Abstract
The potato chloroplast was transformed with codon optimized synthetic hybrid cry gene (SN19) to mitigate crop losses by Colorado potato beetle (CPB). The bombarded explants (leaves and internode) were cultured on MS medium supplemented with BAP (2.0 mg/l), NAA (0.2 mg/l), TDZ (2.0 mg/l) and GA3 (0.1 mg/l); spectinomycin 50 mg/l was used as a selection agent in the medium. Leaf explants of cultivar Kuroda induced highest percentage (92%) of callus where cultivar Santae produced the highest percentage (85.7%) of transplastomic shoots. Sante and Challenger showed 9.6% shoot regeneration efficiency followed by cultivar Simply Red (8.8%). PCR amplification yielded 16 postive transplastomic plantlets out of 21 spectinomycin resistant ones. Target gene integration was confirmed by PCR and Southern blot, whereas RT-qPCR was used to assess the expression level of transgene. The localization of visual marker gene gfp was tracked by laser scanning confocal microscopy which confirmed its expression in chloroplasts of leaf cells. The transplastomic plants ensured high mortality to both larvae and adult CPB. Foliage consumption and weight gain of CPB fed on transplastomic leaves were lower compared to the control plants. Sucessful implementation of current research findings can lead to a viable solution to CPB mediated potato losses globally.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Potato (Solanum tuberosum L.) is a non-cereal staple food crop that is ranked at 3rd position in production (Vincent et al. 2013; FAOSTAT 2022). It has a crucial role to ensure food security (Thiele et al. 2010; Bagri et al. 2018) as well as can mitigate malnutrition and poverty status proficiently (Brown 2005; Çaliskan et al. 2010). Insect pests from different insect orders incur significant losses to the potato crop and economic damages have been estimated as high as 40% (FAOSTAT 2020). Among these insect pests, the Colorado potato beetle (CPB, Leptinotarsa decemlineata) can defoliate the whole plant in the field. CPB is by far the most widely distributed and most destructive insect pest among the coleopteran (EPPO/CABI 1997). As a big potato defoliator, CPB significantly damages the yield of potatoes across the northern hemisphere (Visser 2005; Rondon 2010; Alyokhin et al. 2013).
The heavily defoliated plants with uncontrolled CPB have reduced yields that can exceed to 85% (Maharijaya and Vosman 2015) with an estimated loss of 2–2.5 billion USD in Russia in 2010 (Skryabin 2010). The beetles can develop resistance to newly introduced insecticides within a short time by the dint of its complex, yet unique biological make up. CPB is known to have developed resistance to 56 different chemical compounds belonging to all major classes of insecticides with various modes of action (Raspor and Cingel 2021). The narrow genetic base of crops is a potential barrier for conventional breeding practices and to date no CPB resistance genotype is available (Lindhout et al. 2011). In these circumstances, it is crucial to explore some alternate, sustainable and viable control measures against this devastating insect pest (Zhu et al. 2011).
Transgenic technology may be considered a reliable and effective component of eco-friendly management of potato insect pests (Kos et al. 2009). The insertion of target gene(s) into chloroplast genome has successfully been adopted to impart valuable agronomic traits and production of insect resistant transplastomic plants. Because of several lucrative advantages over nuclear transformation, the plastid transformation has become a potent tool for genetic manipulation of economically important crop plants (Daniell et al. 2021). High ploidy level of plastome facilitates the high level of transgene expression (Cry protein was expressed up to 45.3% of total soluble proteins) (De Cosa et al. 2001). The transplastomic plants also can ensure the multiple gene expression from polycistronic mRNA, avoiding the epigenetic effects and gene silencing. Hence, this technology offers enormous utility over nuclear genome engineering (Bock and Khan 2004; Yu et al. 2020).
Here we report the incorporation of a hybrid cry gene (SN-19, comprised with the domains I and III of cry1Ba and domain II of cry1Ia) into the chloroplast genome. SN-19 can ensure a durable resistance against insect species belonging to the orders coleoptera and lepidoptera as established by earlier researchers (Naimov et al. 2006; Ahmed et al. 2017; Salim et al. 2021). In the current study, the insect resistant transplastomic potato lines expressing SN-19 gene into chloroplast genome exhibited increased resistance enough to ensure 100% mortality of larvae and adult CPB.
Materials and methods
Vector construction, genetic transformation, in-vitro regeneration and primary screening of putative transformants
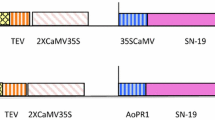
The potato chloroplast transformation vector (PCTV) was developed using potato chloroplast genome sequences from trnA–trnI region to help precise transgene integration into potato chloroplast genome via homologous recombination. SN-19 gene was amplified from already available construct pTF101.1 (Ahmed et al. 2017) in the laboratory and cloned in PCTV. PCTV also harbors FLARE-S as marker gene, which were amplified along with its promoter and terminator sequences from pMSK49 vector (Khan and Maliga 1999). The FLARE-S marker gene confers resistance to spectinomycin as well as streptomycin (Fig. S1). Earlier the shoot cultures of four potato cultivars i.e., Kuroda, Simply Red, Challenger and Sante were established from tuber sprouts and propagated in vitro using single node stem explants on basal MS medium (Murashige and Skoog 1962) to be used as explants for transformation experiments.
For biolistic transformation, explants from potato cultivars were arranged on sterilized whattman no.1 filter paper and placed on regeneration medium. The gold particles (0.6 µm) coated with PCTV DNA were bombarded at 1100 psi He pressure using biolistic gun PDS-1000/He (Bio-Rad Laboratories, Hercules). Methodology required for biolistics transformaton is already described by Khan and Maliga (1999) was followed. Plates containing bombarded explants were placed at 25 ± 2 °C. In-vitro regeneration of bombarded potato tissues with appropriate selection agent, callogenesis, emergence of shoot primordia, their multiplication and primary screening by PCR have been described by Hossain et al. 2021. DNA from transplastomic shoots was extracted folowing Zhang and Stewart (2000) with some modifications. The PCR analysis was conducted using gene-specific primer pairs for the SN-19, aadA and gfp genes (Table 1).
Microscopic detection of transformation events in calli
An inverted microscope Leica dmil led equipped with a CCD camera and gfp detection system was used to track gfp fluorescence in bombarded calli. The system consisted of gfp filters on detector channels ranging between 488 and 568 nm. Channels for green fluorescence images can be adapted. Images were observed from several fields in normal white fields and fluorescent light, and images from gfp fluorescence using led inverted microscopy were carefully recorded. This microscope was used for the analysis of transgenic plastids in well-developed calli tissues. The fundamental idea is to distinguish between transformed and untransformed cells in callus mass. Transplastomic segments in the callus tissues were visually recognized and thus the time and attempts to achieve genetically stable plants were significantly reduced. After having the confirmation of gfp gene integration and expression, gfp positive calli were cultured on the fresh regeneration media with same hormonal combination. Later, embryogenic growth was observed in developed calli after 2–3 weeks of callus formation. The, shoot formation was noticed from those embryogenic cells, and the number of transplastomic candidate shoots were recorded. Once, the spectinomycin resistance transplastomic shoots were regenerated and confirmed through PCR, they were subjected to multiple rounds of selection. Then, plantlets were acclimatized and again subjected to gfp detection using Laser Scanning Confocal Microscope (LSCM, Registration No. 1953) available at the Middle East Technical University, Ankara, Türkiye at 488 nm/514 nm/543 nm excitation, BP: 505–550 nm/LP: 585 nm emission and 20X objective depth.
Further rounds of selection (2nd and 3rd)
Once the plants were confirmed for transgene integration by gfp detetction and PCR, they were subjected to further 2nd and 3rd rounds of selection. For the 2nd round of selection, 2 mm2 pieces of leaves were cut from PCR-positive leaves and placed on selection medium. Those leaf sections were grown in culture room under white, fluorescent lamps (1,900 lx) with 16 h light/8 h dark cycle at 26 °C. After every 15 days, culture was transferred into a new fresh media with the same combination of hormones and selection agent. These leaf sections produced transgenic shoots. Regeneration data from 2nd round of selection was recorded carefully and compared with 1st round of selection. Regenerated shoots were cut again, and these shoots were retransferred to selection medium containing spectinomycin. This step was termed as the third round of selection.
Acclimatization and growth of putative transplastomic plants
After having adequate roots, transgenic plants (approximately 3–6 cm long) were transferred into the growth chamber for acclimatization. Plantlets were shifted to plastic culture vessels containing peat moss and perlite (2:1) and placed into a controlled growth chamber for the acclimatization under 25 ± 2 °C under 16 h light photoperiod in a fully controlled growth cabinet (60% relative humidity, 47 µmol m−2 s−1 irradiance and 58 W fluorescent light). After 10 days, the pots were uncovered for a short time each day to adapt to the environment.
Southern blot analyses
At first, DNA dot blot was performed for the assessment of gene integration in transplastomic plants following protocol as described by Skarnes (2000). Total genomic DNA was isolated from transplastomic plants. The SN-19 probe was prepared using the instructions provided in Biotin Decalabel DNA Labeling Kit (Cat. No. k0652). The membrane was first soaked in pre-hybridization buffer and then hybridized with the generated SN-19 probe at 60 °C overnight. The detection of transgene DNA as a result of hybridization was carried out following the protocol of the commercial detection kit (Fermentas Biotin Chromogenic Detection Kit; Cat# K0661).
Following this analysis, Southern blot analysis of genomic DNA was performed according to the method as described earlier by Southern (1975) to determine the integration of transgene into the chloroplast genome. About 20 μg of transgenic plant DNA was digested with HindIII restriction enzyme and electrophoresed overnight at low voltage. The rest of the procedure was followed as per protocol. SN-19 probe (PCR amplfied) was prepared following the protocol as described in Thermo Scientific Biotin Deca Label DNA labeling kit (Cat. No.: K0651). Following hybridization, the detection procedure was followed as described in Thermo Scientific Biotin Chromogenic Detection kit (Cat No.: K0661).
Quantitative real time PCR
Quantitative Real time PCR was conducted to quantify the relative expression of transgenes in transplastomic plants. Total RNA was isolated from PCR positive plants using RiboZol™ RNA Extraction Reagent following the manufactrer’s instructions. RNA integrity and concentration was measured in 1% agarose gel electrophoresis and nano-drop as well. Furthermore, 1 μg of RNA was used for cDNA synthesis using Fermentas cDNA synthesis kit (Cat. No. 1621). RT-qPCR assay was performed following protocol as described by Hussain et al. (2019). The ingredients of PCR reaction included Syber green master mix (1X), forward Primer (1 μM), reverse Primer (1 μM), RNase-free water and diluted cDNA as template (1:10). The temperature cycle was set up as 95 °C for 15 min, 40 cycles at 95 °C for 10 s, 55 °C for 15 s, 72 °C for 20 s and the melting curve analysis at 70 °C to 99 °C with an increment of 1.0 °C/min. For normalization, elongation factor 1-α (ef1α) was selected as reference gene (Nicot et al. 2005). The threshold values of samples in target gene expression analysis was analyzed by Software of Rotor-Gene Q (QIAGEN) RT-PCR instrument. According to realtime PCR analysis results, the standard deviations of Ct values of the samples were calculated by using Microsoft Excel program and the expression level of the genes were determined according to the 2−ΔΔCt proportional calculation method (Livak and Schmittgen 2001).
Confirmation of transgene expression by SDS-PAGE
SDS-PAGE was performed to confirm the expression of SN-19 protein in transplastomic plants. SDS-PAGE was carried out following the protocol as described by Laemmli (1970a; b) with minör modifications. Protein samples were prepared with protein loading dye with 5 min incubation at 90 °C and stored at − 20 °C before running the gel. After that, 12% separating gel and 4% stacking gel were prepared. The proteins samples were loaded on the gel and run in mini BioRad gel apparatus at 50 V. The gel was stained with Coomassie Blue and destained with a solution containing 10% ethanol and 7.5% acetic acid. The visualization of destained gel was done with Gel DOC™ XR+ Imaging System of Bio-Rad.
CPB culture and rearing
About 100 adult CPB with equal male–female ratio were collected from potato research fields of Nigde Omer Halisdemir University, Turkey during mid June. The entire life cycle pattern was studied under lab conditions. Favourable growth condition for stock CPB culture was ensured as at 25 ± 1 °C under a 16:8 h light–dark photoperiod. The relative humidity was maintained as 60–65% in the growth chamber and all parameters were maintained as described by Gökçe et al. (2006) with few modifications. CPB rearing was carried out at 25 ± 1 °C, 16:8 h light–dark photoperiod and 60–65% relative humidity. Sensitive and resistant potato beetle (CPB) colonies prepared for the use in foliar feeding (bioassay) experiments and its continuity was ensured (Hussain et al. 2019; Naqqash et al. 2020; Salim et al. 2021).
Leaf bio-toxicity assays of primary transformants against CPB
The efficacy of introduced insecticidal genes in transplastomic potato lines to combat CPB was evaluated by subjecting transplastomic potato plants to leaf bio-toxicity assays under laboratory condition. Transplastomic plants from the first round of selection were acclimatized and bioassay was conducted with first and second instar CPB larvae and mortality rate was recorded up to 48 h. After that, another bioassay experiment was done with transplastomic plants from second-round of selection using both larvae (first and second instar) and adult CPB.
To feed CPB on transplastomic potato leaves, the petioles of detached leaves were wrapped and placed into 1.5 ml Eppendorf tube containing water to avoid its withering. Leaf trifoliate from each transplastomic and control plants were placed on cleaned filter paper in 100 × 20 mm diameter petri plates and 10 neonate pre starved CPB larvae were released on each leaf for feeding to check the efficacy of lines. The experiments were conducted in triplicate along with their controls. The efficacy of transplastomic lines against adults CPB was also tested individually using same experimental treatments. Leaves were replaced with fresh ones at 12 h intervals. The toxicity potential of transplastomic potato plants as well as control were evaluated against the larvae and adults CPB and recorded properly as given below.
The leaf areas consumed by various stages of CPB were measured using computer-based software imaging program. Photo of consumed portion of each leaf were taken using a flat background with contrast to the potato leaves with no flash. Leaf region was automatically identified and consumed area was calculated by the BioLeaf software (Machado et al. 2016; Salim et al. 2021). The experiments were repeated thrice and average values were used.
Determination of net weight changes of CPB larvae feeding on transplastomic potato leaves
To check the effect of transplastomic potato leaves harboring insecticidal gene on the growth and development of adult CPB, 1st and 2nd instar larvae were fed on the transplastomic potato leaves as well as control leaves for 72 h. Weights of insect larvae were taken before and after feeding the leaves according to Swift (1997). The increase/decrease in weight of the larvae was recorded. For each treatment, 10 larvae were used. The experiment was carried out in triplicate.
Statistical analysis
In-vitro regeneration data of studied cultivars were recorded and calculated with standard statistical tools. Statistically significant differences between the means of the data were calculated by one-way ANOVA (5% significant level). To measure whether the means are significantly different from each other, Tukey HSD test was applied. Mean and Standard error (mean ± S.E) of data were calculated from 3 replications (n = 3). All statistical analysis was done by Statistix 8.1 software.
Results
In-vitro regeneration, microscopic detection of gfp expression in transformed calli and primary screening of putative transformants
Bombarded explants were cultured on regeneration medium enriched with growth hormones along with selection agent (antibiotics) and callogenesis was recorded after 4–6 weeks. Cultivar Kuroda revealed the highest callus induction proficiency from both internodes (89.0%) and leaves (92.0%) compared to cultivar Simply Red (internodes—87.0%, leaf—82.0%). Lower callus induction proficiency was exhibited by cultivar Challenger (leaf: 74.0% and internodes (71.66%) compared to cultivar Kuroda and Simply Red. Developed calli were manifested by several colors and more prevalence of light brown (LB) color of calli was recorded although Yellowish Green (YG), Light Green (LG), Green (G), Dark Green (DG), and Brown (B) color calli were also observed. From all cultivars, internodal explants induced light brown (LB) color calli, whereas leaf explants induced Greener (G) calli. As callus morphology, Compact (C), Granular (G), and Friable (F) type of calli were induced. Consequently, internodal segments induced more compact and granular type calli whereas; leaf tissues induced friable type of calli. In sum, as a high frequency, Kuroda induced a total 181 calli and cultivar Simply Red and Sante induced 169 and 166 calli, respectively.
Well-developed, mature calli from four potato cultivars were chosen to check the integration and subsequent expression of the repoter green fluorescent protein (gfp) gene. The expression of gfp protein was monitored right from its early developmental stage. After two weeks of incubation on the selection medium, the flourescene of gfp was tracked in calli microscopically. The highest rate of gfp protein accumulation was recorded in Kuroda cultivar. Calli with observed fluorescence were selected, transferred to a new medium, and subcultured. By this means, rapid selection of positive calli was achieved. After that, gfp positive calli were cultured on the fresh regeneration media with same hormonal combination. Three cytokinin’s (TDZ, TR and TZ) showed various responses in cases of shoot regeneration proficiencies and days required. Concerning shoot formation, TDZ produced the highest percentage of shoot formation (41%) requiring 49 days, whereas TR resulted in 40.3% shoot formation in same number of days. The lowest shoot production with TZ's was recorded as 39.6% in 49 days.
Shoot production proficiencies also varied depending on cultivar. Cultivar Kuroda and Sante produced a higher number of shoots, whereas cultivar Kuroda and Challenger respectively yielded the lowest number of shoots. The transformation efficiency was calculated accordingly. The bombarded explants of different cultivars showed varying responses on regeneration media containing spectinomycin. After that, the total number of shoots in cultivar were calculated and recorded. Cultivar Sante and Challenger showed 9.6% shoot regeneration efficiency followed by cultivar Simply Red (8.8%). On the other hand, cultivar Kuroda produced the lowest (6%) shoot regeneration efficiency (Fig. 1). Proliferated shoots were sub-cultured for further rounds of selection to achieve homoplasmic condition on MS media supplemented with 50 mg/l spectinomycin. After successive selection, cultivar Sante produced the highest number of Spectinomycin resistant shoots (43.7%) followed by cultivar Challenger (42.8%). Cultivar Simply Red and Kuroda produced 33.3% and 28.0% Spectinomycin resistant shoots, respectively (Fig. 1).
We screened our putative spectinomycin resistant shoots by PCR with gene specific primers to check the integration of gene of interest (SN-19) and bi-functional visual and selectable marker FLARE-S’ (gfp + aadA). Standard PCR revealed a required band of 480 bp for SN-19 gene and 398 bp for gfp gene. According to PCR findings, highest numbers of PCR positive shoots were recorded from cultivar Santee (85.7%) and cultivar Kuroda produced lowest number of PCR positive shoots (66.6%). Although the cultivar Kuroda produced a smaller number of spectinomycin resistant shoots, its percentage of PCR positive shoots (66.6%) was highest followed by the cultivar challenger (66.0%) (Fig. 1). Finally, we got a total of 16 PCR positive shoots and those shoots were used for further analysis. The regeneration in next round of selection has been presented in Fig. 2.
Successive steps of 2nd round of selection. Leaves from PCR positive transplastomic lines on selection media regenerated from previous selection round (A), Calli after 4 weeks of incubation (B, C), micro shoots from well-developed calli (D, E), microscopic view of micro shoots (F), growth of spectinomycin resistant micro shoots on MS media supplemented with Spectinomycin 50 mg/L−1 (G), rooting of micro shoots under selection (G+H) and transplastomic plants in soil condition (I)
Molecular evaluation of transplastomic plants after acclimatization
Spectinomycin resistant transgenic potato plants grown in in-vitro culture were acclimatized in growth chamber and subjected to various molecular analyses to confirm the integration of the gene of interest (SN-19), the bifunctional marker (gfp + aadA). The required bands of 480 bp for the SN-19 gene, 540 bp for aadA gene, and 398 bp for gfp gene, were confirmed by standard PCR reactions (Fig. S2).
Confirmation of gfp expression in transplastomic plants after acclimatization using confocal laser scanning microscopy
In order to confirm the expression of gfp in acclimatized transplastomic plants, the expression of gfp in transplastomic leaf cells was visualized under Laser Scanning Confocal Microscope. In this study, no gfp expression was obtained from the leaf sample of the control plants, as expected, whereas gfp expression was observed in the leaf cells of transplastomic plants (Fig. 3). The analysis was repeated three times on the same plants and an almost similar pattern of of gfp fluorescence was observed each time.
Microscopic visualization of gfp expression in transplastomic shoots. Microscopic visualization of gfp expression of transplastomic and control plants under laser scanning confocal microscope. A, C gfp image under FITC (Fluorescein isothiocyanate) detector at 488–514 nm wavelength, B, D and leaf samples under white light. E Expression of gfp in leaf section of Sante 6 transplastomic plant was demonstrated under fluorescent light microscopy. White arrows indicate the gfp localization within the cell. The left figure is the gfp expression under UV light, the right figure is the leaf image under white light
Southern DNA dot blot and southern blot analysis
Integration of SN-19 gene into the plant genome was confirmed by DNA dot blot and southern blot analysis. The spots on the membrane of DNA samples from transgenic plants showed the expected hybridization signal when subjected to hybridization with the PCR-amplified biotin-labeled SN-19 probe (Fig. S3). The intensity of signal was higher in K3, Ch3, Sa6 and Sa7 plants followed by SR4 and SR5. No signal was detected in the non-transgenic plant.
Southern blot analysis was performed with genomic DNA extracted from PCR positive transplastomic lines (from Kurado, Simply Red, Sante cultivars, two lines of each) to confirm the integration of SN-19 gene into the chloroplast genome. The results showed specific bands of SN-19 gene as expected of approximately 1.981 kb (Fig. 4).
Southern blot analysis of transplastomic plants. The digested DNA was transferred on nylon membrane and hybridized with gene specific probe. Lane 1: Lambda HindIII marker (marker not visible on nylon membrane), Lane 2: PCTV plasmid used as positive control, Lane 3–4: Kuroda, Lane 5–6: Sante, Lane 7: Challenger, Lane 8: Simply Red
Determination of gene expression levels by qRT-PCR analysis
The expression levels of SN-19 and gfp genes in acclimatized plants were determined following total RNA isolation and cDNA synthesis. The results revealed high expression of the SN-19 gene in transplastomic plants. While the expression level of the gene increased approximately 2.5 times in Sante (Sa6), this increase was found between 1.5 and 2 times in Simply Red (SR1 and SR5) (Fig. 5). In addition, high levels of gfp gene were also detected in the same plants. Accordingly, the expression level of gfp was approximately 4 times higher in Sa6 and Sa7 plants compared to SR5, followed by K3 and SR4 (Fig. 5). It is pertinent to note that the transplastomic plant with the least expression was considered as 1 for calculating fold expression since the gfp as well as SN-19 are heterologously expressed in potato.
qRT-PCR analysis of SN-19 (left blue bar) and gfp (right red bar) genes in soil-grown transplastomic plants. Coefficients of variation of SN-19 and gfp mRNA compared to control plants. For SN-19, an increase in expression level was observed in other plants compared to K1 (control). For gfp, an increase in gfp expression level was observed in other plants compared to SR5 (control). K: Kuroda, Sa: Sante, SR. Simply Red. Values represents mean (± SE) of 3 replicates and the experiment was repeated three times. Upper end of each bar diagram contain cap that represents standard error (± SE)
Determination of SN-19 protein presence in transplastomic plants by SDS-PAGE
As we couldn’t find a commercial ELISA kit specific to SN-19 protein or antibodies, SDS-PAGE was conducted to determine the presence of this protein in transplastomic plants. The expected 73 kDa band for SN-19 protein was detected in the transplastomic plants (Fig. 6).
Determination of the presence of SN-19 protein (73 kDa) in transplastomic plants by SDS-PAGE. M: Molecular protein marker (Blue Plus® V Protein Marker, DM141-01, 10–190 kDa), +Ve: Total protein isolated from E. coli with plasmid pET22b. K1, K3, Sa6, Sa7, Ch3, Ch4,SR4 and SR4 are transplastomic lines harboring the target gene SN-19
Toxicity potential of transplastomic potato plants
First and second instar larvae and adult CPB were allowed to feed on PCR positive transplastomic plant leaves to determine their insecticidal potential. The results did not show any mortality of larvae fed on non-transgenic leaves, whereas larvae fed on transplastomic plants showed remarkable mortality during the incubation period (as depicted in Fig. 7A, B, C). About 100% mortality was recorded in 1st instar and 2nd instar CPB larvae. After 48 h of incubation, K3, Sa6 and Sa7 showed maximum mortality (100%) followed by cultivar Simply Red and Challenger plants. No mortality was recorded during the first 24 h of incubation in 1st instar CPB larvae fed on these plants (Fig. 8A). Similarly, mortality rates up to 65% were recorded from 2nd instar larvae feed on Sa6 and Sa7 following 24 h of incubation (Fig. 8B).
Mortality rates and net weight changes of CPB larvae fed on transplastomic plants. Mortality rates were calculated as a result of leaf biotoxicity tests against first (A) and second (B) instar CPB larvae. No mortality or morbidity was observed in the control plants. Net weight changes of first instar (C) 2nd (C) instar larvae CPB larvae feeding on transplastomic and control potato leaves. Mortality and weight change values consist of mean from 3 replications. Ch: Challenger, K: Kuroda, Sa: Sante, SR. Simply Red
While feeding on transplastomic and control leaves, the larvae gained weight. Therefore, the CPB larvae were weighed after leaf bioassays to determine the sub-lethal effects of insecticidal target gene in insect feeding on transplastomic leaves. There was prominent difference in the increase of weight in different larval instars when fed on transplastomic plants compared to the control plants. In cases of first instar CPB larvae, a significant increase of up to 6.13 mg in weight was observed (Fig. 8C). On the other hand, the insects feeding on SR4 and Ch3 showed 3.30 mg and 2.53 mg of higher body weight, respectively. Transplastomic plants SR5, Ch3, and Sa6 did not show any differences among their weight gain. Besides these, transplastomic plants K1, K3, Ch4 and Sa7 showed the same level of weight gain that was ranges between 1.70 mg to 2.10 mg.
Similarly, 2nd instar larvae feeding on transplastomic plants SR4 showed 3.60 mg of increase in weight, whereas insects feeding on control plants gained 6.87 mg more weight (Fig. 8D). Transplastomic plants Ch3, Sa6 and Sa7 showed the same level of weight gain without having any significant differences and ranges between 3.33 gm to 3.56 gm. These results suggest that the insects feeding on control plants gained more weight while the ones feeding on transplastomic plants gained less weight.
Foliage consumption of CPB on transplastomic potato plants
The foliage parts of transplastomic and control plants were allowed to feed by the 1st and 2nd instar larvae and adult CPB for a specific period (24 h for 1st instar and 2nd instar larvae respectively, 72 h for adult). The consumed parts/area of transplastomic leaves were measured by mentioned software and compared with respective control leaves.
The data showed differences in the feeding behavior of 1st instar CPB larvae on transplastomic potato plants when compared to non-transgenic control. The more leaf consumption was recorded in control plant (0.92 cm2) whereas transplastomic plant Sa7 ensured lowest (0.78 cm2) consumption area of leaves. Among the transplastomic plants, SR4 and SR5 showed the highest consumption rate (0.88 and 0.87 cm2) less than the control. In terms of foliage consumption, after 24 h of feeding, transplastomic plants did not show any prominent differences among them (Fig. 9A).
Foliage consumption by individual larvae (A 1st instar, B 2nd instar) and adult (C) CPB fed on transplastomic and control potato lines. Consumption area was measured up to 72 h after treatment and values consist of means from 3 replications. The experiment was repeated three times. The upper end of each bar diagram contains cap that represents standard error (± SE). K1: Kuroda1, K3: Kuroda 3, SR4: Simply Red 4, SR5: Simply Red 5, Ch3: Challenger 3, Ch4: Challenger 4, Sa6: Sante 6, and Sa7: Sante 7
Foliage consumption rate (area) by 2nd instar CPB larvae feeding on transplastomic and control potato plants for 24 h showed interesting results. Compared with transplastomic plants, control plants showed a higher consumption rate (1.8 cm2). Transplastomic plants K1 (1.56 cm2) and Sa6 (1.56 cm2) exhibited a higher consumption rate among the transplastomic plants. Transplastomic plant K3 (1.46 cm2), SR4 (1.46 cm2), SR5 (1.33 cm2) and Ch3 (1.46cm2) showed an equal level of consumption rate without having any prominent differences. Transplastomic plants Sa7 revealed the lowest foliage consumption rate (Fig. 9B).
Foliage consumption by individual adult CPB fed on transplastomic and control potato have been presented in Fig. 9C. Although among the transplastomic plant SR4 showed a higher foliage consumption rate (8.8 cm2), compared with rest of transplastomic plants, all transplastomic plants showed the same level of consumption rate (Fig. 9C).
Discussion
Genetic transformations of crop plants with Bt genes to develop durable, ecofriendly and financially feasible insect resistance transgenic plants have remained prominent since the last few decades. In recent past, an extensive cultivation of 3rd generation of Bt cotton harboring cry1Ac + cry2Ab + Vip3Aa genes have been performed in Australia (Chen et al. 2017; Tabashnik and Carrière 2017). With the passage of time, insect pests, especially CPB, have developed resistance against Bt crops and the longevity of Bt crops is a burning question. In this study, we have attempted to incorporate hybrid SN-19 gene into the potato plastome aimed to develop insect resistant transplastomic potato lines. PCR based screening of putative shoots revealed that cultivar Sante produced the highest number of PCR positive shoots (85.7%). Expression of visual marker gene gfp in calli and shoots were visualized by inverted microscopy and the observed expression level of potato cultivars did not show any statistically significant differences and these differences were probably because of heteroplastomic status of plastome. In comparison, Sidorov et al. (1999) documented transplastomic event in potato with a single step in-vitro culture on which they reported 6.67% (1 transformant out of 15 shoots) and 2.85% (1from 35 shots) regeneration performance. These findings are much lesser than the regeneration proficiencies of four potato cultivars used in our study. On the other hand, Nguyen et al. (2005) adopted a two-step process tissue culture procedure to produce transplastomic potato on which they recorded 5.5% (1 out of 18 shoots) and 4.0% (1 out of 25 shoots) regeneration performance. However, Nguyen et al. (2005) reported PCR based evidence of integration of aadA and gfp in transplastomic potato. Adopting the same genotype and underlying regeneration protocol of Nguyen et al. (2005), Valkov et al. (2011) roughly doubled (11 shoot per 100 bombardments) the efficiency of transformation compared to previous results documented by Sidorov et al. (1999) and Nguyen et al. (2005). This finding obtained by Valkov et al. (2011) is similar to our current findings regarding the cultivars Challenger and Sante. Khan et al. (2015) developed 9 salinity and drought tolerant transplastomic tobacco from 35 bombardments but decreased transformation efficiency was recorded for the petunia plastome (DeGray et al. 2001). The presence of homologous flanking sequences in a transformation event can instigate the improved frequency of transformation (Scotti et al. 2011) that was proven by Valkov et al. (2014) on potatoes (one transplastomic shoot per bombardment). Improved transplastomic events were recorded by several authors in tobacco (Svab and Maliga 1993; Langbecker et al. 2004) and both of these findings were similar to our current procedure and achievement, although lower efficacy of transformation in Arabidopsis, potato (Solanum tuberosum) and tomato (Lycopersicon esculentum) also reported (Sidorov et al. 1999; Ruf et al. 2001).
Integration and expression of target genes into the potato chloroplast genome were confirmed by PCR, southern blotting, qRT-PCR, and SDS-PAGE. Localization of our visual marker gfp gene was also confirmed by Laser scanning confocal microscopy (LSCM). Our findings are in line with several researchers including Sidorov et al. (1999), Ruf et al. (2001), Langbecker et al. (2004), Nguyen et al. (2005), Valkov et al. (2011, 2014) and Yu et al. (2017). Southern blot analysis of PCR positive transplastomic plants was conducted to ensure integration of transgene in potato plastome. The analysis results are compatible with the integration of the plasmid into DNA as a single copy. Southern blot was widely utilized to detect transgene integration in plant genomes (Wu et al. 2019; He et al. 2020; Xu et al. 2020; Salim et al. 2021). qRT-PCR analysis of transplastomic plants was performed to diagnose expression levels of SN-19 and gfp genes. PCR positive plants were subjected to the qRT-PCR analysis and as result, high expression of SN-19 and gfp genes were noticed in transplastomic plants. The expression level of gene was different in various transplastomic plants. Our results agree with previous studies of Li et al. (2017) and Occhialini et al. (2020).
After successful confirmation of target gene (s) integration into the chloroplast genome, leaf bioassays with transplastomic and control plants were conducted. The transplastomic plants exhibited promising resistance against both CPB larvae and adults compared to non-transgenic control plants. After 12 h of feeding, the toxic effect of cry protein on CPB was manifested by avoiding feeding, slow motion of the insect pests, discoloring insect body appearance and growth retardation. Under continuous feeding, insect feeding behavior and physiological status were recorded from close observation within 12 h’ intervals. No mortality was recorded from insects feeding on non-transplastomic control plants. Time required for 100% mortality for 1st instar, 2nd instar and adult CPB was a range between 5.33 and 24.67 days. Our findings can be compared with the findings of Salim et al. (2021), Ahmad et al. (2017). These studies confirmed 100% lethal effects on 1st instar to adult CPB on resistant potato plants with nuclear transformation whereas in our current study target gene was integrated into the plastome. Genetic makeup and promoter types and confined expression of target genes within the green part of potato facilitated more lethal effect within less time. The insect weight gain feeding on control and transplastomic plants was measured and compared to understand the lethal effects of transplastomic plants on CPB larvae and adults. Larvae and adult CPB feeding on control plants gained more weight compared with transplastomic plants. More consumption and less suppression of insect was recorded from control foliage part while insect pests were suppressed more by transplastomic plants and consumption was less. Our findings were compared with the findings of Cingel et al. (2015) and Salim et al. (2021).
Conclusion
The current research work was conducted to develop insect-resistant transplastomic potato plants to combat the notorious insect pest CPB. Several studies have been conducted with single cry gene integration into the nuclear genomes and even into the chloroplast genome. With the purpose to express cry protein in plant chloroplast and keeping in view, the extraordinary and tremendous adaptive malleability of CPB against insecticides, the current study was designed to incorporate synthetic hybrid gene (SN-19) in potato plastome to encode durable resistance against CPB. Earlier, we introduced an efficient in-vitro regeneration protocol for transplastomic potato, and we got our 16 PCR positive transplastomic shoots. These putative transplastomic plants were screened with molecular tools (PCR, southern blot, qRT-PCR, and SDS-PAGE), and stable integrations of target gene were confirmed, and leaf bioassay was conducted with 1st and 2nd instar larvae and adult CPB. Transplastomic plants showed high toxicity to the CPB population. The weight changes and foliage consumption during feeding on transplastomic and control plants showed that transplastomic plants exhibited resistance against CPB. The successful implementation of this transplastomic technology in potato will help mitigate the CPB mediated potato loss proficiently.
Data availability
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
References
Ahmed HA, Onarıcı S, Bakhsh A, Akdoğan G, Karakoç ÖC, Özcan SF, Aydın G, Aasım M, Ünlü L, Sancak C, Naimov S (2017) Targeted expression of insecticidal hybrid SN19 gene in potato leads to enhanced resistance against Colorado potato beetle (Leptinotarsa decemlineata Say) and tomato leafminer (Tuta absoluta Meyrick). Plant Biotechnol Rep 11:315–329. https://doi.org/10.1007/s11816-017-0453-8
Alyokhin A, Vincent C, Giordanengo P (2013) Insect pests of potato: global perspectives on biology and management. Academic Press, Oxford, p 613
Bagri DS, Upadhyay DC, Jain SK (2018) Upadhyay CP (2018) Biotechnological improvement of nutritional and therapeutic value of cultivated potato. Front Biosci 10:217–228. https://doi.org/10.2741/S510
Bock R, Khan MS (2004) Taming plastids for a green future. Trends Biotechnol 22(6):311–318
Brown CR (2005) Antioxidants in potato. Am J Potato Res 82:163–172. https://doi.org/10.1016/j.tibtech.2004.03.005
Çaliskan ME, Onaran H, Arıoğlu H (2010) Overview of the Turkish potato sector: challenges, achievements, and expectations. Potato Res 53:255–266. https://doi.org/10.1007/s11540-010-9170-1
Chen WB, Lu GQ, Cheng HM, Liu CX, Xiao YT, Xu C, She ZC, Wu KM (2017) Transgenic cotton coexpressing Vip3A and Cry1Ac has a broad insecticidal spectrum against lepidopteran pests. Int J Agric Res 149:59–65. https://doi.org/10.1016/j.jip.2017.08.001
Cingel A, Savić J, Vinterhalter B, Vinterhalter D, Kostić M, Jovanović DŠ, Smigocki A, Ninković S (2015) Growth and development of Colorado potato beetle larvae, Leptinotarsa decemlineata, on potato plants expressing the oryzacystatin II proteinase inhibitor. Transgenic Res 24:729–740. https://doi.org/10.1007/s11248-015-9873-9
Daniell H, Jin S, Zhu X-G, Gitzendanner MA, Soltis DE, Soltis PS (2021) Green giant—a tiny chloroplast genome with mighty power to produce high-value proteins: history and phylogeny. Plant Biotechnol J. https://doi.org/10.1111/pbi.13556
De Cosa B, Moar W, Lee SB, Miller M, Daniell H (2001) Overexpression of the Bt cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat Biotechnol 19(1):71–74. https://doi.org/10.1038/83559
DeGray G, Rajasekaran K, Smith F, Sanford J, Daniell H (2001) Expression of an antimicrobial peptide via the chloroplast genome to control phytopathogenic bacteria and fungi. Plant Physiol 127:852–862. https://doi.org/10.1104/pp.010233
EPPO/CABI (1997) Leptinotarsa decemlineata. In: Smith IM, Scott PR and Holderness. https://www.eppo.int/RESOURCES/eppo_publications
FAOSTAT (2020) Statistical data. Food and Agriculture Organization Statistical Database, United Nations, Rome
FAOSTAT (2022) https://www.fao.org/newsroom/detail/doubling-global-potato-production-in-10-years-is-possible/en. Dublin
Gökçe A, Isaacs R, Whalon ME (2006) Behavioral response of Colorado potato beetle (Leptinotarsa decemlineata) larvae to selected plant extracts. Pest Manag Sci 62:1052–1057. https://doi.org/10.1002/ps.1271
He W, Xu W, Xu L, Fu K, Guo W, Bock R, Zhang J (2020) Length-dependent accumulation of double-stranded RNAs in plastids affects RNA interference efficiency in the Colorado potato beetle. J Exp Bot 71:2670–2677. https://doi.org/10.1093/jxb/eraa001
Hossain MJ, Aksoy E, Gökçe ZNÖ, Joyia FA, Khan MS, Bakhsh A (2021) Rapid and efficient in vitro regeneration of transplastomic potato (Solanum tuberosum L.) plants after particle bombardment. Turk J Agric for 45:313–323. https://doi.org/10.3906/tar-2010-103
Hussain T, Aksoy E, Çalışkan ME, Bakhsh A (2019) Transgenic potato lines expressing hairpin RNAi construct of molting associated EcR gene exhibit enhanced resistance against Colorado potato beetle (Leptinotarsa decemlineata Say). Transgenic Res 28:151–164. https://doi.org/10.1007/s11248-018-0109-7
Khan MS, Maliga P (1999) Fluorescent antibiotic resistance marker for tracking plastid transformation in higher plants. Nat Biotechnol 17:910–915. https://doi.org/10.1038/12907
Khan MS, Kanwal B, Nazir S (2015) Metabolic engineering of the chloroplast genome reveals that the yeast ArDH gene confers enhanced tolerance to salinity and drought in plants. Front Plant Sci 6:725. https://doi.org/10.3389/fpls.2015.00725
Kos M, van Loon JJ, Dicke M, Vet LE (2009) Transgenic plants as vital components of integrated pest management. Trends Biotechnol 27:621–627. https://doi.org/10.1016/j.tibtech.2009.08.002
Laemmli UK (1970a) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. https://doi.org/10.1038/227680a0
Laemmli UK (1970b) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 15:680–685. https://doi.org/10.1038/227680a0
Langbecker CL, Ye GN, Broyles DL, Duggan LL, Xu CW, Hajdukiewicz PT, Armstrong CL, Staub JM (2004) High-frequency transformation of undeveloped plastids in tobacco suspension cells. Plant Physiol 135:39–46. https://doi.org/10.1104/pp.103.035410
Li T, Wang J, Lu M, Zhang T, Qu X, Wang Z (2017) Selection and validation of appropriate reference genes for qRT-PCR analysis in isatis indigotica fort. Front Plant Sci 8:1139. https://doi.org/10.3389/fpls.2017.01139
Lindhout P, Meijer D, Schotte T, Hutten RC, Visser RG, van Eck HJ (2011) Towards F1 hybrid seed potato breeding. Potato Res 54:301–312. https://doi.org/10.1007/s11540-011-9196-z
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Machado BB, Orue JP, Arruda MS, Santos CV, Sarath DS, Goncalves WN, Silva GG, Pistori H, Roel AR, Rodrigues-Jr JF (2016) BioLeaf A professional mobile application to measure foliar damage caused by insect herbivory. Comput Electron Agric 129:44–55. https://doi.org/10.1016/j.compag.2016.09.007
Maharijaya A, Vosman B (2015) Managing the Colorado potato beetle; the need for resistance breeding. Euphytica 204:487–501. https://doi.org/10.1007/s10681-015-1467-3
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497. https://doi.org/10.1111/j.1399-3054.1962.tb08052.x
Naimov S, Zahmanova G, Boncehva R, Kostova M, Minkov I, Dukiandjiev S, De Maagd R (2006) Expression of synthetic SN 19 hybrid delta-endotoxin encoding gene in transgenic potato. Biotechnol Biotechnol Equip 20:38–41. https://doi.org/10.1080/13102818.2006.10817378
Naqqash MN, Gökçe A, Aksoy E, Bakhsh A (2020) Downregulation of imidacloprid resistant genes alters the biological parameters in Colorado potato beetle, Leptinotarsa decemlineata Say (Chrysomelidae: Coleoptera). Chemosphere 240:124857. https://doi.org/10.1016/j.chemosphere.2019.124857
Nguyen TT, Cardi Nugent GT, Dix PJ (2005) Generation of homoplasmic plastid transformants of a commercial cultivar of potato (Solanum tuberosum L.). Plant Sci 168:1495–1500. https://doi.org/10.1016/j.plantsci.2005.01.023
Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56:2907–2914. https://doi.org/10.1093/jxb/eri285
Occhialini A, Pfotenhauer AC, Frazier TP, Li L, Harbison SA, Lail AJ, Mebane Z, Piatek AA, Rigoulot SB, Daniell H, Stewart CN (2020) Generation, analysis, and transformation of macro-chloroplast Potato (Solanum tuberosum) lines for chloroplast biotechnology. Sci Rep 10:1–13. https://doi.org/10.1038/s41598-020-78237-x
Raspor M, Cingel A (2021) Genetically modified potato for pest resistance: thrift or threat? Solanum tuberosum—a promising crop for starvation problem. https://doi.org/10.5772/intechopen.98748
Rondon SI (2010) The potato tuberworm: a literature review of its biology, ecology, and control. Am J Potato Res 87:149–166. https://doi.org/10.1007/s12230-009-9123-x
Ruf S, Hermann M, Berger IJ, Carrer H, Bock R (2001) Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nat Biotechnol 19:870–875. https://doi.org/10.1038/nbt0901-870
Salim M, Bakhsh A, Gökçe, (2021) A Stacked insecticidal gene in potatoes exhibit enhanced toxicity against Colorado potato beetle, Leptinotarsa decemlineata (Coleoptera: Chrysomelidae). Plant Biotechnol Rep 15:197–215. https://doi.org/10.1007/s11816-021-00668-3
Scotti N, Valkov VT, Cardi T (2011) Improvement of plastid transformation efficiency in potato by using vectors with homologous flanking sequences. GM Crops 2:89–91. https://doi.org/10.4161/gmcr.2.2.17504
Sidorov VA, Kasten D, Pang SZ, Hajdukiewicz PT, Staub JM, Nehra NS (1999) Stable chloroplast transformation in potato: use of green fluorescent protein as a plastid marker. Plant J 19:209–216. https://doi.org/10.1046/j.1365-313x.1999.00508.x
Sikdar SR, Serino G, Chaudhuri S, Maliga P (1998) Plastid transformation in Arabidopsis thaliana. Plant Cell Rep 18:20–24. https://doi.org/10.1007/s002990050525
Skarnes WC (2000) Gene trapping methods for the identification and functional analysis of cell surface proteins in mice. Methods Enzymol 328:592–615. https://doi.org/10.1016/s0076-6879(00)28420-6
Skryabin K (2010) Do Russia and Eastern Europe need GM plants? New Biotechnol 27:593–595. https://doi.org/10.1016/j.nbt.2010.07.019
Southern EM (1975) Detection of specific sequence among DNA fragments separated by gel electrophoresis. J Mol Biol 98:503–517
Svab Z, Maliga PAL (1993) High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. PNAS 90:913–917. https://doi.org/10.1073/pnas.90.3.913
Swift ML (1997) GraphPad prism, data analysis, and scientific graphing. J Chem Inf Model 37:411–412. https://doi.org/10.1021/ci960402j
Tabashnik BE, Carrière Y (2017) Surge in insect resistance to transgenic crops and prospects for sustainability. Nat Biotechnol 35:926–935. https://doi.org/10.1038/nbt.3974
Thiele G, Theisen K, Bonierbale M, Walker T (2010) Targeting the poor and hungry with potato science. Potato J 37:75–86
Valkov VT, Gargano D, Manna C, Formisano G, Dix PJ, Gray JC, Scotti N, Cardi T (2011) High efficiency plastid transformation in potato and regulation of transgene expression in leaves and tubers by alternative 5′ and 3′ regulatory sequences. Transgenic Res 20:137–151. https://doi.org/10.1007/s11248-010-9402-9
Valkov VT, Gargano D, Scotti N, Cardi T (2014) Plastid transformation in potato: Solanum tuberosum. Methods Mol Biol 1132:295–303. https://doi.org/10.1007/978-1-62703-995-6_18
Vincent C, Alyokhin A, Giordanengo P (2013) Potatoes and their pests—setting the stage. Insect pests of potato 3–8. https://doi.org/10.1016/B978-0-12-821237-0.00005-6
Visser D (2005) Guide to potato pests and their natural enemies in South Africa. ARC-Roodeplaat Vegetable and Ornamental Plant Institute, Pretoria
Wu Y, Xu L, Chang L, Ma M, You L, Jiang C, Li S, Zhang J (2019) Bacillus thuringiensis cry1C expression from the plastid genome of poplar leads to high mortality of leaf-eating caterpillars. Tree Physiol 39:1525–1532. https://doi.org/10.1093/treephys/tpz073
Xu S, Zhang Y, Li S, Chang L, Wu Y, Zhang J (2020) Plastid-expressed Bacillus thuringiensis (Bt) cry3Bb confers high mortality to a leaf eating beetle in poplar. Plant Cell Rep 39:317–323. https://doi.org/10.1007/s00299-019-02492-0
Yu Q, Lutz KA, Maliga P (2017) Efficient plastid transformation in Arabidopsis. Plant Physiol 175:186–193. https://doi.org/10.1104/pp.17.00857
Yu Y, Yu PC, Chang WJ, Yu K (2020) Plastid Transformation: how does it work? Can it be applied to crops? What can it offer? Int J Mol Sci 21(14):4854. https://doi.org/10.3390/ijms21144854
Zhang J, Stewart JM (2000) Economical and rapid method for extracting cotton genomic DNA. J Cotton Sci 4:193–201
Zhu F, Xu J, Palli R, Ferguson J, Palli SR (2011) Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Manag Sci 67:175–182. https://doi.org/10.1002/ps.2048
Acknowledgements
This research is part of the PhD thesis of Md Jakir Hossain, who completed research work out of project funded by the Scientific and Technological Research Council of Turkey (Tübitak, Project 216O027), Turkey and Pakistan Science Foundation (PSF). We highly acknowledge Tübitak-PSF for financial support to conduct our current research.
Author information
Authors and Affiliations
Contributions
The research was planned and designed by AB and FAJ. JH did all experiments, collected, and analyzed data. FAJ, EA, NZOG and MSK participated in project activities, critically reviewed, and revised the manuscript. AB supervised the overall research activities of the project.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hossain, M.J., Bakhsh, A., Joyia, F.A. et al. Engineering of insecticidal hybrid gene into potato chloroplast genome exhibits promising control of Colorado potato beetle, Leptinotarsa decemlineata (Coleoptera: Chrysomelidae). Transgenic Res 32, 497–512 (2023). https://doi.org/10.1007/s11248-023-00366-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-023-00366-6