 41 and 79% and three genotypes AA, AB, BB with the frequency of 11, 25, and 84% in the Ghezel sheep. There are two reasons why only two genotypes observed in the Makouei sheep, including it may be due to either the small size of the samples or the selection of the allele in the population studied because the frequency of the B allele is much higher and exists in both genotypes.
In the statistical analysis of IGFALS-HinfI marker site, the association between genotypes and BW, BW3, and CG traits in Makouei sheep and the BW trait in Ghezel sheep was significant (P < 0.05). The mutation in this position likely leads to change in the conserved domain of ALS protein, since only the BB genotype had a higher trait value in the abovementioned traits. The structural features of ALS are conserved in almost all species, including 12–13 amino acids of cysteine, six or seven asparagine with glycosylated positions, and 18–20 leucine-rich domains from 24 amino acids. Since these leucine-rich domains form approximately 75% of the mature protein (Janosi et al. 1999), mutations in these regions reduce the level of ALS as well as the formation of the triple complex. According to Butler and Gluckman (1986), the level of ALS-mRNA in the spleen tissue of perinatal lamb was high and increased for 7 days after birth. This expression pattern in sheep was due to the fact that IGFs primarily form 50-kDa complex before birth and then form a 150-kDa ternary complex after 1 week of birth. The mutation in the gene structure and consequently the change in its protein function prevent the formation of a ternary complex, and thus, the effects of growth hormones are not properly induced (Butler and Gluckman 1986).
Fisher’s exact and chi-squared tests showed that allele frequencies between the two breeds were not significantly different in both marker sites, while the difference of genotype the frequency between Makouei and Ghezel sheep was significant for IGFALS-HinfI. Association between gender and BW6, BW9, CG, and HR was also significant. Some studies have shown the importance of this gene as well as its significant association with growth traits in humans and other mammals (Liu et al. 2014; Tseng et al. 2014). Liu et al. (2014) investigated the association of IGFALS gene polymorphisms with growth traits in Chinese beef cattle using the PCR-RFLP method and showed that this position has a notable association with hip width and stature. In the present study, sheep within AB and BB in IGFALS-MscI and IGFALS-HinfI marker sites had a broader chest, respectively, that is consistent fully with Liu et al. (2014) findings. Heath et al. (2008) identified two new recessive mutations in the IGFALS gene. The N276S mutation in two patients led to changes in the conserved amino acid, and another mutation (Q320X) was generated a severely truncated ALS protein. Both mutations caused a severe primary ALS deficiency in the individuals, thereby short-stature patients with very low circulating IGF-I and IGFBP-3 levels. They concluded that the initial deficiency of ALS caused by the mutations in IGFALS would reduce the formation of triple complexes, thereby reducing growth after birth in these patients (Heath et al. 2008). In this study, this gene can be significantly associated with weight and biometric traits due to the presence of SNPs, which altered the structure and function of ALS protein. Tseng et al. (2014) showed that birth weight had a significant association with the levels of ALS, IGF-I, and IGFBP-3 of the umbilical cord plasma. Also, the size of the head of the newborns had a positive association with the level of the ALS cord plasma, while no significant association with IGF-I and IGFBP-3 was found. Their results showed that plasma levels of ALS, IGFBP-3, and IGF-I are highly associated with each other in the pregnancy, and ALS can have a pivotal role in increasing the half-life in the circulation and bioavailability of IGF-I before birth, thereby increasing birth weight. They concluded that ALS and IGF levels in the embryo can be criteria for birth weight and head circumference (Tseng et al. 2014). Based on the statistical results of the present study, two polymorphisms observed in the IGFALS gene were significantly associated with some growth traits. Therefore, in order to confirm the results of this study, it is necessary to study the larger populations along with functional studies to further explore the mechanisms involved in this process. After confirming these mutations, they can be introduced for use in large panels for the selection of superior sheep for the desired traits.
Conclusion
According to our knowledge, no research has been conducted on the IGFALS gene in sheep. Our findings have shown a significant association between different genotypes in polymorphic sites with some weight and biometric traits. However, more research with a larger population is needed to confirm the results of this study and using these marker sites as suitable tools in breeding programs of Iranian sheep.
Abbreviations
- BW:
-
Birth weight
- HW:
-
Height of withers
- CG:
-
Chest girth
- TG:
-
Thigh girth
- BL:
-
Body length
- HR:
-
Height of rump
- WW:
-
Wool weight
References
Boisclair, Y., Seto, D., Hsieh, S., Hurst, K. R. and Ooi, G. T., 1996. Organization and chromosomal localization of the gene encoding the mouse acid labile subunit of the insulin-like growth factor binding complex. Proc. Natl. Acad. Sci., 93, 10028-10033.
Butler, J. H., Gluckman, P. D., 1986. Circulating insulin-like growth factor-binding proteins in fetal, neonatal and adult sheep. J. Endocrinol., 109, 333-338.
Courtland, H. W., DeMambro, V., Maynard, J., Sun, H., Elis, S., Rosen, C. and et al., 2010. Sex specific regulation of body size and bone slenderness by the acid labile subunit. J. Bone Mineral Res., 25, 2059-2068.
Duan, J., Wainwright, M.S., Comeron, J.M., Saitou, N., Sanders, A.R., Gelernter, J. and et al., 2003. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum. mol. genetics, 12, 205-216.
FAO. 2016. FAOSTAT (Food and Agricultural Organization of the United Nations) statistical database. http://www.fao.org/statistics/en/
Heath, K. E., Argente, J., Barrios, V., Pozo, J., Diaz-Gonzalez, F., Martos-Moreno, G. A. and et al., 2008. Primary acid-labile subunit deficiency due to recessive IGFALS mutations results in postnatal growth deficit associated with low circulating insulin growth factor (IGF)-I, IGF binding protein-3 levels, and hyperinsulinemia. J. Clin. Endocr. Metab., 93, 1616-1624.
Holman, S.R., Baxter, R.C. 1996. Insulin-like growth factor binding protein-3: factors affecting binary and ternary complex formation. Growth Regula., 6, 42-47.
Iniguez, G., Argandona, F., Medina, P., Gonzalez, C., San Martin, S., Kakarieka, E. and et al., 2011. Acid-labile subunit (ALS) gene expression and protein content in human placentas: differences according to birth weight. J. Clin. Endocr. Metab., 96, 187-191.
Janosi, J.B.M., Ramsland, P.A., Mott, M.R., Firth, S.M., Baxter, R. C. and Delhanty, P. J. D., 1999. The acid-labile subunit of the serum insulin-like growth factor-binding protein complexes structural determination by molecular modeling and electron microscopy. J. Biol. Chem., 274, 23328-23332
Karimzadeh S., Farhadi A., Tanha T. 2016. Introduction and evaluation of livestock breeds (Iran and World). Parto Vaghe Publication, Tehran.
Leong, S. R., Baxter, R. C., Camerato, T., Dai, J. and Wood, W. I., 1992. Structure and functional expression of the acid-labile subunit of the insulin-like growth factor-binding protein complex. Mol. Endocrinol., 6, 870-876.
Liu, Y., Duan, X., Liu, X., Guo, J., Wang, H., Li, Z. and et al., 2014. Genetic variations in insulin-like growth factor binding protein acid labile subunit gene associated with growth traits in beef cattle (Bos taurus) in China. Gene, 540, 246-250.
Miller, S. A., Dykes, D.D. and Polesky, H., 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res., 16, 1215.
Pagani, F., Raponi, M. and Baralle, F. E., 2005. Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution. Proc. Nat. Acad. Sci., 102, 6368-6372.
Poukoulidou, T., Kowalczyk, J., Metherell, L., De Schepper, J. and Maes, M., 2014. A novel homozygous mutation of the IGFALS gene in a female adolescent: indirect evidence for a contributing role of the circulating IGF-I pool in the pubertal growth spurt. Horm. Res. paediat., 81, 422-427.
Thornton, P. K. 2010. Livestock production: recent trends, future prospects. Philosophical Transactions of the Royal Society B: Biological Sciences, 365(1554), 2853-2867.
Tseng, Y.M., Hwang, Y.S., Lu, C.L., Lin, S.J. and Tsai, W.H., 2014. Association of umbilical cord plasma acid-labile subunit of the insulin-like growth factor ternary complex with anthropometry in term newborns. Pediatr. Neonatol., 55, 139-144.
Twigg, S.M., Kiefer, M.C., Zap, F.J.R. and Baxter, R.C., 1998. Insulin-like growth factor-binding protein 5 complexes with the acid-labile subunit role of the carboxyl terminal domain. J. Biol. Chem., 273, 28791-28798.
Valipour-Koutanaee, O., Farhadi, A., Hafezian, S. H., Gholizadeh, M. 2019. Molecular and bioinformatics analysis of allelic diversity in IGFBP2 gene promoter in indigenous Makuee and Lori-Bakhtiari sheep breeds. Iranian Journal of Applied Animal Science, 9(2), 283-289.
Zapf, J. 1995. Physiological role of the insulin-like growth factor binding proteins. European J Endocrinol., 132(6), 645-654.
Zapf, J. 1997. Total and free IGF serum levels. European J Endocrinol., 136 (2), 146-147.
Acknowledgments
The present study was conducted at the Laboratory for Molecular Genetics and Animal Biotechnology at Sari Agricultural Sciences and Natural Resources University. We thank the University for providing financial and laboratory facilities.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Alizadeh, F., Moradian, F. & Farhadi, A. Association of allelic polymorphisms of IGFALS gene with growth traits in Makouei and Ghezel sheep breeds. Trop Anim Health Prod 52, 3027–3034 (2020). https://doi.org/10.1007/s11250-020-02321-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11250-020-02321-7