Abstract
Viruses constitute a major constraint to soybean production worldwide and are responsible for significant yield losses every year. Although varying degrees of resistance to specific viral strains has been identified in some soybean genetic sources, the high rate of mutation in viral genomes and mixed infections of different viruses or strains under field conditions usually hinder the effective control of viral diseases. In the present study, we generated transgenic soybean lines constitutively expressing the double-strand RNA specific ribonuclease gene PAC1 from Schizosaccharomyces pombe to evaluate their resistance responses to multiple soybean-infecting virus strains and isolates. Resistance evaluation over three consecutive years showed that the transgenic lines displayed significantly lower levels of disease severity in field conditions when challenged with soybean mosaic virus (SMV) SC3, a prevalent SMV strain in soybean-growing regions of China, compared to the non-transformed (NT) plants. After inoculation with four additional SMV strains (SC7, SC15, SC18, and SMV-R), and three isolates of bean common mosaic virus (BCMV), watermelon mosaic virus (WMV), and bean pod mottle virus (BPMV), the transgenic plants exhibited less severe symptoms and enhanced resistance to virus infections relative to NT plants. Consistent with these results, the accumulation of each virus isolate was significantly inhibited in transgenic plants as confirmed by quantitative real-time PCR and double antibody sandwich enzyme-linked immunosorbent assays. Collectively, our results showed that overexpression of PAC1 can increase multiple virus resistance in transgenic soybean, and thus provide an efficient control strategy against RNA viruses such as SMV, BCMV, WMV, and BPMV.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soybean (Glycine max L.) is one of the most important cultivated crops, representing the largest source of vegetable protein and edible oil for human and livestock consumption. Among the factors limiting soybean production, viruses represent one of the most serious threats affecting soybean yield and quality worldwide. For example, soybean mosaic virus (SMV), which is the most prevalent and destructive viral pathogen in soybean, causes approximately 10–35% yield losses every year under field conditions, and even total crop losses have been reported when severe outbreaks occurred (Ross 1983; Yang et al. 2013). SMV-infected soybean plants usually show mosaic patterns, chlorosis, curling, and necrosis of leaves, malformed pods, mottled seeds, and reduced size (Gao et al. 2015; Yang et al. 2014). SMV is mainly disseminated by infected soybean seeds, which are the primary infection source, and transmitted further by aphids leading to the secondary spread of SMV in soybean fields (Gao et al. 2015; Halbert et al. 1981; Hill et al. 1980). Numerous SMV isolates identified to date have been classified into seven strains (G1–G7) in North America (Cho and Goodman 1979, 1982) and 22 strains (SC1–SC22) in China (Shi et al. 2009; Zhang et al. 2011), based on the different phenotypic responses of specific soybean genotypes. Moreover, novel SMV strains (SMV-R) resulting from the recombination of SMV and other soybean-infecting viruses have also been reported, which usually exhibit different pathogenicity in soybean compared to conventional SMV strains (Yang et al. 2014). Other soybean-infecting viruses include bean pod mottle virus (BPMV), soybean dwarf virus (SbDV), alfalfa mosaic virus (AMV), bean common mosaic virus (BCMV), and watermelon mosaic virus (WMV) (Desbiez and Lecoq 2004; Zhang et al. 2011; Zhou et al. 2014). Like SMV, both BCMV and WMV are members of the genus Potyvirus, with a single-stranded, positive-sense RNA. These three viruses also share the same modes of transmission (seeds or aphids) and cause similar symptoms in soybean plants (Yang et al. 2014, 2017; Zhou et al. 2014). It was reported that WMV emerged from an ancestral recombination of BCMV and SMV in the N-terminus of the genome (Desbiez and Lecoq 2004; Shukla et al. 1994). Although no yield losses caused by BCMV and WMV were reported to date, these two viruses still represent a potential threat to soybean production in many soybean-growing areas of China (Zhang et al. 2011; Zhou et al. 2014), especially when synergistic interactions between SMV and these soybean-infecting viruses occur (Yang et al. 2014, 2018). As a member of the genus Comovirus, the genome of BPMV consists of two positive strand RNAs (RNA1 and RNA2), and uses a different strategy from that of SMV to achieve successful reproduction in soybean (Zhang et al. 2011). In the US, BPMV is widespread in many soybean-growing areas, and 40% of yield losses resulting from BPMV infection have been reported (Sinclair 1989).
The main control strategy of soybean-infecting viruses used in practice has been based on prevention measures such as cultural practices and applications of pesticides for aphid control. Thus far, genetic resistance provides the most effective way to control viruses. Several single-dominant resistance (R) loci such as Rsv1, Rsv3, and Rsv4 have been identified in some genetic sources and used for SMV-resistance breeding (Gore et al. 2002; Hayes et al. 2000; Hill and Whitham 2014; Jeong and Maroof 2004; Maroof et al. 2010; Saghai Maroof et al. 2008; Shi et al. 2009). However, genetic resistance mediated by these R genes is frequently broken under field conditions due to the high rate of mutation in viral genomes (Choi et al. 2005; Gagarinova et al. 2008; Koo et al. 2005), and some SMV isolates can even overcome all known resistance alleles at the Rsv1, Rsv3, and Rsv4 loci (Choi et al. 2005). Moreover, the existence of multiple viruses or strains and synergistic interactions between different soybean-infecting viruses associated with field-grown soybean make it more difficult to control the occurrence of viruses by using single R gene-mediated resistance. Transgenic technologies offer a promising alternative for engineering multiple virus resistance in plants. Several attempts have been made to engineer SMV or other soybean-infecting virus resistance based on host RNA interference (RNAi)-mediated suppression of viral genes and sequences (Furutani et al. 2006; Gao et al. 2015; Reddy et al. 2001; Tougou et al. 2006; Yang et al. 2017, 2018; Zhang et al. 2011), and by overexpression of virus-resistance genes (Liu et al. 2014; Luan et al. 2016; Zhou et al. 2014). Multiple-virus resistance was also explored by silencing different virus-specific genes (Yang et al. 2017, 2018; Zhang et al. 2011), and showed great potential in soybean virus-resistance breeding.
The PAC1 RNase from Schizosaccharomyces pombe belongs to the RNase III family of double-stranded (ds)-specific ribonucleases which plays an important role in RNA processing and post-transcriptional gene regulation in both prokaryotes and eukaryotes (Nicholson 1999; Lamontagne et al. 2001; Cao et al. 2013). Among RNase III family, PAC1 is a well-studied family member. It contains conserved regions for catalysis and a dsRNA-binding domain (Iino et al. 1991; Rotondo et al. 1997), and can degrade ds-replicative forms of single-strand RNA (ssRNA) viruses both in vitro (Ishida et al. 1995; Sano et al. 1997; Yan et al. 2006) and in vivo (Ogawa et al. 2005; Sano et al. 1997; Toguri et al. 2003; Watanabe et al. 1995; Yan et al. 2006; Zhang et al. 2001) with no sequence dependence on viral genomes (Yan et al. 2006). Previous studies showed that PAC1 could potentially act against the replication of many plant viruses and viroids such as tomato mosaic virus, cucumber mosaic virus, potato spindle tuber viroid, potato virus Y, chrysanthemum stunt viroid, tomato spotted wilt virus, barley stripe mosaic virus, and barley yellow dwarf virus (Ishida et al. 1995; Ogawa et al. 2005; Sano et al. 1997; Toguri et al. 2003; Watanabe et al. 1995; Yan et al. 2006; Zhang et al. 2001). During the process of viral replication, single-stranded viral RNAs can form double-stranded structures, at least transiently, and thus provide suitable targets for digestion by PAC1. Moreover, it was reported that overexpression of PAC1 caused no unexpected changes in agronomic phenotypes in transgenic potato and wheat (Sano et al. 1997; Yan et al. 2006). To engineer multiple virus resistance in soybean, we generated transgenic soybean plants constitutively expressing PAC1 and evaluated their resistance to multiple soybean-infecting viruses including SMV, BCMV, WMV, and BPMV. Our results showed that the overexpression of PAC1 significantly increased multiple-virus resistance and inhibited the accumulation of these viruses in transgenic soybean plants.
Materials and methods
Construction of the plant expression vector and Agrobacterium tumefaciens-mediated transformation
The coding sequence of PAC1 was amplified from the plasmid pBI121-PAC1 kindly provided by Prof. Shifang Li (Chinese Academy of Agricultural Sciences, China), using primers PAC1-F1 (5′-ATGGGAAGATTCAAGAGGCATCAC-3′) and PAC1-R1 (5′-TCATCTAGCGAATTTTGAATAATCCTTAGC-3′). The amplified fragment was then cloned into the XbaI/SacI sites of the plasmid pCAMBIA3300 under control of the constitutive 35S promoter of cauliflower mosaic virus (CaMV) to generate the plant expression vector pCAMBIA3300-PAC1 (Fig. 1a). The resulting construct also contained the BAR gene which confers glufosinate resistance. Cultivated soybean genotype Williams82 was used for A. tumefaciens-mediated transformation, as previously described (Yang et al. 2017), and the regenerated plants were transferred to a greenhouse for molecular screening.
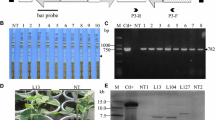
Vector construct and molecular screening of transgenic plants. a Schematic diagram of the vector construct pCAMBIA3300-PAC1. The coding sequence of the PAC1 gene from Schizosaccharomyces pombe was inserted between the constitutive 35S promoter of cauliflower mosaic virus (CaMV) and the nopaline synthase terminator (NOS). The selectable marker BAR, which confers glufosinate resistance, was also contained in the construct. The solid bar represents the probe used for Southern blot analysis. LB, left border; RB, right border. b LibertyLink® strip detection of T0 transgenic plants. NT, non-transformed control; 1–12, T0 transgenic plants. Small arrows indicate the test line. c PCR analysis of transgenic plants. M, DNA marker (2k); +, positive control; −, ddH2O; NT, non-transformed plants, 1–11, LibertyLink® strip-positive plants. d Herbicide tolerance screening of T2 transgenic lines using 1500 mg/L glufosinate spray. NT, non-transformed plants; Q117, independent transgenic line. (E) Southern blot analysis of the transgenic plants. Thirty micrograms of genomic DNA was completely digested with XbaI and hybridized against a digoxigenin-labeled BAR probe. Ctl+, positive control; NT, non-transformed plants; Q110, Q112, Q198, Q72, Q172, Q84, Q117, Q23, T2 transgenic plants. f Western blot analysis of transgenic plants. The polyclonal antibody was raised against a peptide corresponding to 14 amino acids (GRFKRHHEGDSDSS) near the N-terminus of PAC1. Ctl+, recombinant PAC1 protein expressed in Escherichia coli; NT1 and NT2, non-transformed plants; Q117, Q172, Q72, T3 transgenic lines
Molecular screening of transgenic plants and western blot analysis
Transgenic T0 plants were screened using the LibertyLink® strip (QuickStix™ Kit, EnviroLogix Inc., Portland, ME, USA) and polymerase chain reaction (PCR) as described previously (Yang et al. 2017, 2018). Primers PAC1-F2 (5′-GTGGATTGATGTGATATCTCCACTG-3′) and PAC1-R2 (5′-GAAGATGGCTCCTCAATCACAGG-3′), annealing to the CaMV 35S promoter and PAC1 sequences respectively, were used to amplify a 448-bp fragment with genomic DNA as templates to confirm the presence of the transgene. For the screening of subsequent generations (T1–T4), both glufosinate spraying (1500 mg/L) and PCR detection using the above-mentioned primers were performed to analyze the stability of the transgenic plants until homozygous lines was obtained. To demonstrate integration of the transgene into the soybean genome, approximately 30 µg of total DNA from T2 transgenic plants and NT control plants was extracted using a modified cetyltrimethylammonium bromide method (Telzur et al. 1999). After digestion with the restriction endonuclease XbaI, the digested products were separated on a 0.8% agarose gel and then transferred onto positively charged nylon membranes (GE Healthcare, Chicago, IL, USA), according to standard protocols. The hybridization probe was prepared by PCR amplification of the pCAMBIA3300-PAC1 template using primers BAR-F1 (5′-GCACCATCGTCAACCACTACATCGAG-3′) and BAR-R1 (5′-TGAAGTCCAGCTGCCAGAAACCCAC-3′), and labeled with digoxigenin-high prime (Roche, Basel, Switzerland). Hybridization and subsequent washing were performed at 65 °C. Chemical staining was performed using 5-bromo-4-chloro-3′-indolyphosphate (BCIP)/ nitro-blue tetrazolium (NBT) as substrate, until the signal appeared clearly. Western blot hybridization was carried out by GenScript Co., Ltd. (Nanjing, China), using a rabbit polyclonal antibody raised against a 14-amino-acid peptide (GRFKRHHEGDSDSS) near the N-terminus of PAC1. Leaf samples (0.1 g) were collected from T3 transgenic and NT plants and used for total protein extraction in a buffer containing 100 mM NaCl, 10 mM EDTA, 200 mM Tris–HCl, 0.05% Tween-20, 0.1% SDS, 14 mM β-mercaptoethanol, 400 mM sucrose, and 2 mM phenylmethane sulfonyl fluoride. After separation on a 12% polyacrylamide gel by electrophoresis, total protein extract was transferred to a polyvinylidene fluoride membrane (Amersham™ Hybond™, GE Healthcare, USA). The membrane was then blotted with the rabbit polyclonal antibody (1:500 dilution) and horseradish peroxidase (HRP)-labeled goat-anti-rabbit Immunoglobulin G (1:5000 dilution, Abcam, Cambridge, UK) at 25 °C for 4 h. Extensive washing was then performed and protein bands were visualized using the BiodlightTM Western Chemiluminescent HRP substrate (Bioworld Technology, Inc., St. Louis, MN, USA).
Resistance evaluation and agronomic performance of the transgenic lines
Five SMV strains (SC3, SC7, SC15, SC18 and SMV-R) and three isolates of BCMV, WMV and BPMV kindly provided by Prof. Haijian Zhi (Nanjing Agricultural University, China) were prepared for inoculation experiments as described previously (Yang et al. 2017, 2018). For SMV SC3 inoculation, a prevalent strain in China, field-grown transgenic plants were infected by gently rubbing the newly expanded unifoliate leaves with the inoculum. Corresponding non-transformed (NT) plants were also inoculated with SMV SC3 or 0.01 M sodium phosphate buffer (pH 7.2) as positive and negative controls, respectively. The plants were sprayed with insecticides every two weeks to prevent cross-infection via aphids. A randomized block design with three replicates per line was used, and 30 plants were inoculated per replicate. Thirty-five days after inoculation, SMV disease severity and disease index (DI) were evaluated and calculated for fully developed leaves in each plant line, as described previously (Yang et al. 2017, 2018). Disease index was calculated as DI = ∑fi ×Si / (n × Smax), where fi = number of plants with classified disease severity, Si = classified disease severity, Smax = highest disease severity, n = total number of infected plants. Resistance of the transgenic lines to five SMV strains and isolates of BCMV, WMV, and BPMV was also evaluated using the inoculation method mentioned above in an insect-proof greenhouse. Three replicates were performed with 10 plants inoculated per replicate. Disease severity and disease index of the transgenic lines was evaluated and calculated as mentioned above. The agronomic performance of the transgenic lines under field conditions without virus inoculation was also evaluated. At maturity, 10 plants from each transgenic line and their NT counterparts were randomly sampled. Leaf shape, flower color, plant height, maturity period, branch number, node number, pod number, and 100-seed weight, were measured and recorded.
Quantitative real-time PCR (qRT-PCR) analysis
For qRT-PCR analysis, samples from the fully developed three top leaves (upmost, top second and third) were collected for each plants inoculated individually with SC3, SC7, SC15, SC18, SMV-R, BCMV, WMV, and BPMV. The NT plants inoculated with each viral strain or isolate and 0.01 M phosphate-buffered saline were used as both positive and negative controls, respectively. At 28 days after inoculation, total RNA was extracted from the virus-infected plants using the EasyPure PlantRNA Kit (TransGen Biotech, Beijing, China). After treatment with RNase–free DNase, total RNA samples were reverse-transcribed into cDNAs with the ThermoScript RT-PCR system (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. A SYBR Green-Based One-Step qRT-PCR kit (TransGen Biotech) was used for qRT-PCR and the reactions were analyzed on a LightCycler 480 II instrument (Roche). Each virus-specific CP gene was amplified using the primers listed in the Table S1. The soybean GmACT11 (GenBank No. BW652479) was amplified as an internal control. The accumulation of each viral RNA was calculated using the relative quantification (2−ΔCt) method and compared with that of the internal control. The experiments were performed in three biological replicates.
Double antibody sandwich enzyme-linked immunosorbent assay (DAS-ELISA)
The accumulation of each virus stain or isolate (SC3, SC7, SC15, SC18, SMV-R, BCMV, WMV, and BPMV) after inoculation was further analyzed by DAS-ELISA kits with anti-SMV, anti-BCMV, anti-WMV, and anti-BPMV antibodies (AC Diagnostics Inc., Fayetteville, AR, USA). Leaf samples were collected from five randomly selected transgenic plants. The NT plants inoculated with each viral strain or isolate or mock-inoculated (buffer only) were used as positive and negative controls, respectively. The DAS-ELISA analysis was carried out according to the manufacturer’ s protocols and readings were averaged and converted to multiples of the negative controls as previously described (Zhang et al. 2011). Samples with adjusted values ≥2.0 were considered positive for each viral strain or isolate.
Statistical analyzes
Analysis of variance was used to calculate the least significant differences based on t-tests at P < 0.01 by SPSS Statistics software (version 17, IBM Inc., New York, NY, USA). Data were analyzed to evaluate statistical differences between each transgenic line and corresponding NT control.
Results
Generation of transgenic soybean plants overexpressing PAC1
To engineer multiple virus resistance in soybean, we generated transgenic plants constitutively expressing the dsRNA-specific ribonuclease gene PAC1 from S. pombe (Fig. 1a). Ninety-six plants tolerant to glufosinate were produced after A. tumefaciens-mediated genetic transformation. The presence of the transgene in regenerated T0 plants was confirmed by both LibertyLink® strip and PCR analysis (Fig. 1b, c). A 448-bp fragment corresponding to the partial sequence of the promoter cauliflower mosaic virus (CaMV) 35S and the PAC1 gene was amplified by PCR in transgenic plants but not in NT plants (Fig. 1c). Screening of the transgenic progenies in subsequent generations (T1 to T4) by glufosinate spraying and PCR analysis further verified the stable inheritance of the transgene (Fig. 1d). Homozygous transgenic lines were obtained for subsequent virus inoculation assays. Southern blot hybridization was also performed to demonstrate transgene integration into the soybean genome. Each of the eight selected transgenic lines showed single (line Q72), double (lines Q112, Q198, Q172, Q117, and Q123) or triple (lines Q110 and Q84) transfer-DNA inserts, all longer than the expected fragment size of 3.25 kb, which is located between the XbaI site and the left border (Fig. 1e). Different hybridization patterns indicated independent events for these four transgenic lines. To demonstrate the expression of PAC1 in transgenic plants, western blot analysis was performed using a polyclonal antibody against a 14-amino-acid peptide of PAC1. A 41.47-kDa band corresponding to the PAC1 protein was observed in transgenic plants but not in NT plants (Fig. 1f), demonstrating the expression of PAC1 in transgenic plants only. Three independent transgenic lines, Q117, Q172, and Q72, were used for subsequent investigations.
Transgenic plants exhibited stable and enhanced resistance to SMV SC3 under field conditions
A virus inoculation assay was carried out to assess the resistance of the transgenic lines to SMV SC3 under field conditions. After inoculation with the SMV SC3 strain, NT plants showed the typical symptoms of SMV, including serious mosaic pattern, chlorosis, curling, and necrosis of leaves, while transgenic plants displayed no visible or moderate mosaic pattern on the leaves, similar to those of the mock-inoculated controls (Fig. 2a). Similarly, enhanced resistance responses to SMV SC3 were observed in T3 and T4 transgenic lines (data not shown). Quantification of disease severity further revealed that transgenic lines had significantly lower SMV disease indices (19.14–26.60) than NT plants (46.81–53.58) over three consecutive generations (Fig. 2b), demonstrating the stable and enhanced SMV SC3 resistance of transgenic plants overexpressing PAC1 under field conditions. The effects of PAC1 overexpression on agronomic traits were also evaluated without SMV inoculation. No significant differences were observed between transgenic and NT plants on leaf morphology, flower color, hilum color, plant height, podding height, node number, 100-seed weight, or maturity period (Table S2). These results showed that the overexpression of PAC1 significantly enhanced resistance to SMV SC3, but caused no visible changes in agronomic traits in transgenic soybean plants.
Resistance evaluation of the transgenic lines to SMV SC3 under field conditions. a Resistance responses of the transgenic lines to SMV SC3 inoculation. Fully expanded first trifoliolate leaves of soybean seedlings were inoculated with the SMV SC3 strain, and the resistance responses of transgenic lines were assessed at 35 days after inoculation (dai). b Disease indices in three generations of transgenic lines inoculated with SMV SC3. Resistance evaluation of T2, T3, and T4 transgenic lines to the SMV SC3 strain was performed under field conditions. At 35 dai, the disease severity and disease index (DI) of each transgenic line were evaluated. Disease index was calculated as DI = ∑fi × Si/(n × Smax), where fi = number of plants with classified disease severity, Si = classified disease severity, Smax = highest disease severity, n = total number of infected plants. Three replicates for each transgenic line were carried out with 30 plants inoculated per replicate. Average values and standard errors of DIs were calculated. Asterisks in each generation indicate significant differences (least significant differences based on t tests, P < 0.01). NT, non-transformed plants inoculated with SMV SC3; Mock, non-transformed plants inoculated with 0.01 M sodium phosphate buffer (pH 7.2); Q117, Q172, and Q72, transgenic lines.
Transgenic plants showed increased resistance to multiple viruses under greenhouse conditions
To evaluate resistance responses to multiple soybean-infecting viral strains and isolates in transgenic lines, four SMV strains, including SC7, SC15, SC18, and SMV-R, and three BCMV, WMV, and BPMV isolates were used for single inoculation experiments under greenhouse conditions. Thirty-five days after inoculation with different viruses, almost all NT plants exhibited varying degrees of mosaic patterns or leaf curling (Fig. 3a). In contrast, transgenic lines showed significantly compromised symptom development. For SC7, SC15, SC18, SMV-R, BCMV, and WMV, no visible symptoms or milder mosaic patterns were observed on the leaves of the transgenic plants (Fig. 3a). Although the BPMV-infected transgenic plants showed slight leaf curling and mosaic patterns on the leaves, their resistance responses were much stronger than those of NT plants. The disease index (DI) based on disease severity was further calculated to assess the resistance levels of the transgenic lines to different virus strains or isolates. Consistent with the above-mentioned observations, inoculated transgenic lines had significantly lower DIs (ranging from 1.39 to 47.59) than NT plants (23.15–84.26) for all seven virus strains and isolates (Fig. 3b). The transgenic plants inoculated with BPMV showed relatively higher DIs (28.70 to 47.59) than the transgenic plants inoculated with the other six virus strains and isolates (1.39 to 19.44); however, the DIs of these plants were still much lower than that (84.26) of the BPMV-infected NT plants (Fig. 3b). These results proved that transgenic plants expressing PAC1 significantly increased resistance to multiple soybean-infecting viruses.
Resistance evaluation of transgenic plants to multiple viral strains and isolates. a Resistance responses of the transgenic lines to the single inoculation of seven virus strains and isolates. Fully expanded first trifoliolate leaves of soybean seedlings were inoculated singly with four SMV strains (SC7, SC15, SC18, and SMV-R), three isolates of BCMV, WMV, and BPMV, and 0.01 M phosphate-buffered saline (mock). Resistance responses were assessed at 35 days after inoculation (dai). b Disease indices of greenhouse-grown T4 lines of transgenic plants after virus inoculation. Three replicates, each containing 10 inoculated plants, were used for each transgenic line. At 35 dai, the disease severity and the disease index (DI) for each transgenic line were evaluated. Disease index was calculated as DI = ∑fi × Si/(n × Smax), where fi = number of plants with classified disease severity, Si = classified disease severity, Smax = highest disease severity, n = total number of infected plants. Average values and standard errors of the DIs for each line were calculated from three replicates. Asterisks for each viral strain indicate significant differences (least significant differences based on t-tests, P < 0.01). NT, non-transformed plants; Q117, Q172, and Q72, transgenic lines.
The accumulation of virus strains and isolates was significantly lower in transgenic than in NT plants
Virus accumulation in transgenic plants after single inoculations was further determined by both qRT-PCR and DAS-ELISA analysis. As shown in Fig. 4, accumulation of virus RNA was significantly lower in transgenic plants inoculated with single virus strain or isolate than in infected NT plants at 28 days after inoculation. Consistent with the results of resistance evaluation, the BPMV-infected transgenic plants showed higher virus RNA levels than the plants infected with the other seven virus strains and isolates (SC3, SC7, SC15, SC18, SMV-R, BCMV, and WMV), but significantly lower than the BPMV-infected NT plants (Fig. 4). Moreover, all virus strains and isolates (except BPMV) were below the detection limits in the transgenic plants but not in the NT plants after inoculation, as shown by DAS-ELISA analysis (Table 1). Again, BPMV accumulation was detected in the transgenic plants, but the corresponding OD405nm values were much lower than that of NT controls (Table 1). These results indicated that viral RNA replication and accumulation of the eight virus strains and isolates examined here were significantly reduced in transgenic soybean plants.
Relative accumulation levels of viral RNAs in single virus-inoculated transgenic plants. Twenty-eight days after single inoculation with eight viral strains and isolates (SC3, SC7, SC15, SC18, SMV-R, BCMV, WMV, and BPMV), the leaf samples were collected and mixed from the fully developed top three leaves (upmost, top second and third) for each plant and total RNA was extracted. The NT plants inoculated with each viral strain or 0.01 M phosphate- buffered saline were used as both positive and negative controls, respectively. Quantification of the viral RNAs was based on the formula 2ΔCt for the accumulation of viral RNA relative to that of the internal control GmACTIN11. Data represent means of three biological replicates with error bars indicating standard errors. Asterisks indicate significant differences between each transgenic line and the corresponding NT plants at P < 0.01. NT, non-transformed plants; Q117, Q172, and Q72, transgenic lines
Discussion
Viral pathogens are a major limiting factor for soybean production worldwide. Previous studies showed that RNAi-mediated silencing is a powerful strategy for controlling viral diseases in soybean (Furutani et al. 2006; Gao et al. 2015; Reddy et al. 2001; Tougou et al. 2006; Yang et al. 2017, 2018; Zhang et al. 2011). Actually, the resistance mechanism in most transgenic strategies for the control of soybean-infecting viruses is based on RNAi silencing. Several viral genes and sequences such as CP (Zhang et al. 2011), NIb (Yang et al. 2017; Zhang et al. 2011), HC-Pro (Gao et al. 2015; Lim et al. 2007), and P3 (Yang et al. 2018) have been utilized for engineering viral resistance. However, due to the sequence specificity of RNAi-mediated silencing, concerns are raised that the resistance of the transgenic plants engineered to be resistant to typically one virus could be broken by a different, heterologous virus, which could also infect soybean in field conditions. This heterologous virus could repress the RNAi mechanism by silencing suppressor proteins, thereby leading to the loss of the initially engineered resistance. Here, we developed a promising alternative for the control of multiple soybean-infecting viruses by overexpressing a ds-specific ribonuclease gene PAC1 in transgenic soybean. Virus inoculation assays showed that transgenic soybean plants significantly increased resistance to seven potyvirus strains or isolates (SMV SC3, SMV SC7, SMV SC15, SMV SC18, SMV-R, BCMV, and WMV) and one comovirus (BPMV). Compared to NT plants, transgenic plants displayed a significant reduction in disease severity and inhibited replication and accumulation of the viruses. These results demonstrated the effectiveness of engineering resistance to multiple taxonomically different soybean-infecting viruses by constitutively expressing PAC1.
Previous studies showed that PAC1 can specifically degrade dsRNAs, but not ssRNAs, in a sequence-independent manner (Rotondo et al. 1997; Yan et al. 2006). Different viruses or viroids could be efficiently degraded by PAC1 in vitro (Ishida et al. 1995; Sano et al. 1997; Yan et al. 2006) and transgenic plants expressing PAC1 showed enhanced resistance to distantly related viruses (Ogawa et al. 2005; Sano et al. 1997; Toguri et al. 2003; Watanabe et al. 1995; Yan et al. 2006; Zhang et al. 2001). In the present study, transgenic plants displayed significantly enhanced resistance to multiple soybean-infecting virus strains and isolates, which have a single or double RNA genome. The dsRNAs generated during viral replication in the host might provide a suitable substrate for PAC1 degradation. However, it should be noted that transgenic soybean lines expressing PAC1 were not entirely immune to viral infections under our experimental conditions. Yan et al. (2006) reported that there might be a dose-dependent effect of the resistance mediated by PAC1. Increased resistance might be related to the degradation rate of dsRNAs in viral replication and to the replication rates of the viruses. Thus, viral resistance levels might be closely correlated with the activity of PAC1 in transgenic soybean plants. Additionally, the differences in virus multiplication may also affect their resistance level in the transgenic plants overexpressing PAC1. In this study, the BPMV-infected transgenic plants displayed relatively higher levels of disease severity and virus accumulation than those inoculated with other seven potyvirus strains and isolates. The observation might be explained by the different reproduction strategy or replication rate of BPMV from that of potyvirus.
Another interesting question is whether dsRNAs degradation by PAC1 has any unexpected effect on the native soybean RNAs and thus interferes in normal metabolism, changing phenotypes on the PAC1-overexpressing transgenic plants. The extensive presence of double strand structures in inter- and intra-RNAs in plant cells suggests a possible degradation mechanism for these types of native RNAs by the heterologous PAC1. However, in the present study, transgenic soybean plants expressing PAC1 and NT plants showed no visible differences in agronomic performance. Similar results were observed in previous studies (Sano et al. 1997; Yan et al. 2006). On the other hand, if PAC1 can degrade native soybean RNAs, it is still difficult to distinguish between the effects of PAC1 and the positional effects of the transgene insertion in the soybean genome. Further investigations are therefore needed for clarifying this intriguing issue.
References
Cho EK, Goodman RM (1979) Strains of soybean mosaic virus: classification based on virulence in resistant soybean cultivars. Phytopathology 69:467–470
Cho EK, Goodman RM (1982) Evaluation of resistance in soybeans to soybean mosaic virus strains. Crop Sci 22:1133–1136
Choi BK, Koo JM, Ahn HJ, Yum HJ, Choi CW, Ryu KH, Chen P, Tolin SA (2005) Emergence of Rsv-resistance breaking soybean mosaic virus isolates from Korean soybean cultivars. Virus Res 112:42–51
Cao X, Lu Y, Di D, Zhang Z, Liu H, Tian L, Zhang A, Zhang Y, Shi L, Guo B, Xu J, Duan X, Wang X, Han C, Miao H, Yu J, Da Li (2013) Enhanced virus resistance in transgenic maize expressing a dsRBA-specific endoribonuclease gene from E. coli. PLoS ONE 8:60829. https://doi.org/10.1371/journal.pone.0060829
Desbiez C, Lecoq H (2004) The nucleotide sequence of watermelon mosaic virus (WMV, Potyvirus) reveals interspecific recombination between two related potyviruses in the 5′ part of the genome. Arch Virol 149:1619–1632
Furutani N, Hidaka S, Kosaka Y, Shizukawa Y, Kanematsu S (2006) Coat protein gene-mediated resistance to soybean mosaic virus in transgenic soybean. Breed Sci 56:119–124
Gagarinova AG, Babu M, Poysa V, Hill JH, Wang A (2008) Identification and molecular characterization of two naturally occurring soybean mosaic virus isolates that are closely related but differ in their ability to overcome Rsv4 resistance. Virus Res 138:50–56
Gore MA, Hayes AJ, Jeong SC, Yue YG, Buss GR, Maroof MAS (2002) Mapping tightly linked genes controlling potyvirus infection at the Rsv1 and Rpv1 region in soybean. Genome 45:592–599
Gao L, Ding X, Li K, Liao W, Zhong Y, Ren R, Liu Z, Adhimoolam K, Zhi H (2015) Characterization of soybean mosaic virus resistance derived from inverted repeat-SMV-HC-Pro genes in multiple soybean cultivars. Theor Appl Genet 128:1489–1505
Hill JH, Whitham SA (2014) Control of virus diseases in soybeans. Adv Virus Res 90:355–390
Halbert SE, Irwin ME, Goodman RM (1981) Alate aphid (Homoptera: Aphididae) species and their relative importance as field vectors of soybean mosaic virus. Ann Appl Biol 97:1–9
Hayes AJ, Ma G, Buss GR, Maroof MAS (2000) Molecular marker mapping of RSV4, a gene conferring resistance to all known strains of soybean mosaic virus. Crop Sci 40:1434–1437
Hill JH, Lucas BS, Benner HI, Tachibana H, Hammond RB, Pedigo LP (1980) Factors associated with the epidemiology of soybean mosaic virus in Iowa. Phytopathology 70:536–540
Iino Y, Sugimoto A, Yamamoto M (1991) Whose overexpression inhibits sexual development, encodes a ribonuclease. Embo J 10:221–226
Ishida I, Ogawa T, Yoshioka M, Hori T, Ohtani T (1995) Production of plants tolerant to multiple viral infections by genetic manipulations. Acta Hortic 10:927–928
Jeong SC, Maroof MAS (2004) Detection and genotyping of SNPs tightly linked to two disease resistance loci, Rsv1 and Rsv3 of soybean. Plant Breed 123:305–310
Koo JM, Choi BK, Ahn HJ, Yum HJ, Choi CW (2005) First report of an Rsv resistance-breaking isolate of soybean mosaic virus in Korea. Plant Pathol 54:573–573
Lamontagne B, Larose S, Boulanger J, Elela SA (2001) The RNase III family: a conserved structure and expanding functions in eukaryotic dsRNA metabolism. Yeast 45:154–158
Liu JZ, Braun E, Qiu WL, Shi YF, Marcelino-Guimarães FC, Navarre D, Hill JH, Whitham SA (2014) Positive and negative roles for soybean MPK6 in regulating defense responses. Mol Plant-Microbe Interact 27:824–834
Luan H, Shine MB, Cui X, Chen X, Ma N, Kachroo P, Zhi H, Kachroo A (2016) The Potyviral P3 protein targets eukaryotic elongation factor 1A to promote the unfolded protein response and viral pathogenesis. Plant Physiol 172:221–234
Lim HS, Ko TS, Hobbs HA, Lambert KN, Yu JM, McCoppin NK, Korban SS, Hartman GL, Domier LL (2007) Soybean mosaic virus helper component-protease alters leaf morphology and reduces seed production in transgenic soybean. Plants Phytopathol 97:366–372
Maroof MAS, Tucker DM, Skoneczka JA, Bowman BC, Tripathy S, Tolin SA (2010) Fine mapping and candidate gene discovery of the soybean mosaic virus resistance gene Rsv4. Plant Genome 3:14–22
Nicholson AW (1999) Function, mechanism and regulation of bacterial ribonucleases. FEMS Microb Rev 23:371–390
Ogawa T, Toguri T, Kudoh H, Okamura M, Momma T, Yoshioka M, Hagiwara Y, Sano T (2005) Double-stranded RNA-specific ribonuclease confers tolerance against chrysanthemum stunt viroid and tomato spotted wilt virus in transgenic chrysanthemum plants. Breed Sci 55:49–55
Ross JP (1983) Effect of soybean mosaic on component yields from blends of mosaic resistant and susceptible soybeans. Crop Sci 23:343–346
Rotondo G, Huang JY, Frendewey D (1997) Substrate structure requirements of the Pac1 ribonuclease from Schizosaccharmyces pombe. Publ RNA Soc 3:1182–1193
Reddy MSS, Ghabrial SA, Redmond CT, Dinkins RD, Collins GB (2001) Resistance to bean pod mottle virus in transgenic soybean lines expressing the capsid polyprotein. Phytopathology 91:831–838
Sinclair JB (1989) Compendium of soybean diseases. 3rd Ed. APS Press, Saint Paul
Shukla DD, Ward CW, Brunt AA (1994) The Potyviridae. CAB International, Wallingford
Sano T, Nagayama A, Ogawa T, Ishida I, Okada Y (1997) Transgenic potato expressing a double-stranded RNA-specific ribonuclease is resistant to potato spindle tuber viroid. Nat Biotechnol 15:1290–1294
Saghai Maroof MA, Jeong SC, Gunduz I, Tucker DM, Buss GR, Tolin SA (2008) Pyramiding of soybean mosaic virus resistance genes by marker-assisted selection. Crop Sci 48:517–526
Shi A, Chen P, Li D, Zheng C, Zhang B, Hou A (2009) Pyramiding multiple genes for resistance to soybean mosaic virus in soybean using molecular markers. Mol Breed 23:113–124
Telzur N, Abbo S, Myslabodski D, Mizrahi Y (1999) Modified CTAB procedure for DNA isolation from Epiphytic cacti of the Genera Hylocereus and Selenicereus (Cautaceae). Plant Mol Biol Rep 17:249–254
Toguri T, Ogawa T, Kakitani M, Tukahara M, Yoshioka M (2003) Agrobacterium-mediated transformation of chrysanthemum (Dendranthema grandiflora) plants with a disease resistance gene (pac1). Plant Biotechnol 20:121–127
Tougou M, Furutani N, Yamagishi N, Shizukawa Y, Takahata Y, Hidaka S (2006) Development of resistant transgenic soybeans with inverted repeat-coat protein genes of soybean dwarf virus. Plant Cell Rep 25:1213–1218
Watanabe Y, Ogawa T, Takahashi H, Ishida I, Takeuchi Y, Yamamoto M, Okada Y (1995) Resistance against multiple plant viruses in plants mediated by a double stranded-RNA specific ribonuclease. Febs Lett 372:165–168
Yang Y, Lin J, Zheng G, Zhang M, Zhi H (2014) Recombinant soybean mosaic virus is prevalent in Chinese soybean fields. Arch Virol 159:1793–1796
Yan F, Zheng Y, Zhang W, Xiao H, Shifang LI, Cheng Z (2006) Obtained transgenic wheat expressing pac1 mediated by Agrobacterium is resistant against barley yellow dwarf virus. Chin Sci Bull 51:2362–2368
Yang Y, Zheng G, Han L, Dagang W, Yang X, Yuan Y, Huang S, Zhi H (2013) Genetic analysis and mapping of genes for resistance to multiple strains of soybean mosaic virus in a single resistant soybean accession PI 96983. Theor Appl Genet 126:1783–1791
Yang X, Niu L, Zhang W, He H, Yang J, Xing G, Guo D, Du Q, Qian X, Yao Y, Li Q, Dong Y (2017) Robust RNAi-mediated resistance to infection of seven potyvirids in soybean expressing an intron hairpin NIb RNA. Transgenic Res 26:665–676
Yang X, Niu L, Zhang W, Yang J, Xing G, He H, Guo D, Du Q, Qian X, Yao Y, Li Q, Dong Y (2018) RNAi-mediated SMV P3 cistron silencing confers significantly enhanced resistance to multiple Potyvirus strains and isolates in transgenic soybean. Plant Cell Rep 37:103–114
Zhang L, French R, Langenberg WG, Mitra A (2001) Accumulation of barley stripe mosaic virus is significantly reduced in transgenic wheat plants expressing a bacterial ribonuclease. Transgenic Res 10:13–19
Zhang X, Sato S, Ye X, Dorrance AE, Morris TJ, Clemente TE, Qu F (2011) Robust RNAi-based resistance to mixed infection of three viruses in soybean plants expressing separate short hairpins from a single transgene. Phytopathology 101:1264–1269
Zhou GC, Wu XY, Zhang YM, Wu P, Wu XZ, Liu LW, Wang Q, Hang YY, Yang JY, Shao ZQ, Wang B, Chen JQ (2014) A genomic survey of thirty soybean-infecting bean common mosaic virus (BCMV) isolates from China pointed BCMV as a potential threat to soybean production. Virus Res 191:125–133
Acknowledgements
This work was supported by Grants from China National Novel Transgenic Organisms Breeding Project (2016ZX08004-004), Jilin Provincial Agricultural Science & Technology Innovation Project (CXGC2017JQ013) and National Natural Science foundation of China (31671764). We thank Prof. Haijian Zhi (Nanjing Agricultural University in China) for providing virus strains and isolates and Prof. Shifang Li (Chinese Academy of Agricultural Sciences, China) for providing the plasmid pBI121-PAC1. We would also like to thank Editage (www.editage.cn) for English language editing.
Author information
Authors and Affiliations
Contributions
Yingshan Dong and Qiyun Li designed the experiments. Xiangdong Yang and Lu Niu conducted the experiments and drafted the manuscript. Jing Yang, Guojie Xing, Dongquan Guo, Qianqian Zhao, and Haiyun Li performed A. tumefaciens-mediated transformation experiments. Wei Zhang conducted the virus inoculation assays. Hongli He and Xiaofang Zhong participated in the qRT-PCR analyses. All authors participated in the manuscript revision.
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest in the publication of this paper.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, X., Niu, L., Zhang, W. et al. Increased multiple virus resistance in transgenic soybean overexpressing the double-strand RNA-specific ribonuclease gene PAC1. Transgenic Res 28, 129–140 (2019). https://doi.org/10.1007/s11248-018-0108-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-018-0108-8