Abstract
The present study aimed at investigating the percent prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in equines and associated personnel. A total of 150 swabs of equines and 50 nasal swab samples of associated personnel were collected. These samples were processed in mannitol salt broth for enrichment. A total of 175 nasal swab samples changed the broth color from pink to yellow which were detected as samples containing S. aureus. These samples were processed further on specific media, namely mannitol salt agar, Staph-110, and blood agar, for phenotypic and Gram’s staining–based confirmation of S. aureus isolates. Out of these 175 S. aureus–positive samples, 150 were of equine and 25 were of human origin. Identification of MRSA isolates in 175 S. aureus–positive samples was carried out by antimicrobial susceptibility testing by disc diffusion method. Results showed the presence of MRSA in 87 samples, out of which 81 samples were collected from equines and six samples from humans. Results of antibiotic testing revealed that percentage positivity of MRSA was higher (54%) in equines as compared with the associated personnel (24%). Most resistant antibiotics against MRSA isolates were oxacillin and methicillin while linezolid was found to be the most sensitive antibiotic against MRSA. In conclusion, our findings indicated prevalence of MRSA in equines and associated personnel evidencing an occupational risk of contracting MRSA from horses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The transmission dynamics of infectious diseases due to possible interaction of human, animals, and the environment results in public health disease outbreaks (Buttigieg 2015). One Health concept considers the dynamic interaction between animals, humans, and the pathogens living in the same environment. The S. aureus is a Gram-positive bacterium and some strains of S. aureus have developed resistance against certain antibiotics due to their widespread use and improper dosage regime (Grema et al. 2015). Penicillin worked like a wonder drug until the discovery of methicillin in the 1940s. According to one hypothesis, methicillin-susceptible S. aureus attained the methicillin resistance responsible gene, i.e., mecA, from coagulase-negative Staphylococcus through a procedure known as horizontal gene transfer, developing a strain named as methicillin-resistant S. aureus (MRSA) (Stryjewski and Corey 2014). The mecA gene encodes a protein known as penicillin-binding protein (PBP2a) which confers resistance to different beta-lactam antibiotics by producing alterations in bacterial cell wall biosynthesis (Walther et al. 2017). This gene is present in specific staphylococcal cassette chromosome mec (SCCmec) which conferred 11 basic types so far (Cuny et al. 2016).
The MRSA is considered as a widespread pathogen causing serious diseases in human beings. These infections are of both type, i.e., hospital acquired and community acquired (Parisi et al. 2017). The possible emergence of MRSA strains along with the causative agent’s altering epidemiology has impact on veterinary health care workers and animal husbandry (Bitrus et al. 2018). The presence of PVL gene in MRSA strains acts as a virulence factor and causes skin and soft tissue infections (Bhatta et al. 2016). In equines, the MRSA outbreaks were reported for the first time in equine veterinary settings in 1997 in Japan which caused metritis in infected equines (Bortolami et al. 2017). Risk factors responsible for prevalence of MRSA in equines included previous colonization, the presence of infected horses in the close vicinity, contact with MRSA-positive owners, extensive wounds, immuno-suppression, and prolonged hospitalization (Pomba et al. 2016).
There are several ways by which MRSA can be transmitted between human and animals. They are listed as environmental contamination, direct contact, and handling or eating of contaminated meat (Verkade and Kluytmans 2014). Horses which are colonized by MRSA are pathogenic for those humans working in close contact with them. The MRSA was found in nasal swabs of live cockerels in Pakistan, suggesting a reservoir for human transmission (Zaheer et al. 2017). The most prevalent genotype of MRSA known to cause infection in both horses and humans is CC398 reported in various countries (Walther et al. 2018; Haenni et al. 2017). This specific strain can also be transmitted between equine and their associated personnel (Parisi et al. 2017; Bortolami et al. 2017). Environmental contamination by MRSA is also reported. The persistence of MRSA in environment is mainly due to the presence of some biofilm-related genes in them, i.e., icaD and icaA especially in its CC398 complex (Bortolami et al. 2017). Surface contamination by MRSA can cause occupational and nosocomial infections in veterinary settings (Rojas et al. 2017).
The MRSA is known to be prevalent in horses with a varying percent prevalence from 0.6 to 4.7%. In contrary to farm horses, the horses which were admitted to different veterinary hospitals showed a higher prevalence ranging from 5.8 to 12% (Van Balen et al. 2014). The MRSA carriage in nasal samples in North America exhibited a positive prevalence in the range of 0–4.7%. In the UK and Canada, the MRSA occurrence in equines was found between 0 to 6% and 2.7% respectively (Boyle et al. 2017).
In human, three types of nasal colonization were reported, i.e., persistent carriers which showed prevalence of MRSA from 20 to 25%, intermittent carriers which showed the presence of MRSA up to 60% of total population, and non-carriers in which only 20% were positive for MRSA (Bitrus et al. 2018).
In the last decade, the MRSA has been reported in many animal species which were both healthy carriers and infected ones. These animal species included cats, dogs, camels, horses, and cattle. The MRSA existence in equines has produced a serious risk for both animal and public health (Paterson et al. 2014). In Pakistan, data is lacking about the prevalence of MRSA in equines and their associated personnel; therefore, the present study aimed at describing the MRSA prevalence in horses and associated personnel.
Materials and methods
In this study, convenient sampling was carried out from different areas of Punjab, Pakistan. A total of 150 equine nasal swab samples were collected including Race Club Lahore (n = 100); Brooke’s Hospital, University of Agriculture Faisalabad (UAF) (n = 35); and livestock farm (L/S), UAF (n = 15), with prior ethical approval. For collection of the required samples, sterile normal saline–moistened cotton-tipped swabs were rubbed over the anterior portion of both nares of horses as described earlier (Heller et al. 2009). The history of the horses included in the study was noted on a prescribed questionnaire with information of name, specie, breed, age, previous antibiotic treatment, and any previous disease.
A total of 50 human nasal swab samples were collected including Race Course Club, Lahore (n = 35); Brooke’s Hospital, UAF (n = 10); and livestock farm, UAF (n = 5). Samples were collected from those humans, who were closely associated with the sampled horses. For the collection of the required samples, properly sterile normal saline–moistened cotton-tipped swabs were rubbed over the anterior portion of both nares of human with prior consent.
Immediately after collection of samples, moistened nasal swabs were inoculated on mannitol salt broth and the samples were kept at 37 °C for 24 h. After enrichment of those swab samples in mannitol salt broth, streaking on mannitol salt agar (MSA) plates (Oxoid, UK) was carried out. The S. aureus isolates were detected by color change of the agar from pink to yellow. Further, the phenotypic confirmation of S. aureus was carried on Staph-110 medium and blood agar.
Identification of S. aureus isolates was carried out by Gram’s staining and colony morphology. Biochemical tests performed for detection of S. aureus isolates were catalase, sugar fermentation, and coagulase tests as described by Sarwar et al. 2014. The samples showing positive results for these tests were declared as positive for S. aureus isolates.
S. aureus isolates were tested against selected antibiotics to obtain their antimicrobial susceptibility profiles. The antibiotics used were oxacillin, methicillin, linezolid, amoxicillin-clavulanic acid, vancomycin, ciprofloxacin, enrofloxacin, clindamycin, and rifampicin. Briefly, S. aureus isolates were used to prepare inoculum in normal saline and then the turbidity of this inoculum was compared with 0.5 McFarland turbidity standards. The inoculum was swabbed on the surface of Mueller-Hinton agar plates (Oxoid, UK) and different antibiotic discs were placed over it prior to incubation at 37 °C for 24 h. The S. aureus isolate showing resistance to methicillin and oxacillin was declared as MRSA. The MRSA isolates were then checked against different antibiotics to obtain their antibiogram assays according to CLSI standards.
Zones of inhibition in millimeter produced by each antibiotic were measured and were compared with the CLSI-2017 standards. The isolates were declared as sensitive, intermediate, or resistant against each specific antibiotic. The data obtained was analyzed statistically. The variables were analyzed by statistical software named GraphPad Prism 5. T test and one-way ANOVA were applied to analyze the significance rates. Results were analyzed as significant if P < 0.05 or non-significant if P > 0.05.
Results
Out of 200 nasal swab samples collected from equines (n = 150) and associated personnel (n = 50), 175 (n = 175) samples changed the color of mannitol salt broth from pink to yellow after 24 h of incubation at 37 °C. Out of those positive 175 samples, 150 samples were from equine origin and 25 samples were from human individuals that were positive for S. aureus on mannitol salt agar (MSA) and Staph-110 media as shown in Fig. 1a and b. Further, the S. aureus isolates were positive for beta hemolysis on blood agar.
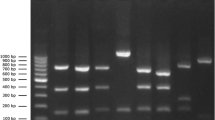
Biochemical characterization of S. aureus isolates was performed by catalase test, coagulase test, and sugar fermentation test. The positive result of these tests indicated the presence of S. aureus isolates in collected nasal swab samples. A total of 175 samples were tested to check resistance against methicillin and oxacillin. In the case of equines, out of 150 tested samples, 81 samples were positive for prevalence of MRSA. In the case of human beings, out of 25 streaked samples, 6 samples were found to be positive for MRSA. Antibiogram of represented S. aureus isolate is shown in Fig. 2. The antibiotic susceptibility of the MRSA isolates against selected antibiotics is shown in Table 1; briefly, the oxacillin, methicillin, and ciprofloxacin were found to be resistant antibiotics against MRSA isolates while linezolid and vancomycin were found to be the most sensitive antibiotics against MRSA. Resistance to methicillin and oxacillin indicated the presence of MRSA strain in collected nasal swab samples from equines and associated human individuals (Table 1).
Out of 87 MRSA positive samples, 81 samples were of equine origin and 6 were of human origin (Fig. 3a and b). The positivity of S. aureus and MRSA isolates and area-wise prevalence in collected samples are presented in Table 2. Briefly, the MRSA was detected in five equine samples at livestock farm, UAF (Fig. 3a), without any positive human sample for MRSA (Table 2). Eighteen equine and two human samples were positive for MRSA at Brook’s Hospital, UAF, while 60 equine and 04 human were positive for MRSA at Race Course Club, Lahore (Table 2, Fig. 3a and b).
Finally, the data was analyzed statistically to find out the significance level. There was a non-significant difference in percent positivity of MRSA in all equine groups. In contrast to equines, the percent positivity data of MRSA was found significant in the case of human swab samples at Brooke’s Hospital and livestock farm, UAF, as shown in Fig. 4. The percent positivity data of MRSA was found non-significant for all other human groups.
Discussion
The menace of antimicrobial-resistant bacteria causes morbidity and mortality and it is associated with health care–acquired infections. The sharing of pathogens at the animal-human-environment interface (One Health concept) described the spread of the diseases including S. aureus–associated diseases shared between animals and humans. The presence of multidrug-resistant, zoonotic pathogens such as methicillin-resistant Staphylococcus aureus (MRSA) in horse clinics poses a biosafety concern to workers and animal patients (Cuny et al. 2016; Van Balen et al. 2014). The S. aureus developed resistance to methicillin (namely MRSA) via mecA and mecC genes encoding penicillin-binding protein. Our study described the occurrence of MRSA in horses and their associated personnel. A total of 200 nasal swab samples were collected from equines (n = 150) and human beings (n = 50) to detect the prevalence of MRSA. The percent prevalence of MRSA in equines and humans was 54% and 24% respectively. The prevalence of MRSA in equine suffering from sinusitis and its associated personnel showed the presence of MRSA Sequence Type 8 through phenotypic and genotypic analyses in Italy (Carfora et al. 2016). In another study in Japan, the MRSA was present in thoroughbred horses and associated veterinarians (Kuroda et al. 2016), suggesting a possible occupational risk of contracting MRSA at the animal-human interface. The equines were found to be an important reservoir of MRSA strains with a percent positivity of 12% in Denmark (Islam et al. 2017). Collectively, our findings in concomitance with other reported data demonstrated plausible risk of sharing of MRSA between horses and associated human. The percent prevalence of MRSA in equines was in the range of 1 to 10% while percent prevalence of MRSA in human beings was 1 to 5%. However, the percent prevalence of MRSA in equines was higher in our study as compared with the previous studies. The possible reasons for this difference could be attributed to environmental temperature and poor hygienic conditions and housing of the animals. The area-wise positivity of MRSA in the present study showed variable percentage which was related with abovementioned housing and environmental factors of study locales.
A horse-specific epidemiology in France and the UK reported a large dissemination of a unique clone (CC398 clone of MRSA); however, the prevalence of MRSA was decreased from 2010 to 2015 in dogs, cats, and horses (Haenni et al. 2017; Bortolami et al. 2017). This study did not report the transmission of MRSA to people associated with the companion animals in contrast to our study. Our study demonstrated the presence of MRSA in associated human at two study locales (Brooke’s Hospital and Race Course Club) without any positivity at L/S farm, UAF. Albeit the fact that MRSA-ST398 harboring different mobile genetic elements are crucial in immune evasion, toxin production, and invasiveness of S. aureus, recent data evidenced spread of equine MRSA-ST398 in different habitats and hosts referring it as “generalist” phenotype (Walther et al. 2018). Apart from horses, there is possible risk of zoonotic transmission of MRSA between mastitic cows and associated workers. The nasal carriage of the cows and workers showed a prevalence of MRSA in 92% of the total collected human samples and 87% of mastitic cow samples in Algeria harboring mecC and mecA genes in MRSA (Akkou et al. 2016). The animal products such as milk and milk products were reported to be sources of MRSA (up to 8%) (Jamali et al. 2015).
The MRSA isolates recovered from horses and human in our study were resistant to methicillin, oxacillin, and ciprofloxacin but sensitive to linezolid. A study in Pakistan from Sindh province found 80 S. aureus isolates from different animals such as sheep, goat, buffalo, camel, horse, cattle, dog, and human and found variable pattern of drug resistance (Habib et al. 2015). Moreover, the food handlers and health care workers have been reported to be a source of drug-resistant S. aureus (Shamebo et al. 2016), indicating animals as reservoirs of MRSA having potential professional and food safety concern (Parisi et al. 2017; Verkade and Kluytmans 2014). The occurrence of MRSA and methicillin-resistant Staphylococcus pseudintermedius (MRSP) among employees and in the environment of a small animal hospital suggested the transfer of these bacteria at the human-animal-hospital environment triad (Feßler et al. 2018).
Appropriate biosafety measures should be adopted to avoid animal to human transmission of MRSA including identification and isolation of infected animals, proper cleaning, and disinfection of contaminated environment (Catry et al. 2010). Based on the results of antimicrobial susceptibility profile of MRSA strains, it is suggested to use linezolid and vancomycin to treat these MRSA-associated infections.
References
Akkou, M., Antri, K., Bachtarzi, M.A., Bes, M., Tristan, A., Dauwalder, O., Kaidi, R., Meugnier, H., Tazir, M., Etienne, J., and Laurent, F., 2016. Phenotypic and genotypic characterization of Staphylococcus aureus strains associated with bovine mastitis and nasal carriage of workers in contact to animals in Algeria. Pakistan Veterinary Journal, 36, 184–188.
Bhatta, D.R., Cavaco, L.M., Nath, G., Kumar, K., Gaur, A., Gokhale, S. and Bhatta, D.R., 2016. Association of Panton Valentine Leukocidin (PVL) genes with methicillin resistant Staphylococcus aureus (MRSA) in Western Nepal: a matter of concern for community infections (a hospital based prospective study). BMC infectious diseases, 16, 199.
Bitrus, A.A., Zunita, Z., Goni, M.D. and Mshelia, I.T., 2018. Dissemination of resistance and virulence determinants in methicillin-resistant Staphylococcus aureus during colonization and disease, A Review. Advances in Animal and Veterinary Sciences, 6, 44–54.
Bortolami, A., Williams, N.J., McGowan, C.M., Kelly, P.G., Archer, D.C., Corrò, M., Pinchbeck, G., Saunders, C.J. and Timofte, D., 2017. Environmental surveillance identifies multiple introductions of MRSA CC398 in an Equine Veterinary Hospital in the UK, 2011-2016. Scientific Reports, 7, 5499.
Boyle, A.G., Rankin, S.C., Duffee, L.A. and Morris, D., 2017. Prevalence of Methicillin-Resistant Staphylococcus aureus from Equine Nasopharyngeal and Guttural Pouch Wash Samples. Journal of Veterinary Internal Medicine, 31,1551–1555.
Buttigieg, M., 2015. A review of the One Health concept: increasing awareness and collaboration between the Maltese medical and veterinary professionals. Malta Medical Journal, 27, 34–37
Carfora, V., Caprioli, A., Grossi, I., Pepe, M., Alba, P., Lorenzetti, S., Amoruso, R., Sorbara, L., Franco, A. and Battisti, A., 2016. A methicillin-resistant Staphylococcus aureus (MRSA) Sequence Type 8, spa type t11469 causing infection and colonizing horses in Italy. FEMS Pathogens and Disease, 74, 1–6.
Catry, B., Van Duijkeren, E., Pomba, M.C., Greko, C., Moreno, M.A., Pyörälä, S., Ružauskas, M., Sanders, P., Threlfall, E.J., Ungemach, F. and Törneke, K., 2010. Reflection paper on MRSA in food-producing and companion animals: epidemiology and control options for human and animal health. Epidemiology and Infection, 138, 626–644.
Cuny, C., Abdelbary, M.M., Köck, R., Layer, F., Scheidemann, W., Werner, G. and Witte, W., 2016. Methicillin-resistant Staphylococcus aureus from infections in horses in Germany are frequent colonizers of veterinarians but rare among MRSA from infections in humans. One Health, 2, 11–17.
Feßler, A.T; Schuenemann, R; Kadlec, K; Hensel, V; Brombach, J; Murugaiyan, J; Oechtering, G; Burgener, I.A; Schwarz, S; 2018. Methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-resistant Staphylococcus pseudintermedius (MRSP) among employees and in the environment of a small animal hospital. Veterinary Microbiology, 221, 153–158.
Grema, H.A., Geidam, Y.A., Gadzama, G.B., Ameh, J.A. and Suleiman, A., 2015. Methicillin resistant Staphyloccus aureus (MRSA): a review. Advances in Animal and Veterinary Sciences, 3, 79–98.
Habib, F., Malhi, K.K., Kamboh, A.A., Rind, R. and Burriro, R., 2015. Antimicrobial susceptibility profile of Staphylococcus aureus isolates recovered from various animal species. Journal of Animal Health and Production, 3, 99–103.
Haenni, M; Châtre, P; Dupieux-Chabert, C; Métayer, V; Bes, M; Madec, J.Y; Laurent, F; 2017. Molecular Epidemiology of Methicillin-Resistant Staphylococcus aureus in Horses, Cats, and Dogs Over a 5-Year Period in France. Frontiers in Microbiology, 13, 8, 2493.
Heller, J., Armstrong, S.K., Girvan, E.K., Reid, S.W.J., Moodley, A. and Mellor, D.J., 2009. Prevalence and distribution of meticillin-resistant Staphylococcus aureus within the environment and staff of a university veterinary clinic. Journal of Small Animal Practice, 50, 168–173.
Islam, M.Z., Espinosa-Gongora, C., Damborg, P., Sieber, R.N., Munk, R., Husted, L., Moodley, A., Skov, R., Larsen, J. and Guardabassi, L., 2017. Horses in Denmark are a reservoir of diverse clones of methicillin-resistant and-susceptible Staphylococcus aureus. Frontiers in Microbiology, 8, 543.
Jamali, H., Paydar, M., Radmehr, B., Ismail, S. and Dadrasnia, A., 2015. Prevalence and antimicrobial resistance of Staphylococcus aureus isolated from raw milk and dairy products. Food Control, 54, 383–388.
Kuroda, T., Kinoshita, Y., Niwa, H., Shinzaki, Y., Tamura, N., Hobo, S. and Kuwano, A., 2016. Meticillin-resistant Staphylococcus aureus colonisation and infection in Thoroughbred racehorses and veterinarians in Japan. Veterinary Record, 178, 473.
Parisi, A., Caruso, M., Normanno, G., Latorre, L., Miccolupo, A., Fraccalvieri, R., Intini, F., Manginelli, T. and Santagada, G., 2017. High Occurrence of Methicillin-Resistant Staphylococcus aureus in Horses at Slaughterhouses Compared with Those for Recreational Activities: A Professional and Food Safety Concern?. Foodborne Pathogens and Disease, 14, 735–741.
Paterson, G.K., Harrison, E.M. and Holmes, M.A., 2014. The emergence of mecC methicillin-resistant Staphylococcus aureus. Trends in Microbiology, 22, 42–47.
Pomba, C., Rantala, M., Greko, C., Baptiste, K.E., Catry, B., Van Duijkeren, E., Mateus, A., Moreno, M.A., Pyörälä, S., Ružauskas, M. and Sanders, P., 2016. Public health risk of antimicrobial resistance transfer from companion animals. Journal of Antimicrobial Chemotherapy, 72, 957–968.
Rojas, I., Barquero-Calvo, E., van Balen, J.C., Rojas, N., Munoz-Vargas, L. and Hoet, A.E., 2017. High Prevalence of Multidrug-Resistant Community-Acquired Methicillin-Resistant Staphylococcus aureus at the Largest Veterinary Teaching Hospital in Costa Rica. Vector-Borne and Zoonotic Diseases, 17, 645–653.
Sarwar, F.Y.M.I., Sherwani, A.H.S.K., Hussain, M.S., Zeb, M. and Sarwar, I., 2014. Identification of Staphylococcus aureus in Pus samples and its Anti-microbial Susceptibility against Imipenem, Tobramycin and Linezolid. International Journal of Basic Medical Sciences and Pharmacy, 4, 9–12.
Shamebo, T., Bacha, K. and Ketema, T., 2016. The growth potential and antimicrobial susceptibility patterns of Salmonella species and Staphylococcus aureus isolated from mobile phones of food handlers and health care workers in Jimma Town, Southwest Ethiopia. African Journal of Microbiology Research, 10, 254–259.
Stryjewski, M.E. and Corey, G.R., 2014. Methicillin-resistant Staphylococcus aureus: an evolving pathogen. Clinical Infectious Diseases, 58,10–19.
Van Balen, J., Mowery, J., Piraino-Sandoval, M., Nava-Hoet, R.C., Kohn, C. and Hoet, A.E., 2014. Molecular epidemiology of environmental MRSA at an equine teaching hospital: introduction, circulation and maintenance. Veterinary Research, 45, 31.
Verkade, E. and Kluytmans, J., 2014. Livestock-associated Staphylococcus aureus CC398: animal reservoirs and human infections. Infection, Genetics and Evolution, 21, 523–530.
Walther, B., Tedin, K. and Lübke-Becker, A., 2017. Multidrug-resistant opportunistic pathogens challenging veterinary infection control. Veterinary Microbiology, 200, 71–78.
Walther, B; Klein, K.S; Barton, A.K; Semmler, T; Huber, C; Merle, R; Tedin, K; Mitrach, F; Lübke-Becker, A; Gehlen, H; 2018. Equine Methicillin-Resistant Sequence Type 398 Staphylococcus aureus (MRSA) Harbor Mobile Genetic Elements Promoting Host Adaptation. Frontiers in Microbiology, 24, 9, 2516.
Zaheer, Z., Hussain, I., Rahman, S.U., Younas, T., Zaheer, I., Abbas, G. and Nasir, M., 2017. Occurrence and antibiotic susceptibility of methicillin-resistant Staphylococcus aureus recovered from oropharynx of live cockerels. Pakistan Veterinary Journal, 37, 108–10.
Acknowledgments
We thank Brooke’s Equine Hospital, Directorate of Farms, University of Agriculture Faisalabad, and Race Course Club, Lahore, for helping in the collection of samples.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical statement
This research work was conducted in compliance with the Institutional Biosafety and Bioethics Committee (IBBC), University of Agriculture Faisalabad. An informed consent was taken from the individuals involved in the study before sampling.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Waqar, N., Amin, Q., Munir, T. et al. A cross-sectional study of methicillin-resistant Staphylococcus aureus at the equine-human interface. Trop Anim Health Prod 51, 1927–1933 (2019). https://doi.org/10.1007/s11250-019-01888-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11250-019-01888-0