Abstract
In the present work it was demonstrated that transgenic Danio rerio overexpressing growth hormone (GH-transgenic) present either altered gene expression at a determined time point, or different expression pattern along the LD cycle, when compared with non-transgenic (NT) animals, in the positive and negative loops of the circadian system. Gene expression of clock paralogs was reduced in GH fish at the beginning of the dark phase, leading to diminished expression amplitude along the LD cycle. Furthermore, although no differences were observed between NT and GH animals for bmal1a and cry2b expression at each time point, only GH fish presented amplitude along the LD cycle. Also, the locomotor activity behavior was evaluated for both groups. GH-transgenic animals presented higher locomotor activity along the whole LD cycle when compared with NT animals. These data suggest that alterations in the gene expression patterns along the LD cycle of the positive and negative loops of the circadian system, could lead to altered locomotor activity behavior in GH-transgenic fish, and GH overexpression could be responsible for these alterations, either affecting the pathways involved in the expression of genes from the circadian system or altering the metabolism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The physiological functioning and behavior of living organisms are driven by environmental cues, mainly the shifting between light and dark (LD) cycles, which allow them to anticipate changes in daylight, temperature and food availability conditions occurring with marked periodicity (Aschoff 1976; Moore-Ede et al. 1976, 1982). Such capability relies on evolutionary events which culminated on the development of an internal, self-sustaining clock, with periodicity near 24 h, which orchestrates their rhythms, according to their geographical localization (De Coursey 2004; Dunlap et al. 2004). This clock is composed by molecular oscillators formed by transcriptional activators and inhibitors of the clock itself and other genes, forming positive and negative loops, respectively (Dunlap et al. 2004).
Many studies involving locomotor activity rhythms of animals kept under constant conditions have been developed aiming to understand the physiological organization of different circadian systems in a wide range of species (Pittendrigh and Daan 1976; López-Olmeda et al. 2012). As in many other organisms, fish present behavioral and physiological rhythms (López-Olmeda et al. 2012; Oliveira et al. 2013) occurring within a determined time interval which, under constant laboratory conditions, have periodicity near 24 h. When organisms are in their natural environment, these rhythms are entrained by specific cues, named Zeitgebers (Aschoff 1960). The main factor capable to synchronize the circadian rhythms in animals is light, which is mediated by photoreceptive organs able to respond to daily variations (Bell-Pedersen et al. 2005).
Regarding fish species, many other environmental factors, besides light (López-Olmeda et al. 2006, 2012; Hurd et al. 1998), can entrain and/or modulate the circadian system of these animals, such as temperature (López-Olmeda et al. 2006; López-Olmeda and Sánchez-Vázquez 2009) and feeding (Blanco-Vives and Sanchez-Vazquez 2009; López-Olmeda et al. 2012; Sanchez et al. 2009; Sanchez and Sanchez-Vazquez 2009). The age (Zhdanova et al. 2008) and light intensity (Li and Dowling 1998) have also been implicated to influence some aspects of behavioral rhythms in fish. Concerning the experimental model zebrafish, the main Zeitgeber, capable to entrain the circadian system is light, which strongly influences the locomotor activity behavior (Hurd et al. 1998; Li and Dowling 1998; Blanco-Vives and Sanchez-Vazquez 2009). Environmental light information reaches the pineal gland and drives the rhythmic secretion of melatonin through a machinery of circadian oscillator (Cahill 2002). However, the role of pineal gland as the central circadian pacemaker has been questioned in non-mammalian vertebrates because these groups of animals appear to have a decentralized circadian clock, once the presence of photoreceptive mechanisms have been observed in peripheral organs and tissues, exhibiting robust patterns of circadian expression of genes known to be activators and repressors of the circadian system (Whitmore et al. 1995; Pando et al. 2001; Amaral and Johnston 2012).
Important advances have been achieved using the experimental model zebrafish (Danio rerio). This animal presents diverse clock paralog genes of the circadian system (Kobayashi et al. 2000), probably due to an evolutionary event leading to genome duplication (Postlethwait et al. 1998). Different paralog genes of the positive loop have been identified in the species allowing different combinations of the resulting proteins, with different efficiency of transcription activation, which are inhibited by CRY proteins isoforms (Hirayama et al. 2003; Ishikawa et al. 2002; Wang 2008, 2009). Besides, differently from what is observed in mammals (Shearman et al. 1999), clock and bmal paralog genes present a similar expression pattern (Cermakian et al. 2000). Regarding the negative loop, six cry genes paralogs have been identified and the resulting proteins, in general, act as transcriptional repressors of clock and bmal (Kobayashi et al. 2000). Some studies have also shown that cry1a expression is directly driven by light and, therefore, this gene expression could be an important mechanism underlying light entrainment to the circadian system (Amaral and Johnston 2012; Kobayashi et al. 2000; Tamai et al. 2007). The transcriptional situation is similar in per paralogs. Three out of four paralogs are clock-regulated and per 2 is light-inducible (Wang 2008). Together, per2 and cry1a appear to have a key role on the light entrainment to the clock machinery and seems to present an important involvement at embryonic stages (Hirayama et al. 2003; Tamai et al. 2007; Ziv et al. 2005). Additionally, robust rhythmic expression of core-clock genes was recently shown in skeletal muscle of zebrafish, including the light-driven cry1a and per2 (Amaral and Johnston 2012).
Some studies have shown the influence of the circadian clock mechanism on hormones release pattern, such as melatonin (Bégay et al. 1998; Cahill and Besharse 1995; Cahill 1996; Gamse et al. 2002), cortisol (Kulczykowska and Sánchez-Vázquez 2010; Montoya et al. 2010) and growth hormone (Canosa and Peter 2005; Gomez et al. 1996; Singh et al. 2009) of fish along the LD cycle. Also, the rhythm observed in peripheral organs could also be adjusted by endogenous factors such cortisol (Dickmeis et al. 2007). Besides, transgenic zebrafish models have been used in the last decade aiming to understand the structures and molecular mechanisms underlying the circadian clock system, principally when entrained by light (Dickmeis et al. 2007; Gothilf et al. 2002; Kaneko and Cahill 2005; Kaneko et al. 2006; Kojima et al. 2008; Vatine et al. 2009).
Interestingly, a couple of studies explored the link between the circadian system and the somatotropic axis, when the expression of insulin like growth-factor binding protein paralogues (IGFBPs) was analyzed in response to feeding/fasting condition (Amaral and Johnston 2011) and along the circadian cycle (Amaral and Johnston 2012). Once the involvement of some IGFBPs in the growth axis is known and circadian expression of genes coding for these proteins was already observed in these last mentioned studies, the use a zebrafish GH-transgenic model could be helpful to understand if and how these two mechanisms are related. For that goal, a transgenic line of zebrafish (F0104), developed by Figueiredo et al. (2007a, b), was used in the present study. These animals have been widely employed by our research group (Figueiredo et al. 2007a, b; Rosa et al. 2008, 2010, 2011; Studzinski et al. 2009) and, along almost a decade, different locomotor activity behavior along the LD cycle has been noticed during the maintenance of GH-transgenic fish. No evidence has been shown, until the present moment, regarding the effects of GH overexpression on the circadian system of fish. Therefore, the aim of the present work was to verify the effects GH overexpression on the locomotor activity behavior, as well as on core-clock genes expression in the skeletal muscle of zebrafish (Danio rerio), along one LD-cycle.
Materials and methods
Transgenic fish
The fish used in the present study were provided by the transgenic fish vivarium from Universidade Federal do Rio Grande (FURG—RS, Brazil) and were cultured in a system with water recirculation, according to Westerfield (1995), kept at room temperature of 28 °C, with a 12 h light: 12 h dark (12:12 LD) photoperiod (lights on at 08:00 h; lights off at 20:00 h; ZT0 = 08:00; lighting provided by fluorescent lamps with luminance measured above the aquaria of approximately 1000 lx) and fed with high-protein (47.5%) commercial flake food (Color Bits—Tetra Gmbh—Germany) using 5% of body weight/day, twice a day at any time during the light phase until satiety. Transgenic and non-transgenic (NT) zebrafish were all from the same brood and were identified by GFP co-expression in transgenic animals. Males and females with age of 9–10 months were used in the procedures.
All experiments were conducted in compliance with the Ethics Committee of Animal Use (Comitê de Ética em Uso Animal—CEUA—process number: 23116.005683/2013-12) from Universidade Federal do Rio Grande—FURG, RS, Brazil.
Locomotor activity assays
Recording apparatus and conditions
Twelve hours before starting the recording of locomotor activity, the animals were transferred from the colony to the recording room. The groups submitted to these assays were not placed in the recording room always at the same period. Some groups were transferred to the recording room during the dark period and some during the light period, assuring that behavior pattern would be the same for all animals in each group, and discharging behavioral differences caused by stress after changing the environment. The room temperature was fixed at 28 °C with photoperiod 12L:12D (Lights on at 08:00 = ZT0). Four white light bulbs, directed one to each wall of the room, aiming to avoid shadows in the arenas, were used as light source. Illuminance during the light period measured right above the water inside the arenas was approximately 460 lx. Animals were individually placed into opaque plastic arenas with diameter of 14 cm. The arenas were riddled, to allow water flow, and placed into a bath under constant aeration and temperature of 26–28 °C with water from the colony system up to 4 cm height, intending to avoid dislocation of animals in the water column. The arenas and filming structure were placed in the center of the room. Four animals, one by arena, were recorded at a time. Lineages were recorded separately in each trial and animals were not fed during the experiment.
A digital video camera recorder (Sony DCR-TRV8), which was equipped with an infrared light source, was set on a tripod and placed above the arenas. The infrared light allowed for the recording during the dark phase of the photoperiod. The camera was coupled to a computer to store the video recording data. Each trial consisted of a 24-h recording along the LD cycle, and the animals were not disturbed while changing the camera settings to optimize the recording during the light or dark period. The recording apparatus scheme is represented in Fig. 1.
Schematic diagram of the recording apparatus seen from camera view and lateral view. 1 Camera; 2 cable connection between the camera and the computer; 3 computer (notebook); 4 infrared lightening attached to the camera; 5 arena (one for each animal); 6 water column (4 cm high); 7 aeration; 8 aquarium heater; 9 animals (one/arena); 10 bath tank with water from the colony system
Gene expression analysis
Animals maintenance and tissue extraction
After locomotor activity assays, animals were taken back to the culture system and acclimated for at least 2 weeks and, following this period, the fish were distributed into different aquaria (NT or GH individuals, n = 25 per group). The aquaria were kept isolated in the culture system to avoid disturbances during the experiment and a dim light was used during night sampling. Five animals from each lineage were collected from the aquaria at different time-points (10:00—ZT02, 16:00—ZT08, 22:00—ZT14 and 04:00— ZT20) along one LD cycle. Subjects were immediately placed on ice for dissection and each animal originated a single sample for total RNA extraction from the skeletal muscle. No sudden changes in animal’s activity were observed during sample collections. Dissection procedures were realized during the period between each time point of sample collection. The fish were killed with a lethal dose of tricaine (MS-222; Ethyl-3 aminobenzoate) and stored in ice until tissue collection. During the night procedures, the animals were taken from the room inside sealed boxes to avoid exposure to light.
Skeletal muscle was dissected from the dorsal epaxial myotomes, directly immersed in TRIZOL Reagent™ (Invitrogen, Brazil) and consecutively stored at −80 °C until total RNA extraction procedures.
Total RNA extraction and cDNA synthesis
Total RNA was isolated by TRIZOL Reagent Solution™ method according to manufacturer’s instructions (Invitrogen, Brazil). RNA integrity was further checked by agarose gel electrophoresis. Consecutively, the obtained total RNA (1 µg) was used as a template for cDNA synthesis using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Brazil) according to manufacturer’s instructions.
Primers design and gene expression analysis
The sequence of ten primer pairs, for genes described as core-clock genes in other vertebrate models (clock1a, clock1b, bmal1a, bmal1b, cry1b, cry2b, per1a, per1b, per2 and per3), were obtained from a previous study (Amaral and Johnston 2012). The primers for two genes, beta-actin (BAC—F:CTGTCCACCTTCCAGCAGAT; R:GATGGACCTGCCTCGTCGTA; product length 60 bp; accession number NM_131031) and beta-2-microglobulin (B2M—F:GCCTTCACCCCAGAGAAAGG; R:GCGGTTGGGATTTACATGTTG; length 101 bp; NM_131163), used as internal controls, were designed using the software Primer Express 3.0 (Applied Biosystems, Brazil) based on sequences available at GenBank (http://www.ncbi.nlm.nih.gov).
Gene expression was analyzed by real time quantitative PCR (qPCR). Each sample (n = 5 fish per time-point per lineage) was analyzed in duplicate and PCR reactions were performed in a 7300 Real-Time System (Applied Biosystems—Brazil) using Platinum SYBR Green qPCR SuperMix-UDG Kit (Invitrogen, Brazil). The thermocycling program consisted of 2 min at 95 °C and 40 cycles of 15 s at 95 °C and 30 s at 60 °C. Serial dilutions of a skeletal muscle sample obtained from NT fish were realized for all primer pairs to determine the efficiency of qPCR reaction.
Data analysis
Data analysis from locomotor activity
To collect data from the video recording experiments, a software capable of capturing the bi-dimensional positioning coordinates of the subjects along the time was developed. The software, developed in language C, analyses the recording data frame by frame. The positioning coordinates are captured at each frame analyzed. The analysis interactively applied to the video frames is realized according to the type of environmental light (including night vision resources). Before analysis, the frame is treated with a low-pass spatial filter to improve its quality in the analysis process. Subsequently, if the frame is from a diurnal video, it is segmented by threshold method. Each pixel is labeled either as background or the body of the animal. The same is applied in the nocturnal video recording data but, in this case, the pixels labeling is applied based on the result of the subtraction of the frame by a background image. After the pixels labeling by segmentation, the pixels identified as part of the fish’s body are re-evaluated. Those united by connectivity-of-8, in consistent portions with the size of the animal body, are considered as a representation of the subject body. This re-evaluation avoids the dispersed residuals, generated by the fish, or artifacts originated from the recording process, which are considered in the identification of the position coordinates. The process of data collection using this software can be applied in experiments involving controlled environments, with uniform background and illumination, providing contrast and visibility of the fish. By the end of the analysis, an archive with the position coordinates of the fish along time is generated. These data are treated in MatLab software, aiming to eliminate the noise by hysteresis. Also, the dislocation of each fish at each time point was calculated, allowing measurement of the traveled distance in pixels along each 5 min.
The following variables, according to Witting et al. (1990), were obtained using the data extracted from the video recordings: 10 most active hours (M10), 5 least active hours (L5), beginning of M10 (F10), beginning of L5 (F5), relative amplitude (RA), responsiveness to light on (RL), relative activity (Act) and relative rest (RR). The variable M10 was calculated by summing the distance covered by each animal each 5 min along the LD cycle using a fixed window of 10 h to find the highest value. The variable L5 was calculated by summing the distance covered by each animal each 5 min along the LD cycle using a fix window of 5 h to find the lower value. The variable RA was calculated by the equation (M10-L5/M10 + L5). The variable RL was calculated by normalizing the sum of the distance covered by each animal, each 5 min, from 08:35 to 08:45 (10 min of interval after 35 min from lights on—ZT0), by the total time of recording (1440 min). The variable Act was calculated by normalizing the sum of the distance covered by each animal, each 5 min, from 00:10 am (ZT16) to 04:20 (ZT20—middle period of night phase) by the total minutes of recording (1440 min). The variable RR was calculated by normalizing the sum of the distance covered by each animal, each 5 min, from 12:40 (ZT04) to 16:50 (ZT08—middle period of light phase), by the total minutes of recording (1440 min).
Each of the mentioned variables calculated for locomotor activity was submitted to Kolmogorov–Smirnov and Levene’s tests confirming that 95% of the data followed the assumptions of normality and homogeneity of variance. Consecutively, one-way ANOVA was performed followed by Newman–Keuls post hoc test and mathematical transformations were applied when necessary. Besides, additional analysis of locomotor activity data was realized concerning the sum of fish dislocation each 30 min. One-way ANOVA, followed by Newman–Keuls post hoc test, was performed intending to verify differences between the locomotor activity of NT and GH-animals at each time point (each 30 min).
Data analysis from gene expression
Results from serial dilutions were analyzed by the software REST57 and significant differences in gene expression data between NT and GH-transgenic animals were accessed by 2−∆Ct method for each paralog at each time point. Consecutively, intending to access possible differences in gene expression between NT and GH animals, one-way ANOVA was performed, at each time point, followed by Neuman-Keuls post hoc test. Data are expressed as mean (±SE). Significant differences were assumed when p < 0.05 for all analyses.
Results
Locomotor activity assays
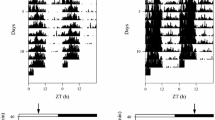
NT and GH-transgenic animals presented higher activity during the dark phase, and significantly higher locomotor activity was observed in GH transgenic fish from 22:00 (ZT14) to 05:00 h (ZT21) at almost all time points (Fig. 2a). Besides, the same differences were also observed during the light phase at almost all time points comprised between 12:30 (ZT04) hours and 15:00 h (ZT07) and at 19:30 (ZT11) and 20:00 h (ZT12).
a Locomotor activity profile of non-transgenic and GH-transgenic animals kept under a 12L:12D LD cycle (460 lx) along 1 day. Each line represents the average locomotor activity from NT (n = 6) and GH (n = 8) fish. Locomotor activity profile is given by the average of locomotor activity values of all animals, in each group, each 30 min. The black bar represents the dark phase and the white bar represents the light phase of the LD cycle. Vertical dashed and dotted lines represent the approximate M10 and L5 interval respectively, given by the average F10 and F5 values from all animals in both groups. b M10 values of NT and GH animals calculated based on the video recording data. Values are represented by average ±SE. Stars indicate significant differences (p < 0.05) between NT and GH animals
Considering the variables calculated based on locomotor activity data, GH-transgenic animals presented significantly higher M10 values when compared with control group (Fig. 2b). No significant differences were observed for the period of 5 h of lower activity (L5), relative amplitude (RA) and relative rest (RR) (data not shown). The absence of significant differences for F10 and F5 values evinces that the most and less active period of locomotor activity, respectively, started near the same time for all animals in each group (data not shown). Also, no statistical differences were observed between the groups concerning the period right after lights were turned on (RL), evincing that the animals were not differently affected by this factor (data not shown).
Interestingly, although differences in M10 values are in agreement with the period where differences were observed at each time point during the period of 10 h of higher activity (Fig. 2a), the same was not observed for L5 values when, despite differences were observed from 12:30 (ZT04) hours and 15:00 h (ZT07) and at 19:30 (ZT11) and 20:00 h (ZT12), no difference was observed for this variable (data not shown).
Gene expression assays
After the locomotor activity assays were realized, the gene expression pattern of the clock machinery that could be underlying the observed behavior was assessed.
Gene expression of each paralog at each time point for both groups
Significant differences were observed between NT and GH animals only at a single time point for clock paralogue genes expression (Fig. 3). Decreased clock1a and clock1b expression was observed in the skeletal muscle of GH animals at ZT14, when compared with NT animals. No significant differences between both groups were observed for the remaining analyzed genes at any time point.
Differences in the expression of genes known to be activators of the circadian system between NT (white bars) and GH animals (grey bars) at ZT 2, 8, 14 and 20. Values are represented as average ±SE, n = 5 per time point. Stars represent significant differences between both groups at each time point and different letters represent significant differences along the time for each gene in each group (p < 0.05)
Gene expression of each paralog along the time for each group
Concerning the analyzed paralogues from the positive loop of the circadian system, significant differences were observed in gene expression along time for bmal1a (GH), and clock 1a (NT) for at least one group (Fig. 3). Only GH animals showed significant increase in bmal1a gene expression at ZT08, while only NT fish evinced significant increase in clock1a gene expression at ZT14, when compared with ZT02 and ZT08, and this increase was maintained at the last time point (ZT20).
Except for cry1b, significant differences were observed for gene expression along the time in all paralogues for at least one group, regarding the analyzed genes from the negative loop of the circadian system (Fig. 4). The paralogue cry2b presented increased expression at ZT14 when compared with the first (ZT02) and the last time point. A similar pattern was observed for the paralogues per1a and per1b, presenting decreased expression at ZT08 and ZT14, when compared with the first and the last time point, for both groups. Non-transgenic animals evinced significantly increased per2 expression at the first time point, and GH fish presented, for this same gene, two peaks of expression at ZT02 and ZT14, which were not significantly different between them but from the remaining time points (ZT08 and ZT20). Similar expression pattern was also observed in both groups for the paralogue per3. Significant decreased expression was noticed, for NT and GH fish, from ZT08 to ZT14, with the last time point evincing an expression level similar to that observed at the first.
Differences in the expression of genes known to be repressors of the circadian system between NT (white bars) and GH animals (grey bars) at time points at ZT 2, 8, 14 and 20. Values are represented as average ±SE, n = 5 per time point. Different letters represent significant differences along the time for each gene in each group (p < 0.05)
Discussion
Zebrafish exhibit typical diurnal locomotor activity rhythm. Since the pioneer experiments of Hurd et al. (1998), involving locomotor activity rhythm response to light and temperature, many other researchers have presented the same locomotor activity pattern while trying to elucidate the strength and mechanisms underlying the response to other Zeitgebers (Blanco-Vives and Sanchez-Vazquez 2009; Del Pozo et al. 2011; Hurd and Cahill 2002; López-Olmeda and Sánchez-Vázquez 2009; López-Olmeda et al. 2010; Sanchez and Sanchez-Vazquez 2009). Interestingly, the present work pictured the opposite situation. Both, non-transgenic (NT) and GH-transgenic animals, presented higher locomotor activity pattern during the dark phase of the LD cycle (Fig. 2a). Such difference can be attributed to the plasticity described in recent studies for zebrafish, which could be considered a dual species and not strictly diurnal (López-Olmeda and Sánchez-Vázquez 2009; López-Olmeda et al. 2010; Del Pozo et al. 2011). Also, strong evidences have been described relating a nocturnal behavior to a constitutive pattern of clock paralogs in mammals, mainly rodents (Oishi et al. 2003; Shearman et al. 2000; Dardente et al. 2004), and a diurnal behavior to a rhythmic pattern of these genes, as observed in sheep and mouse species (Lincoln 2006; Chakir et al. 2015). Although these data is consistent for the clock 1b expression pattern observed in the present work, it should be considered that fish species present decentralized circadian clock machinery, differently from what is observed in mammals, as it will be discussed further in the text. Therefore, more evidence should be gathered to trace a parallel between clock expression pattern and a diurnal or nocturnal behavior in fish species as it has been already speculated in mammals.
Although the opposite pattern of locomotor activity along the LD cycle was observed in the present work, differences were observed between the dark and light phases in and between the groups (Fig. 2). Both groups evinced differences between light and dark phase, as confirmed by the locomotor activity values observed during M10 and L5 periods (Fig. 2a) and M10 (Fig. 2b), L5, RA, Act and RR (Data not shown) values. These data characterized the dark phase as the activity period and the light phase as the resting period. Such differences in the amplitude of locomotor activity along the LD cycle are commonly observed in animals undergoing normal LD cycles (Blanco-Vives and Sánchez-Vázquez 2009; Del Pozo et al. 2011; Hurd and Cahill 2002; López-Olmeda and Sánchez-Vázquez 2009; López-Olmeda et al. 2010; Sanchez and Sanchez-Vazquez 2009).
Despite no differences were observed in L5 values between NT and GH fish, increased activity was punctually observed during the period of time comprising the L5 interval (Fig. 2a). Also, M10 values were significantly different between both groups (Fig. 2b) and significant differences were also punctually observed along almost the whole period comprising M10 interval (Fig. 2a). Therefore, although data analysis from the locomotor activity variables indicate a solid period of activity and rest for both groups, GH-transgenic animals appear to present a higher locomotor activity, when compared with NT, along the LD cycle.
Direct effect of GH on the signaling pathways controlling the expression of genes involved in the circadian system could also be occurring in the transgenic fish. However, evidence of higher metabolic level have been already documented for GH-transgenic animals, such as higher growth rate and gene expression levels of GHR and IGF-I in liver (Figueiredo et al. 2007b), higher levels of oxygen consumption and reactive species of oxygen production (Rosa et al. 2008, 2011), the energetic cost of a downregulation mechanism in the somatotropic axis (Studzinski et al. 2009), early senility (Rosa et al. 2008) and higher food intake (Dalmolin et al. 2015). Therefore, the differences in the locomotor activity behavior observed in GH-transgenic animals could have been caused by alterations in the metabolism of these animals, triggered by the excess of GH.
Differently from mammals, zebrafish appears to have a decentralized circadian clock and many peripheral tissues can be directly responsive to light, the strongest Zeitgeber capable to set the circadian machinery in the species by adjusting the expression pattern of genes known to be activators and repressors of the circadian system (Amaral and Johnston 2012; Mracek et al. 2012; Pando et al. 2001; Vallone et al. 2004; Vatine et al. 2009; Whitmore et al. 1995). Concerning the genes from the positive loop of the circadian system, some studies, involving clock and bmal paralogs, were able to identify a strong expression pattern in the central pacemaker and peripheral tissues of zebrafish such as brain, eye, pineal, heart, kidney, spleen (Cermakian et al. 2000; Whitmore et al. 1998) and muscle (Amaral and Johnston 2012) along the LD cycle, peaking at the beginning of the evening. In this last, the acrophase of all clock and bmal paralogs occurred 2 h after the beginning of the dark phase and, except for bmal1a, the scotophase occurred 2 h after the beginning of the light phase.
Data regarding gene expression of the positive loop in the present study raises two interesting possibilities. At first sight, the decreased expression of clock paralogues (clock1a and clock1b) (Fig. 3), observed in GH-transgenic animals when compared with the control group at ZT14, and the absence of differences between both groups for bmal paralogs (bmal1a and bmal1b), points to a possible control of circadian rhythm by clock paralogs in the transgenic animals. Such affirmation makes sense, once the expression levels of clock paralogues were supposed to be peaking at the mentioned time point, and the CLOCK/BMAL complex is the central transcriptional activator of the circadian system. On the other hand, a slight but significant increase in bmal1a expression was noticed at ZT08 when analyzing GH fish along the time, but the same was not observed for NT animals along the LD cycle (Fig. 3) and, similarly, the same was observed in the expression of clock1a from NT fish at ZT14. Therefore, these data taken together could also suggest a possible control of the circadian rhythm by bmal1a in GH fish, once the amplitude of clock paralogues expression appears to be diminished in these animals due to the observed decreased expression at ZT14, while the circadian system of NT animals would be controlled by the variation observed in clock1a expression along the LD cycle.
The negative loop of the molecular machinery, composed by PER and CRY proteins, presents a robust and well defined circadian rhythm depending on gene expression of per paralog genes in different tissues, which appears to be driven by the period of the day (Delaunay et al. 2000; Vallone et al. 2004; Ziv et al. 2005; Lahiri et al. 2005; Vatine et al. 2011; Mracek et al. 2012; Amaral and Johnston 2012). On the other hand, except for cry1a, the remaining cry paralogs present a quite variable expression pattern (Kobayashi et al. 2000; Vatine et al. 2011). Interestingly, almost all genes from the negative loop were expressed almost in anti-phase in the skeletal muscle of zebrafish (Amaral and Johnston 2012).
No significant transcriptional alteration of genes composing the negative loop of the circadian system was observed when comparing both groups of fish (Fig. 4) at each time point. Considering the expression patterns previously observed for skeletal muscle (Amaral and Johnston 2012), although no significant differences were found between both groups at each time point for all analyzed genes, cry2b appears to be already downregulated at the beginning of the light phase in GH fish, once significantly lower expression levels were observed at ZT02, but the same was not observed for NT group. Thus, similarly to what was observed for bmal1a and clock1a (Fig. 3), it might be suggested that a control of the negative loop in GH-transgenic fish could be driven by cry2b expression, once this was the only gene evincing expression variation along the LD cycle and the same was not observed in NT animals. Besides, the remaining analyzed genes from the negative loop of the circadian system exhibited no variation at all (cry1b) or similar expression pattern (per1a, per1b, per2 and per3) along the LD cycle in both groups of animals, as shown in Fig. 4, suggesting no possible control or modulation by these genes.
These differences observed in the expression of genes from the circadian machinery, in GH-transgenic animals, could be related with the evidence of higher metabolic level characteristics already documented, such as higher growth rate and gene expression levels of GHR and IGF-I in liver (Figueiredo et al. 2007b), higher levels of oxygen consumption and reactive species of oxygen production (Rosa et al. 2008, 2011), the energetic cost of a downregulation mechanism in the somatotropic axis (Shearman et al. 1999), early senility (Rosa et al. 2008) and increased hexokinase gene expression, besides higher food intake (Dalmolin et al. 2015). On the other hand, as a possible correlation between the circadian system and somatotropic has been already documented (Amaral and Johnston 2011, 2012), the excess of growth hormone could be directly altering the expression pattern of genes involved in the circadian system in the skeletal muscle of GH-transgenic zebrafish, by triggering different pathways involved in the control of genes from the circadian machinery.
Despite all the altered responses of transgenic animals to the LD cycle observed in the present study, it should be considered that zebrafish present physiological changes caused by aging, such as diminished sleep-like state, probably related to the decrease in melatonin production (Tsai et al. 2007; Zhdanova et al. 2008). Besides, many studies have reported that zebrafish circadian system can be influenced by other factors, such as temperature (López-Olmeda et al. 2006; López-Olmeda and Sánchez-Vázquez 2009), feeding time (Blanco-Vives and Sánchez-Vázquez 2009; Del Pozo et al. 2011; López-Olmeda et al. 2010; Sanchez and Sánchez-Vázquez 2009) and age (Zhdanova et al. 2008). Therefore, the altered responses to light observed in GH-transgenic animals shown in this work could be associated with early senility, as previously reported (Rosa et al. 2010), caused by the decrease of antioxidant defenses.
The present work shows evidence that GH transgenesis in zebrafish causes altered responses in the expression pattern of genes from the circadian system machinery in the skeletal muscle along the LD cycle. This fact suggests that GH could be an endogenous factor involved in the regulation of the circadian system, once GH overexpression, or even the consequences over other systems caused by this factor, appears to be leading to higher locomotor activity along the LD cycle in zebrafish.
References
Amaral IPG, Johnston IA (2011) Insulin-like growth factor (IGF) signalling and genome-wide transcriptional regulation in fast muscle of zebrafish following a single-satiating meal. J Exp Biol 214(Pt 13):2125–2139. doi:10.1242/jeb.053298
Amaral IPG, Johnston IA (2012) Circadian expression of clock and putative zebrafish clock-controlled genes in skeletal muscle of the zebrafish. Am J Physiol Regul Integr Comp Physiol 302(1):193–206. doi:10.1152/ajpregu.00367.2011
Aschoff J (1960) Exogenous and endogenous components in circadian rhythms. Cold Spring Harb Symp Quant Biol 25:11–28. doi:10.1101/SQB.1960.025.01.004
Aschoff J (1976) Circadian systems in man and their implications. Hosp Pract 11(5):51–57
Bégay V, Falcón J, Cahill GM, Klein DC, Coon SL (1998) Transcripts encoding two melatonin synthesis enzymes in the teleost pineal organ: circadian regulation in pike and zebrafish, but not in trout. Endocrinology 139(3):905–912. doi:10.1210/en.139.3.905
Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE (2005) Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 6(7):544–556. doi:10.1038/nrg1633
Blanco-Vives B, Sanchez-Vazquez FJ (2009) Synchronisation to light and feeding time of circadian rhythms of spawning and locomotor activity in zebrafish. Physiol Behav 98(3):268–275. doi:10.1016/j.physbeh.2009.05.015
Cahill GM (1996) Circadian regulation of melatonin production in cultured zebrafish pineal and retina. Brain Res 708(1–2):177–181. doi:10.1016/0006-8993(95)01365-2
Cahill GM (2002) Clock mechanisms in zebrafish. Cell Tissue Res 309(1):27–34. doi:10.1007/s00441-002-0570-7
Cahill GM, Besharse JC (1995) Circadian rhythmicity in vertebrate retinas: regulation by a photoreceptor oscillator. Prog Retinal Eye Res 14(1):267–291. doi:10.1016/1350-9462(94)00001-Y
Canosa LF, Peter RE (2005) Pre-pro-somatostatin-III may have cortistatin like function in fish. Ann NY Acad Sci 1040(1):253–256. doi:10.1196/annals.1327.036
Cermakian N, Whitmore D, Foulkes NS, Sassone-Corsi P (2000) Asynchronous oscillations of two zebrafish CLOCK partners reveal differential clock control and function. Proc Natl Acad Sci USA 97(8):4339–4344. doi:10.1073/pnas.97.8.4339
Chakir I, Dumont S, Pévet P, Ouarour A, Challet E, Vuillez P (2015) The circadian gene clock oscillates in the suprachiasmatic nuclei of the diurnal rodent Barbary striped grass mouse, Lemniscomys barbarous: a general feature of diurnality? Brain Res 1594:165–172. doi:10.1016/j.brainres.2014.10.0630006-8993
Dalmolin C, Almeida DV, Azevedo MA, Marins LF (2015) Food intake and appetite control in a GH-transgenic zebrafish. Fish Physiol Biochem 41(5):1131–1141. doi:10.1007/s10695-015-0074-5
Dardente H, Menet JS, Challet E, Tournier BB, Pévet P, Masson-Pévet M (2004) Daily and circadian expression of neuropeptides in the suprachiasmatic nuclei of nocturnal and diurnal rodents. Brain Res Mol Brain Res 124:143–151
De Coursey PJ (2004) The behavioral ecology and evolution of biological timing systems. In: Dunlap JC, Loros JJ, De Coursey PJ (eds) Chronobiology: biological timekeeping. Sinauer Associates, England, pp 27–65
Del Pozo A, Sánchez-Férez JA, Sánchez-Vázquez FJ (2011) Circadian rhythms of self-feeding and locomotor activity in Zebrafish (Danio rerio). Chronobiol Int 28(1):39–47. doi:10.3109/07420528.2010.530728
Delaunay F, Thisse C, Marchand O, Laudet V, Thisse B (2000) An inherited functional circadian clock in zebrafish embryos. Science 289(5477):297–300. doi:10.1126/science.289.5477.297
Dickmeis T, Lahiri K, Nica G, Vallone D, Santoriello C, Neumann CJ et al (2007) Glucocorticoids play a key role in circadian cell cycle rhythms. PLoS Biol 5(4):e78. doi:10.1371/journal.pbio.0050078
Dunlap JC, Loros JJ, De Coursey PJ (2004) Chronobiology: biological timekeeping. Sinauer Associates, England
Figueiredo MA, Lanes CFC, Almeida DV, Marins LF (2007a) Improving the production of transgenic fish germline: in vivo mosaicism evaluation by GFP transgene co-injection strategy. Genet Mol Biol 30(1):31–36. doi:10.1590/S1415-47572007000100008
Figueiredo MA, Lanes CFC, Almeida DV, Proietti MC, Marins LF (2007b) The effect of GH overexpression on GHR and IGF-I gene regulation in different genotypes of GH-transgenic zebrafish. Comp Biochem Physiol D Gen Prot 2(3):228–233. doi:10.1016/j.cbd.2007.04.004
Gamse JT, Shen YC, Thisse C, Thisse B, Raymond PA, Halpern ME et al (2002) Otx5 regulates genes that show circadian expression in the zebrafish pineal complex. Nat Genet 30(1):117–121
Gomez JM, Boujard T, Fostier A, Le Bail PY (1996) Characterization of growth hormone nycthemeral plasma profiles in catheterized rainbow trout. Oncorhynchus mukiss. J Exp Zool 274(3):171–180. doi:10.1002/(SICI)1097-010X(19960215)274:3<171:AID-JEZ4>3.0.CO;2-L
Gothilf Y, Toyama R, Coon SL, Du SJ, Dawid IB, Klein DC (2002) Pineal-specific expression of green fluorescent protein under the control of the serotonin-N-acetyltransferase gene regulatory regions in transgenic zebrafish. Dev Dyn 225(3):241–249. doi:10.1002/dvdy.10152
Hirayama J, Fukuda I, Ishikawa T, Kobayashi Y, Todo T (2003) New role of zCRY and zPER2 as regulators of sub-cellular distributions of zCLOCK and zBMAL proteins. Nucleic Acids Res 31(3):935–943. doi:10.1093/nar/gkg174
Hurd MW, Cahill GM (2002) Entraining signals initiate behavioral circadian rhythmicity in larval zebrafish. J Biol Rhythms 17(4):307–314. doi:10.1177/074873002129002618
Hurd MW, Debruyne J, Straume M, Cahill GM (1998) Circadian rhythms of locomotor activity in zebrafish. Physiol Behav 65(3):465–472. doi:10.1016/S0031-9384(98)00183-8
Ishikawa T, Hirayama J, Kobayashi Y, Todo T (2002) Zebrafish CRY represses transcription mediated by CLOCK-BMAL heterodimer without inhibiting its binding to DNA. Genes Cells 7(10):1073–1086. doi:10.1046/j.1365-2443.2002.00579.x
Kaneko M, Cahill GM (2005) Light-dependent development of circadian gene expression in transgenic zebrafish. PLoS Biol 3(2):e34. doi:10.1371/journal.pbio.0030034
Kaneko M, Hernandez-Borsetti N, Cahill GM (2006) Diversity of zebrafish peripheral oscillators revealed by luciferase reporting. Proc Natl Acad Sci USA 103(39):14614–14619. doi:10.1073/pnas.0606563103
Kobayashi Y, Ishikawa T, Hirayama J, Daiyasu H, Kanai S, Hi Toh et al (2000) Molecular analysis of zebrafish photolyase/cryptochrome family: two types of cryptochromes present in zebrafish. Genes Cells 5(9):725–738. doi:10.1046/j.1365-2443.2000.00364.x
Kojima D, Torii M, Fukada Y, Dowling JE (2008) Differential expression of duplicated VAL-opsin genes in the developing zebrafish. J Neurochem 104(5):1364–1371. doi:10.1111/j.1471-4159.2007.05093.x
Kulczykowska E, Sánchez-Vázquez FJ (2010) Neurohormonal regulation of feed intake and response to nutrients in fish: aspects of feeding rhythm and stress. Aquac Res 41(5):654–667. doi:10.1111/j.1365-2109.2009.02350.x
Lahiri K, Vallone D, Gondi SB, Santoriello C, Dickmeis T, Foulkes NS (2005) Temperature Regulates Transcription in the Zebrafish Circadian Clock. PLoS Biol 3(11):e351. doi:10.1371/journal.pbio.0030351
Li L, Dowling JE (1998) Zebrafish visual sensitivity is regulated by a circadian clock. Vis Neurosci 15(5):851–857. doi:10.1017/S0952523898155050
Lincoln GA (2006) Melatonin entrainment of circannual rhythms. Chronobiol Int 23(1):301–306
López-Olmeda JF, Sánchez-Vázquez FJ (2009) Zebrafish temperature selection and synchronization of locomotor activity circadian rhythm to ahemeral cycles of light and temperature. Chronobiol Int 26(2):200–218. doi:10.1080/07420520902765928
López-Olmeda JF, Madrid JA, Sánchez-Vázquez FJ (2006) Light and temperature cycles as zeitgebers of zebrafish (Danio rerio) circadian activity rhythms. Chronobiol Int 23(3):537–550. doi:10.1080/07420520600651065
López-Olmeda JF, Tartaglione EV, de la Iglesia HO, Sánchez-Vázquez FJ (2010) Feeding entrainment of food-anticipatory activity and per1 expression in the brain and liver of zebrafish under different lighting and feeding conditions. Chronobiol Int 27(7):1380–1400. doi:10.3109/07420528.2010.501926
López-Olmeda JF, López-García I, Sánchez-Muros MJ, Blanco-Vives B, Aparicio R, Sánchez-Vázquez FJ (2012) Daily rhythms of digestive physiology, metabolism and behavior in the European eel (Anguilla anguilla). Aquacult Int 20(6):1085–1096. doi:10.1007/s10499-012-9547-z
Montoya A, López-Olmeda JF, Garayzar ABS, Sánchez-Vázquez FJ (2010) Synchronization of daily rhythms of locomotor activity and plasma glucose, cortisol and thyroid hormones to feeding in Gilthead seabream (Sparus aurata) under a light-dark cycle. Physiol Behav 101(1):101–107. doi:10.1016/j.physbeh.2010.04.019
Moore-Ede MC, Schmelzer WS, Kass DA, Herd JA (1976) Internal organization of the circadian timing system in multicellular animals. Fed Proc 35(12):2333–2338
Moore-Ede M, Sulzman FM, Fuller CA (1982) The clocks that time us: physiology of the circadian timing system. Commonwealth Fund Publications. Harvard University Press, England
Mracek P, Santoriello C, Idda ML, Pagano C, Ben-Moshe Z, Gothilf Y et al (2012) Regulation of per and cry genes reveals a central role for the D-Box enhancer in light-dependent gene expression. PLoS ONE 7(12):e51278
Oishi K, Miyazaki K, Kadota K, Kikuno R, Nagase T, Atsumi G, Ohkura N, Azama T, Mesaki M, Yukimasa S, Kobayashi H, Litaka C, Umehara T, Horikoshi M, Kudo T, Shimizu Y, Yano M, Monden M, Machida K, Matsuda J, Horie S, Todo T, Ishida N (2003) Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J Biol Chem 278:41519–41527
Oliveira CCV, Aparício R, Blanco-Vives B, Chereguini O, Martín I, Sánchez-Vazquez FJ (2013) Endocrine (plasma cortisol and glucose) and behavioral (locomotor and self-feeding activity) circadian rhythms in Senegalese sole (Solea senegalensis Kaup 1858) exposed to light/dark cycles or constant light. Fish Physiol Biochem 39(3):479–487. doi:10.1007/s10695-012-9713-2
Pando MP, Pinchak AB, Cermakian N, Sassone-Corsi P (2001) A cell-based system that recapitulates the dynamic light dependent regulation of the vertebrate clock. Proc Natl Acad Sci USA 98(18):10178–10183. doi:10.1073/pnas.181228598
Pittendrigh CS, Daan S (1976) A functional analysis of circadian pacemakers in nocturnal rodents V. Pacemaker structure: a clock for all seasons. J Comp Physiol 106(3):333–355. doi:10.1007/BF01417860
Postlethwait JH, Yan Y-L, Gates M, Horne S, Amores A, Brownlie A et al (1998) Vertebrate genome evolution and the zebrafish gene map. Nat Genet 18(4):345–349. doi:10.1038/ng0498-345
Rosa CE, Figueiredo MA, Lanes CFC, Almeida DV, Monserrat JM, Marins LF (2008) Metabolic rate and reactive oxygen species production in different genotypes of GH-transgenic zebrafish. Comp Biochem Physiol B: Biochem Mol Biol 149(1):209–214. doi:10.1016/j.cbpb.2007.09.010
Rosa CE, Kuradomi RY, Almeida DV, Lanes CFC, Figueiredo MA, Dytz AG et al (2010) GH overexpression modifies muscle expression of anti-oxidant enzymes and increases spinal curvature of old zebrafish. Exp Gerontol 45(6):449–456. doi:10.1016/j.exger.2010.03.012
Rosa CE, Figueiredo MA, Lanes CFC, Almeida DV, Marins LF (2011) Genotype-dependent gene expression profile of the antioxidant defense system (ADS) in the liver of a GH-transgenic zebrafish model. Transgenic Res 20(1):85–89. doi:10.1007/s11248-010-9395-4
Sanchez JA, Sanchez-Vazquez FJ (2009) Feeding entrainment of daily rhythms of locomotor activity and clock gene expression in zebrafish brain. Chronobiol Int 26(6):1120–1135. doi:10.3109/07420520903232092
Sanchez JA, Lopez-Olmeda JF, Blanco-Vives B, Sanchez-Vazquez FJ (2009) Effects of feeding schedule on locomotor activity rhythms and stress response in sea bream. Physiol Behav 98(1–2):125–129. doi:10.1016/j.physbeh.2009.04.020
Shearman LP, Zylka MJ, Reppert SM, Weaver DR (1999) Expression of basic helix-loop-helix/PAS genes in the mouse suprachiasmatic nucleus. Neuroscience 89(2):387–397. doi:10.1016/S0306-4522(98)00325-X
Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, Reppert SM (2000) Interacting molecular loops in the mammalian circadian clock. Science 288:1013–1019
Singh AK, Lal B, Pati AK (2009) Variability in the characteristics of ultradian and circadian rhythms in plasma levels of growth hormone in the Indian walking catfish, Clarias batrachus. Biol Rhyt Res 40(3):211–221. doi:10.1080/09291010701875054
Studzinski AL, Almeida DV, Lanes CF, Figueiredo MA, Marins LF (2009) SOCS1 and SOCS3 are the main negative modulators of the somatotrophic axis in liver of omozygous GH-transgenic zebrafish (Danio rerio). Gen Comp Endocrinol 161(1):67–72. doi:10.1016/j.ygcen.2008.10.008
Tamai TK, Young LC, Whitmore D (2007) Light signaling to the zebrafish circadian clock by Cryptochrome 1a. Proc Natl Acad Sci USA 104(37):14712–14717. doi:10.1073/pnas.0704588104
Tsai SB, Tucci J, Uchiyama J, Fabian NJ, Lin MC, Zhdanova IV et al (2007) Differential effects of genotoxic effects on both concurrent body growth and gradual senescence in adult zebrafish. Aging Cell 6(2):209–224. doi:10.1111/j.1474-9726.2007.00278.x
Vallone D, Gondi SB, Whitmore D, Foulkes NS (2004) E-box function in a period gene repressed by light. Proc Natl Acad Sci USA 101(12):4106–4111. doi:10.1073/pnas.0305436101
Vatine G, Vallone D, Appelbaum L, Mracek P, Ben-Moshe Z, Lahiri K et al (2009) Light directs zebrafish period2 expression via conserved D and E boxes. PLoS Biol 7(10):e1000223. doi:10.1371/journal.pbio.1000223
Vatine G, Vallone D, Gothilf Y, Foulkes NS (2011) It’s time to swim! Zebrafish and the circadian clock. FEBS Lett 585(10):1485–1494. doi:10.1016/j.febslet.2011.04.007
Wang H (2008) Comparative analysis of teleost fish genomes reveals preservation of different ancient clock duplicates in different fishes. Mar Genomics 1(2):69–78. doi:10.1016/j.margen.2008.06.003
Wang H (2009) Comparative genomic analysis of teleost fish bmal genes. Genetica 136(1):149–161. doi:10.1007/s10709-008-9328-9
Westerfield M (1995) The zebrafish book: a guide for the laboratory use of zebrafish Danio rerio. University of Oregon Press, Oregon
Whitmore D, Foulkes NS, Sassone-Corsi P (1995) Light acts directly on organs and cells in culture to set the vertebrate circadian clock. Nature 404(6773):87–91. doi:10.1038/35003589
Whitmore D, Foulkes NS, Strahle U, Sassone-Corsi P (1998) Zebrafish clock rhythmic expression reveals independent peripheral circadian oscillators. Nat Neurosci 1(8):701–707. doi:10.1038/3703
Witting W, Kwa IH, Eikelenboom P, Mirmiran M, Swaab DF (1990) Alterations in the circadian rest-activity rhythm in aging and alzheimer’s disease. Biol Psych 27(6):563–572. doi:10.1016/0006-3223(90)90523-5
Zhdanova IV, Yu L, Lopez-Patino M, Shang E, Kishi S (2008) Aging of the circadian system in zebrafish and the effects of melatonin on sleep and cognitive performance. Brain Res Bull 75(2–4):433–441. doi:10.1016/j.brainresbull.2007.10.053
Ziv L, Levkovitz S, Toyama R, Falcon J, Gothilf Y (2005) Functional development of the zebrafish pineal gland: light-induced expression of period 2 is required for onset of the circadian clock. J Neuroendocrinol 17(5):314–320. doi:10.1111/j.1365-2826.2005.01315.x
Acknowledgements
The authors would like to thank Carlos Frederico Ceccon Lanes, Daniela Volcan Almeida, Carolina Peixoto and Fábio Everton Maciel, who gently helped during tissue extraction and samples processing, and also Glaucio Magalhães Sequeira and Guilherme Granada, who kindly conceded the recording equipment. This work was supported by Brazilian CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) and CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior). L. F. Marins is a research fellow from CNPq (Proc. No. 304675/2011-3).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that no competing financial interests exists.
Rights and permissions
About this article
Cite this article
Cruz, B.P., Brongar, L.F., Popiolek, P. et al. Clock genes expression and locomotor activity are altered along the light–dark cycle in transgenic zebrafish overexpressing growth hormone. Transgenic Res 26, 739–752 (2017). https://doi.org/10.1007/s11248-017-0039-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-017-0039-9